Abstract
This systematic review aimed to identify different gut microbiome profiles across the human lifespan and to correlate such profiles with the body composition. PubMed, Scopus, and Cochrane were searched from inception to March 2022. Sixty studies were included in this systematic review. Overall, the gut microbiome composition in overweight participants exhibited decreased α-diversity, decreased levels of the phylum Bacteroidetes and its taxa, and increased levels of the phylum Firmicutes, its taxa, and the Firmicutes/Bacteroidetes ratio, in comparison to normal-weight participants. Other body composition parameters showed similar correlations. Fat mass and waist circumference were found to correlate positively with the Firmicutes taxa and negatively with the Bacteroidetes taxa. In contrast, lean body mass and muscle mass demonstrated a positive correlation with the Bacteroidetes taxa. Notably, these correlations were more pronounced in athletes than in obese and normal-weight individuals. The composition of the gut microbiome is evidently different in overweight individuals or athletes of all age groups, with the former tending towards decreased Bacteroidetes taxa and increased Firmicutes taxa, while a reversed relationship is observed concerning athletes. Further studies are needed to explore the dynamic relationship between energy intake, body composition, and the gut microbiome across the human lifespan.
1. Introduction
The gut microbiome is involved in multiple essential functions responsible for the normal functioning of the intestine and the host [1,2], but its composition is unique to each person. In fact, there is less than 10% similarity between any two individuals [3]. Its formation is determined early from birth through adulthood and modified by genetic and environmental factors, such as diet, physical activity, age, gender, sleep, smoking, and antibiotics [1,4].
The brain participates dynamically in energy balance regulation via its ability to communicate with the peripheral organs through various nerve and chemical signals, most of them coming from the gastrointestinal tract, a relationship called the gut–brain axis [5]. The activation of neuropeptide Y/agouti-related peptide (NPY/AGRP) neurons in the hypothalamus of the brain by the hormone ghrelin has an orexigenic effect by stimulating an increase in appetite and a decrease in energy expenditure [6,7]. Ghrelin is negatively correlated with the genera Bifidobacterium, Lactobacillus, Blautia coccoides, and Eubacterium rectale and positively correlated with the genera Bacteroides and Prevotella [8,9]. The activation of pro-opiomelanocortin/cocaine-amphetamine-related transcript (POMC/CART) and the suppression of NPY/AGRP neurons by the hormones insulin, leptin, cholecystokinin (CCK), peptide YY (PYY), glucagon-like-peptide 1 (GLP-1), and oxyntomodulin (OXM) leads to the opposite, anorexigenic effects [6,7].
Moreover, the specific pathways through which the gut microbiome communicates with the brain and interacts with energy expenditure, body weight, and body composition are well known. The first mechanism involves lipopolysaccharides (LPS), found in the cell walls of Gram-negative bacteria on macrophages and adipose tissue. LPS activate a cascade of pro-inflammatory responses that are accountable for a chronic state of underlying inflammation [8,9,10,11,12,13]. The second mechanism involves short-chain fatty acids (SCFAs), which are produced by the fermentation of fiber. In addition to contributing to approximately 10% of energy intake, they participate in other metabolic pathways, such as promoting hepatic lipogenesis and gluconeogenesis, inflammatory reduction, and an increase in GLP-1 and PYY production [8,10,11,12]. The latter mechanism involves bile acids, which are fermented by the colon microbiome to produce secondary bile acids. Secondary bile acids later promote increased energy expenditure and the production of GLP-1 [8].
As a result, the gut microbiome profile appears to be different in overweight and obese compared to normal-weight individuals, demonstrated through various studies [14,15,16]. Dysbiosis, or the imbalance of the gut microbiota, has been associated with inflammatory responses observed in clinical conditions, underlying the microbiota’s influence on gastrointestinal health and disease mechanisms [17]. Recent findings underscore the potential of monitoring the gut microbiome for diagnostic and therapeutic strategies for inflammatory bowel diseases (IBD), irritable bowel syndrome (IBS), and colorectal cancer, highlighting the microbiota’s integral role in gastrointestinal health [18]. Notably, it is suggested that gut microbiome modification could be a potential strategy for the early prevention and treatment of relevant conditions and obesity, through improving dietary habits; taking probiotics, prebiotics, and synbiotics; and fecal microbiota transplantation from healthy individuals [10,19,20].
However, based on the literature review, the formation of the gut microbiome in relationship to the body composition is poorly systematized. Specifically, there are no summarized gut microbiome profiles across the human lifespan according to age groups in healthy individuals. Thus, the current systematic review focused on identifying different gut microbiome profiles in healthy individuals, from children to older adults, and to correlate such profiles with the body composition.
2. Materials and Methods
2.1. Information Sources and Search Strategy
This systematic review was based on the updated Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 guidelines [21]. The literature search of the systematic review was carried out on 18 March 2022 in the PubMed, Scopus, and Cochrane databases using the following keywords: (((healthy individual* OR human* OR obes*) NOT (disease* OR disorder* OR syndrome OR diabetes OR cancer)) AND (“gut microbio*” OR “intestinal microbio*” OR “fecal microbio*” OR “cecal microbio*” OR microflora OR “gut bacteria” OR “intestinal bacteria”)) AND (“body composition” OR “fat-free mass” OR “fat mass” OR “body fat” OR “body mass” OR “body mass index” OR BMI OR “energy expenditure” OR “basal metabolic rate” OR BMR OR “resting metabolic rate” OR RMR). A supplementary search for relevant studies was conducted from the reference lists of the screened manuscripts.
2.2. Eligibility Criteria
The research question and inclusion and exclusion criteria were determined using the PICO strategy (Patient, Intervention, Controls, Outcome). The inclusion criteria were (1) primary research; (2) studies that presented the gut microbiome in the large intestine; (3) studies written in the English language; (4) studies that had as a population healthy children, adults, older adults, and postmenopausal women; (5) studies that intervened by providing probiotic, prebiotic, and symbiotic supplements; (6) studies that performed an intervention by modifying the diet or physical activity or both. The exclusion criteria were (1) non-primary research (i.e., reviews and case studies); (2) studies not written in English; (3) studies with a non-healthy population (except obese); (4) studies that presented the gut microbiome in other areas, such as the mouth; (5) studies that involved twins, infants, pregnancy, or breastfeeding; (6) studies that performed an intervention by providing medication. The PICO criteria for the inclusion and exclusion of studies are presented in Table 1.

Table 1.
The PICO criteria for inclusion and exclusion of studies.
2.3. Data Collection Process
Primary screening was conducted by two independent researchers (I.K., A.V.) using Microsoft Excel, according to the established eligibility criteria. Full-text secondary screening for the selection of the final articles was also conducted by these two independent researchers, while, where there were conflicts, a third independent researcher (CDG) resolved them.
2.4. Data Extraction and Quality Assessment
Data extraction from the final articles was conducted by one researcher (I.K.), who presented the results in four tables based on age groups (children, adults, older adults, and whole age range). Extracted data included the name of the first author and publication date, sample size, gender, age, BMI category, body composition, and results.
The quality assessment was conducted by one researcher using the Newcastle–Ottawa Scale (NOS) tool, adapted according to the study design [22]. The NOS tool consists of three sections regarding sample selection, a search for confounding factors, and study outcomes. As confounding factors, in the present systematic review, a check for the exclusion of antibiotic and/or probiotic intake was determined. The tool involves eight or nine questions and ten is the maximum score achieved. Due to the final score, studies were classified as “low quality” if the score was <5, as “moderate quality” if the score was 5–7, and as “high quality” if the score was >7.
Extracted data were categorized into four tables based on age groups: (i) children, <18 years, (ii) adults 18–65 years, (iii) older adults >65 years, and (iv) whole age range, children to older adults. The extracted data in each table were further categorized according to (1) sex—males and females; (2) BMI category—children were classified according to growth charts and adults according to BMI into (i) normo-weight, (ii) overweight, and (iii) obese, also considering the ethnicity-specific BMI cutoffs, as provided by the original articles; (3) body composition—some studies included information regarding body composition besides BMI, such as body fat percentage and muscle mass; (4) athletes—athletes were included in some studies as part of the sample to observe differences between them and non-athlete individuals. Athletes’ competition levels were determined based on the description provided by each study.
3. Results
3.1. Study Selection
During the search process using the keywords in the PubMed, Scopus, and Cochrane databases, 995 potentially relevant studies were found, with 614 studies remaining after duplicates were removed and the filters “human” and “humans” were applied. After the primary screening, which included the title and abstract reading, 188 studies were selected for full-text screening. The final studies that met the eligibility criteria and were included in this systematic review totaled 60. Figure 1 shows the study selection process in detail, according to the PRISMA 2020 guidelines.
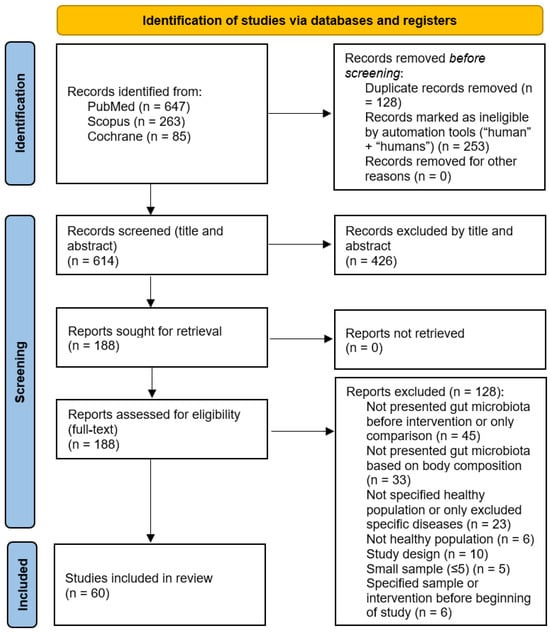
Figure 1.
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram for study selection.
3.2. Study Characteristics
The main characteristics of the studies included in this systematic review are presented in four categories based on age. Full summaries of the study characteristics, BMI categories, body composition, and results are provided in Table 2, Table 3, Table 4, Table 5 and Table 6. The gut microbiome was presented in all groups by stool collection, and, in the majority of the studies, the 16S rRNA amplicon sequencing method was applied [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54], while the quantitative PCR (qPCR) method was applied to determine the bacterial abundance [16,25,27,30,31,33,42,44,47,48,50,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. The body composition assessment was achieved by using WHO growth charts or BMI z-scores in children, while the BMI was used for both adults and older adults. Apart from the BMI, eight studies in children [23,24,27,31,55,58,60,69], 19 studies in adults [33,34,35,36,37,40,41,42,45,61,63,64,65,66,68,70,71,72,73], four studies in older adults [46,48,74], and one study in the whole age range [49] conducted further body composition measurements, such as the body fat percentage, visceral fat, lean body mass percentage, waist circumference, and waist/hip ratio.

Table 2.
Characteristics of studies investigating the gut microbiome composition in children.
The 18 studies with children as the target group were published between 2010 and 2022, with sample sizes ranging from 21 to 502 participants, while countries worldwide were included. None of the 18 studies included athletes. Fourteen studies were cross-sectional, one was a case–control study, and three were clinical trials. The 32 studies with adults as the target group were published between 2011 and 2021, with sample sizes ranging from 20 to 263 participants, while countries from all over the world were included. Three studies included athletes in their samples [34,39,64]. Nineteen studies were cross-sectional, one was longitudinal, 11 were clinical trials, and one was a comparative study. The four studies with older adults as the target group were published between 2017 and 2021, with sample sizes ranging from 22 to 201, with participants from Japan, Slovakia, and Italy. One study of this category included athletes in its sample [46]. Three studies were cross-sectional and one was a clinical trial. The six studies with participants from 14 to 88 years old were published between 2010 and 2021, with sample sizes ranging from 26 to 528, with participants from European and Asian countries. One study included athletes in its sample [49] and all six were cross-sectional.
The gut microbiome analysis was presented in each age group in terms of α-diversity, β-diversity, and bacterial taxonomy, including dominant phyla and genera and correlations of the gut microbiome composition with the BMI and other body composition measurements.
According to the Newcastle–Ottawa Scale (NOS), regarding evidence quality, two studies were rated as “low quality” (score < 5), all from the adult age group category [36,62]. Twenty-three studies were rated as “moderate quality” (score 5–7): eight from the children category [24,30,56,57,59,60,69,75], 11 from the adult category [33,35,37,38,39,42,61,63,73,76,77], three from the older adult category [47,48,74], and one from the whole age range category [53]. Thirty-five studies were rated as “high quality” (score > 7): 10 from the children category [23,25,26,27,28,29,31,32,55,58], 19 from the adult category [16,34,40,41,43,44,45,64,65,66,67,68,70,71,72,78,79,80,81], one from the older adult category [46], and five from the whole age range category [49,50,51,52,54].

Table 3.
Characteristics of studies investigating the gut microbiome composition in adults.
Table 3.
Characteristics of studies investigating the gut microbiome composition in adults.
| Author(s), Date | N | Sex | Age (Years) | BMI Category (kg/m2) | Body Composition | Results |
|---|---|---|---|---|---|---|
| Allen et al., 2018 [61] | 32 | M and F | 20–45 | Lean: 22.21 ± 2.76 Obese: 35.71 ± 5.11 | Lean (body fat % = 26.04 ± 6.12, lean mass % = 71.52 ± 6.18, bone density = 1.11 ± 0.08) Obese (body fat % = 38.42 ± 4.98, lean mass % = 59.42 ± 5.03, bone density = 1.21 ± 0.12) | Gut microbiota composition was different between lean and obese adults at baseline (p = 0.034) |
| Assmann et al., 2020 [33] | 103 | M and F | Eutropic: 44.7 ± 9.1 Obesity: 46.6 ± 9.4 | Eutropic: 18.6 ± 2.1 Obesity: 32.9 ± 2.4 | Eutropic (WC cm = 75.2 ± 7.6, fat mass % = 13.6 ± 5.7, lean mass % = 47.6 ± 12.2) Obesity (WC cm = 104.9 ± 10.2, fat mass % = 34.7 ± 6.5, lean mass % = 57.0 ± 11.7) | Bacterial genera: 18 were statistically different between obese and normal-weight individuals (p < 0.05) → ↑ Mogibacterium, Mitsuokella, Megamonas, Howardella, Anaerovibrio, Bacteroides, Allisonella, Adlercreutzia, Abiotrophia. ↓ Victivallis, Succinivibrio, Rothia, Parvimonas, Intestimonas, Haemophilus, Faecalibacterium, Dorea, Anaerococcus Bacterial species: 12 were statistically different between obese and normal-weight individuals (p < 0.02) → ↑ Abiotrophia defectiva, Actinomyces odontolyticus, Allisonella histaminiformans, Barnesiella intestinihominis, Dorea longicatena, Howardella ureilytica, Lactobacillus curvatus, Megamonas funiformis, Mitsuokella jaladudinii, Odoribacter laneus. ↓ Bacteroides eggerthii, Haemophilus parainfluenzae. Shannon index (α-diversity) was not different between obese and normal-weight groups. Β-diversity was statistically different. |
| Barnes et al., 2019 [62] | 32 | M and F | 18–50 | Lean control: 22.1 (1.6) Lean mango: 22.9 (2.2) Obese mango: 34.6 (4.9) | NR | Day 0: Obese → ↑ Clostridium leptum (p = 0.0264), Bacteroides thetaiotaomicron (p = 0.0359). ↓ Lactococcus lactis (p = 0.443). |
| Basciani et al., 2020 [63] | 48 | M and F | 56.2 ± 6.1 | Obese: 35.9 ± 4.1 | WPG (WC = 110.0 ± 9.4 cm, HC = 123.6 ± 12.1 cm, TC = 63.6 ± 5.3 cm, arm circumference = 36.6 ± 3.9 cm) VPG (WC = 108.2 ± 8.5 cm, HC = 123.3 ± 9.3 cm, TC = 64.1 ± 5.3 cm, arm circumference = 36.3 ± 3.7 cm) APG (WC = 105.3 ± 9.1 cm, HC = 122.5 ± 10.6 cm, TC = 65.4 ± 7.2 cm, arm circumference = 37.7 ± 3.0 cm) | TO: Obese → dominant phyla: Firmicutes, Bacteroidetes, Proteobacteria, Verrucomicrobia, Fusobacteria, Actinobacteria. Firmicutes: 80–90%, Bacteroidetes: 0–10%. |
| Bezek et al., 2020 [70] | 200 | M and F | 35.4 ± 7.0 (25–50) | 24.2 ± 3.5 (18.5–35) | WHR: 0.87 ± 0.07, visceral fat index: 4.7 ± 2.9 | All participants: Phylum (%) → Firmicutes (71.02 ± 11.45), Bacteroidetes (13.85 ± 10.20), Proteobacteria (3.52 ± 3.33), Actinobacteria (2.80 ± 3.25), Verrucomicrobia (0.28 ± 2.87). Genus (%) → Blautia (11.79 ± 5.84), Faecalibacterium (8.59 ± 5.09), Bacteroides (7.97 ± 8.05), Ruminococcus (6.51 ± 3.17), Clostridium (4.79 ± 3.48). Clusters (most prevalent): C1 → Phylum = Bacteroidetes, Genus = Bacteroides, Prevotella. C2 → Phylum = Firmicutes, Genus = Blautia, Clostridium. C3 → Phylum = Actinobacteria, Genus = Bifidobacterium. C4 → Phylum = Proteobacteria, Verrucomicrobia, Genus = Erysipelothrix. C2: higher obesity measures → ↑ Firmicutes, Firmicutes/Bacteroidetes (F/B) ratio, ↓ Bacteroidetes. |
| Bielik et al., 2020 [64] | 24 | M | Lean athletes (LA): 27.3 (23.5–31.0) Control athletes (CTRLs): 30.0 (25.1–34.9) | LA: 20.14 (19.31–20.97) CTRLs: 24.1 (22.9–25.2) | LA: body fat % = 11.73 (9.9–13.6) CTRLs: body fat % = 13.1 (11.2–14.9) | Phylum: Actinobacteria (p ≤ 0.01). Class: LA → ↓ Gamma proteobacteria (Proteobacteria) (p = 0.04), Shewanella (p = 0.04), Xanthomonas (p = 0.03). Order: LA → ↓ Alteromonadales (Proteobacteria) (p = 0.04). Genus: LA → ↑ Roseburia spp. (Firmicutes) (p = 0.03), Barnesiella spp. (Bacteroidetes) (p = 0.05). Family: LA → ↓ Coriobacteriaceae (Actinobacteria) (p = 0.04). |
| Bloemendaal et al., 2021 [78] | 56 | F | 18–40 | Probiotics group: 21.9 ± 0.32 Control group: 21.7 ± 0.30 | NR | Phylum before intervention: Firmicutes (68.0%), Bacteroidetes (19.5%), Actinobacteria (8.7%), Proteobacteria (1.5%), Verrucomicrobiota (1.4%), Euryarcheota (0.4%), Tenericutes (0.29%), Cyanobacteria (0.25%). |
| Borgo et al., 2018 [71] | 40 | M and F | NW (M: 48.7 ± 10.2, F: 51.7 ± 8.3) O (M: 53.8 ± 7.7, F: 51.3 ± 6.7) | NW: 22.8 ± 1.8 O: 35.8 ± 8.3 | NW (M: 83.1 ± 2.4, F: 82.9 ± 3.2) O (M: 112.1 ± 8.5, F: 109.3 ± 9.8) | Lumen-associated microbiota (LAM): Obese → ↓ α-diversity, Oscillospira genus. ↑ Veillonellaceae, Dialister spp. Flavonifractor plautii + Faecalibacterium prausnitzii negatively associated with BMI. Mucosal-associated microbiota (MAM): no significant differences between BMI groups. |
| Brignardello et al., 2010 [72] | 24 | M and F | 18–50 | Normal-weight: 23.5 ± 2.4 Obese: 35.9 ± 5.0 | Normal-weight (waist circumference = 78.7 ± 7.5 cm, body fat = 25.1 ± 7.3%, fat body mass = 15.6 ± 3.8 kg, lean body mass = 47.2 ± 11.3 kg) Obese (waist circumference = 112.5 ± 9.6 cm, body fat = 48.9 ± 9.3%, fat body mass = 43.1 ± 11.2 kg, lean body mass = 54.9 ± 10.6 kg) | Obese: ↑ relative abundance of bacteria with 23–37% G + C content in their DNA, ↓ bacteria with 40–47% and 57–61% G + C content in their DNA. Dominant bacteria regarding G + C content: obese → 36.2 ± 1.0%, normal-weight → 41.7 ± 1.4%. |
| Clarke et al., 2014 [34] | 86 | M | Elite athletes: 28.8 ± 3.8 Low BMI controls: 28.1 ± 5.1 High BMI controls: 30.8 ± 5.6 | Elite athletes: 29.1 ± 3.0 Low BMI controls: 22.7 ± 1.8 High BMI controls: 31.2 ± 3.0 | Elite athletes (body mass = 101.3 ± 13.8 kg, body fat = 16.9 ± 6.1 kg, lean body mass = 80 ± 8.9 kg, waist/hip ratio = 0.8 ± 0.04) Low BMI controls (body mass = 74.3 ± 6.3 kg, body fat = 15 ± 4.6 kg, lean body mass = 55.4 ± 5.6 kg, waist/hip ratio = 0.8 ± 0.05) High BMI controls (body mass = 103.1 ± 13.8 kg, body fat = 33.9 ± 8.8 kg, lean body mass = 65 ± 8 kg, waist/hip ratio = 0.9 ± 0.07) | α-diversity: ↑ Elite athletes compared with both control groups, no difference between the control groups. Elite athletes—High BMI controls: ↑ 48 taxa (top 6 → Firmicutes, Ruminococcaceae, S24-7, Succinivibrionaceae, RC9, Succinivibrio), ↑ Family Akkermansiaceae (p = 0.049) + Genus Akkermansia (p = 0.035), ↓ Bacteroidetes (p = 0.022). Elite athletes—Low BMI controls: ↑ 40 taxa (top 6 → Prevotellaceae, Erysipelotrichaceae, S24-7, Succinivibrionaceae, Prevotella, Succinivibrio), ↓ Lactobacillaceae (p = 0.001), Bacteroides (p = 0.035), Lactobacillus (p = 0.001). High BMI controls—Low BMI controls: difference in 7 taxa, ↑ Dorea (p = 0.026), Pseudobutyrivibrio (p = 0.022), ↓ Ruminococcaceae Incertae Sedis (p = 0.021), Akkermansia (p = 0.006). |
| Dekker Nitert et al., 2020 [35] | 36 | M and F | No back pain: 34 (25–42) Back pain: 30 (27–36) | ≥25. No back pain: 29.9 (28.0–32.4) Back pain: 30.9 (28.2–34.5) | No back pain: WHR = 1.1 (0.8–1.4) Back pain: WHR = 1.1 (0.9–1.2) | Adlercreutzia: positively correlated with BMI (p = 0.03). |
| Durk et al., 2019 [65] | 37 | M and F | 25.7 ± 2.2 (22–32) | 23.7 ± 3.6 (17.9–31.4) | Body fat % = 23.1 ± 9.1 (7.0–38.0), fat mass kg = 16.2 ± 8.0 (4.1–40.2), fat-free mass kg = 53.0 ± 11.4 (33.7–80.1) | F/B: statistically correlated only with VO2max (p < 0.003) No other BMI or body composition variables were significantly correlated. |
| F S Teixeira et al., 2013 [66] | 32 | F | Lean: 28.05 ± 6.9 Obese: 30.7 ± 5.7 | Lean: 20.6–21.9 Obese: 32.8–36.7 | Lean (waist circumference cm = 66.5–72.0, body fat % = 18.0–23.8) Obese (waist circumference cm = 89.5–97.0, body fat % = 36.7–38.9) | Obese: ↓ Lactobacillus plantarum, Akkermansia muciniphila (p = 0.06), Bifidobacterium genus, Bifidobacterium longum, Clostridium coccoides, Clostridium leptum (p < 0.05) → negative correlations with BMI and waist circumference (p < 0.05). Body fat %: correlated inversely with Bifidobacterium genus, Bifidobacterium longum, Clostridium leptum, Clostridium coccoides, Lactobacillus plantarum (p < 0.05). |
| Fernandes et al., 2014 [67] | 94 | M and F | LN: 32.0 ± 1.8 OWOB: 37.9 ± 2.0 | LN: 21.8 ± 0.3 OWOB: 30.3 ± 0.7 | NR | Obese: ↓ Escherichia coli (p = 0.005). F/B: not significantly different between 2 groups. Combined 2 groups: BMI → inversely related to Bacteroidetes (r = −0.21, p = 0.04) and E. coli (r = −0.34, p = 0.002), no association with F/B. |
| Gallè et al., 2020 [79] | 140 | M and F | 22.5 ± 2.9 (18–36) | 22.4 ± 2.8 (15.2–33.8) | NR | Phyla: 28 different phyla detected—the most abundant → Firmicutes (61.6 ± 14.6) and Bacteroidetes (30.7 ± 13.3). BMI (underweight/normal-weight—overweight/obese): No significant differences in Shannon index, Firmicutes, Bacteroidetes, and F/B. Genera → ↑ Selemonas (p = 0.02), Megasphaera (p = 0.001), Streptococcus (p = 0.001), Dorea (p = 0.001), Lachnobacterium (p = 0.007), Jannaschia (p = 0.02), Dialister (p = 0.001), Eubacterium (p = 0.01), ↓ Paraprevotella (p = 0.01) in overweight/obese compared with underweight/normal-weight participants. |
| Henning et al., 2019 [36] | 63 | M and F | CTRL: 36.4 ± 10.8 AVO: 42.5 ± 12.7 | CTRL: 30.0 ± 3.7 AVO: 30.1 ± 3.2 | CTRL: Total body fat % = 38.3 ± 8.5 AVO: Total body fat % = 41.2 ± 5.1 | Baseline bacteria: Phylum (CTRL, AVO) → Firmicutes (61.29 ± 11.00, 53.91 ± 10.02), Bacteroidetes (26.94 ± 9.83, 34.88 ± 14.41), Actinobacteria (7.24 ± 6.07, 7.59 ± 7.86), Euryarcheota (1.76 ± 2.95, 1.05 ± 2.42), Verrucomicrobia (0.75 ± 1.90, 1.23 ± 1.73), Proteobacteria (1.09 ± 1.61, 0.89 ± 1.22). Family (CTRL, AVO)—Top 3 → Bacteroidaceae (Bacteroidetes) (17.27 ± 11.31, 23.37 ± 12.55), Ruminococcaceae (Firmicutes) (20.03 ± 6.02, 18.54 ± 7.33), Lachnospiraceae (Firmicutes) (16.56 ± 5.89, 15.37 ± 4.82). Genus (CTRL, AVO)—Top 3 → Bacteroides (Bacteroidetes) (17.27 ± 11.31, 23.37 ± 12.55), Clostridium (Firmicutes) (8.75 ± 3.17, 8.20 ± 3.41), Dialister (Firmicutes) (0.39 ± 0.61, 0.63 ± 1.01). |
| Hjorth et al., 2019 [37] | 52 | M and F | 0-P: 47.9 ± 6.8 Low P/B: 43.4 ± 8.7 High P/B: 41.8 ± 11.5 | 0-P: 30.7 ± 1.1 Low P/B: 29.7 ± 2.2 High P/B: 31.9 ± 2.8 | 0-P: Body fat % = 48.7 ± 3.9 Low P/B: Body fat % = 44.9 ± 4.1 High P/B: Body fat % = 44.4 ± 5.0 | Baseline: High P/B group → statistically significant ↑ body weight, BMI, relative abundance of Prevotella spp. and ↓ relative abundance of Bacteroides spp. |
| Janssens et al., 2016 [73] | 58 | M and F | Green tea: 28.2 ± 10.8 Placebo: 28.1 ± 10.5 | Green tea: 23.0 ± 4.0 Placebo: 23.6 ± 4.6 | Green tea (FMI kg/m2 = 6.9 ± 3.1, FFMI kg/m2 = 16.1 ± 1.9, WHR = 0.76 ± 0.09, FM kg = 19.9 ± 8.9, FFM kg = 46.9 ± 9.1, body fat % = 29.1 ± 8.2) Placebo (FMI kg/m2 = 7.2 ± 3.5, FFMI kg/m2 = 16.3 ± 2.0, WHR = 0.73 ± 0.08, FM kg = 20.4 ± 9.0, FFM kg = 47.2 ± 9.1, body fat % = 29.5 ± 8.7) | Participants categorized based on their BMI as normal-weight (18–25 kg/m2) and overweight (≥25 kg/m2). Baseline: Overweight → ↓ Shannon diversity index (α-diversity) for all phyla combined compared with normal-weight subjects (r = −0.39; p = 0.002). |
| Joller et al., 2020 [76] | 26 | F | 25–35 | 30–35 | NR | Baseline: 3 different enterotypes (most common to less common) → Enterotype 3—Firmicutes/Ruminococcus observed enriched in 21 females, Enterotype 2—Prevotella observed enriched in 3 females, Enterotype 1—Bacteroides observed enriched in 2 females. F/B ratio: ↑ (>1.6) in 12 females. |
| Kasai et al., 2015 [80] | 56 | M and F | N-Ob: 45.6 ± 9.6 Ob: 54.4 ± 8.2 | Non-obese: BMI < 20 Obese: BMI ≥ 25 | NR | Phylum: Obese → ↓ Bacteroidetes, ↑ F/B ratio, bacterial diversity and richness. Species: Obese → significantly associated with Blautia hydrogenotorophica (Firmicutes), Coprococcus catus (Firmicutes), Eubacterium ventriosum (Firmicutes), Ruminococcus bromii (Firmicutes), Ruminococcus obeum (Firmicutes); Non-obese → Bacteroides faecichinchillae, Bacteroides thetaiotaomicron, Blautia wexlerae, Clostridium bolteae, Flavonifractor plautii |
| Kobayashi et al., 2015 [38] | 92 | M | 21–59 | Lean: <18.5 Obese: >25.0 (17.3–30.2) | NR | Bacillus spp., Erysipelothrix spp., Holdemania spp. → related to lean group. Microbacteriaceae, Actinobacterium → related to obese group → Presence of Firmicutes and Actinobacteria may be related to BMI. |
| Koliada et al., 2017 [77] | 61 | M and F | 20–60+ | Underweight: <18.5 Normal: 18.5–24.9 Overweight: 25.0–29.9 Obese: ≥30 | NR | Phylum: ↑ BMI → ↑ Firmicutes, F/B ratio, ↓ Bacteroidetes |
| Million et al., 2013 [16] | 263 | M and F | 50 ± 17 | Anorexic: 13.5 (11.7–14.6) Lean: 22.4 (20.7–23.7) Overweight: 27.1 (25.9–28.6) Obese: 40.0 (36.4–46.8) | NR | Positive correlation with BMI: Lactobacillus reuteri (p = 0.02). Negative correlation with BMI: Bifidobacterium animalis (p = 0.03), Methanobrevibacter smithii (p = 0.08), Escherichia coli (p < 0.001). |
| Most et al., 2017 [68] | 37 | M and F | 37.8 ± 1.6 | 29.6 ± 0.5 | EGCG + RES (waist/hip ratio = 0.88 ± 0.02, body fat % = 29.7 ± 1.9) F (waist/hip ratio = 0.87 ± 0.02, body fat % = 31.6 ± 1.4) | Baseline bacteria: Genus (PLA—EGCG + RES) → Bacteroidetes % (82.5 ± 2.9–84.3 ± 2.9), Firmicutes % (12.6 ± 2.1–12.5 ± 2.7), Actinobacteria % (2.8 ± 1–2 ± 0.5), γ-Proteobacteria % (1.7 ± 0.4–1.1 ± 0.3), Akkermansia muciniphila % (0.4 ± 0.2–0 ± 0). Males compared with Females → ↑ Bacteroidetes (p < 0.001), ↓ Firmicutes (p < 0.001), Actinobacteria (p = 0.04). |
| Murtaza et al., 2019 [39] | 21 | M | 20–35 | 16.91–23.03 | NR | Baseline bacteria: 3 distinct clusters (genus) → Cluster 1—Prevotella dominant, Cluster 2—Bacteroides dominant, Cluster 3—Firmicutes dominant. Cluster 1 and Cluster 2 were more common. Shannon diversity → no significant differences between 3 clusters. |
| Palmas et al., 2021 [40] | 92 | M and F | NW: 49 ± 11 OB: 50 ± 12 | NW: 21.6 ± 2.1 OB: 36.0 ± 6.0 | NW (waist circumference cm = 73.7 ± 5.7) OB (Fat mass kg = 39.1 ± 11.9, fat mass % = 42.3 ± 5.7, muscle mass kg = 48.5 ± 11.3, waist circumference cm = 111 ± 15) | Richness and diversity: α-diversity → ↓ in obese group, although no significant difference in Shannon index (p = 0.833). β-diversity → significant difference between 2 groups (p = 0.002). Bacterial abundance: Obese → ↑ F/B ratio (p = 0.007), Firmicutes and Firmicutes taxa (main biomarkers: Lachnospiraceae, Megasphaera spp. + Gemellaceae, Paenibacilleae, Streptococcaceae, Thermicanaceae, Gemella, Mitsuokella, Streptococcus, Acidaminococcus spp., Eubacterium spp., Ruminococcus spp., Megamonas spp., Streptococcus, Thermicanus, Veillonella spp.), Proteobacterium taxa (main biomarkers: Escherichia, E. albertii), ↓ Bacteroidetes and Bacteroidetes taxa (main biomarkers: Flavobacteria, Flavobacterium, Bacteroides spp. + Porphyromonadaceae, Sphingobacteriaceae, Rikenella spp., Pedobacter spp., Parabacteroides spp.). Body fat and waist circumference → negatively correlated with Bacteroidetes taxa. Body fat → positively correlated with Firmicutes taxa. Muscle mass and physical activity → negatively correlated with Firmicutes taxa. |
| Resende et al., 2021 [41] | 24 | M | 20–45 | CG: 23.68 ± 3.29 EG: 25.28 ± 4.11 (18.5–29.9) | CG (%FM = 21.87 ± 12.18, %FFM = 78.12 ± 12.18) EG (%FM = 23.59 ± 11.63, %FFM = 76.40 ± 11.63) | Baseline bacteria. 10 phyla were detected → most abundant: Bacteroidetes, Firmicutes, Proteobacteria—no statistical difference between 2 groups. BMI: negative correlation with Desulfovibrio. Body fat: negative association with Faecalibacterium. Fat-free mass %: positive association with Faecalibacterium. |
| Sergeev et al., 2020 [42] | 20 | M and F | Placebo: 47.0 ± 15.4 Synbiotic: 47.8 ± 8.99 | Placebo: 32.77 ± 4.51 Synbiotic: 34.20 ± 5.60 | Placebo (body mass kg = 97.6 ± 23.1, WC = 106.9 ± 12.47, body fat mass kg = 40.66 ± 6.92, body fat % = 40.97 ± 5.02, body lean mass kg = 57.39 ± 17.76, BMC kg = 2.66 ± 0.64, body lean mass + BMC kg = 60.05 ± 18.38) Synbiotic (body mass kg = 90.6 ± 11.9, WC = 109.6 ± 8.07, body fat mass kg = 36.97 ± 11.35, body fat % = 40.51 ± 8.96, body lean mass kg = 51.13 ± 8.87, BMC kg = 2.38 ± 0.48, body lean mass + BMC kg = 53.52 ± 9.35) | Baseline bacteria: Firmicutes and Bacteroidetes → the 2 most abundant phyla, Bacteroides → the most abundant genus. |
| Valeriani et al., 2020 [43] | 59 | M and F | 23.1 ± 3.14 (20–36) | 22.2 ± 2.6 (16.6–29.7) | NR | Phylum: Most abundant → Firmicutes (61.6 ± 14.6), Bacteroidetes (30.7 ± 13.3). Correlation analysis: BMI → positive but not significant correlation with Firmicutes (r = 0.22; p = 0.08), Bacteroidetes (r = 0.06; p = 0.63), F/B ratio (r = 0.11; p = 0.38). |
| Whisner et al., 2018 [44] | 82 | M and F | 18.4 ± 0.6 | <18.5 18.5–24.9 25.0–29.9 ≥30 | NR | F/B ratio: 0.65 (0.39–1.23) → no statistically significant difference by BMI (p = 0.413). |
| Yang et al., 2017 [45] | 71 | F | 19–49 | Low VO2max: 31.7 (30.2–33.1) Moderate VO2max: 27.9 (26.7–29.1) High VO2max: 24.6 (23.0–26.2) | Low VO2max (fat % = 40.6 (38.1–43.0)) Moderate VO2max (fat % = 35.5 (33.2–37.8)) High VO2max (fat % = 28.0 (25.0–31.0)) | Eubacterium rectale–Clostridium coccoides: positively correlated with fat% → ↑ in low VO2max, followed by moderate and high VO2max. |
| Zuo et al., 2011 [81] | 104 | M and F | Normal-weight: 33.02 ± 10.37 Obese: 34.65 ± 11.91 | Normal-weight: 20.26 ± 1.50 (18.5–24) Obese: 30.79 ± 2.80 (≥28) | NR | Obese: ↓ Bacteroides (p = 0.012), Clostridium perfringens (p = 0.001). No other statistically significant differences in Escherichia coli, Enterococci, Lactobacilli, Bifidobacteria between groups → Enterococci: tendency to be ↑ in the obese group. |
APG = Animal Protein Group; AVO = Avocado Group; BMI = Body Mass Index; CG = Control Group; CTRL = Control Group; EG = Exercise Group; F/B = Firmicutes to Bacteroidetes Ratio; F = Female; FFM = Fat-Free Mass; FM = Fat Mass; HC = Hip Circumference; LN = Lean; M = Male; NR = Not Reported; NW = Normal-Weight; O = Obese; OB = Obese; OW = Overweight; TC = Thigh Circumference; VO2 = Volume of Oxygen; VPG = Vegetable Protein Group; WC = Waist Circumference; WHR = Waist-to-Hip Ratio; WPG = Whey Protein Group.

Table 4.
Characteristics of studies investigating the gut microbiome composition in older adults.
Table 4.
Characteristics of studies investigating the gut microbiome composition in older adults.
| Author(s), Date | N | Sex | Age (Years) | BMI Category (kg/m2) | Body Composition | Results |
|---|---|---|---|---|---|---|
| Morita et al., 2019 [74] | 29 | F | 70 (66–75) | 21.4 (18.8–23.1) | Body fat % = 29.0 (23.6–32.7) | Baseline bacteria: Genus (TM group—AE group) → Bacteroides (40.7%–43.0%), Clostridium subcluster XIVa (16.6%–17.9%), Bifidobacterium (not available %), Clostridium cluster IV (not available %). |
| Šoltys et al., 2021 [46] | 22 | M | LA: 63.5 (61.4–65.7) CTRL: 64.9 (62.1–67.7) | LA: 24.8 (24.0–25.6) CTRL: 27.3 (24.9–29.7) | LA (total body fat % = 19.4 (17.3–21.5), visceral body fat = 9.5 (8.3–10.6), muscle mass % = 37.44 (34.9–40.0)) CTRL (total body fat % = 26.2 (21.9–30.5), visceral body fat = 14.1 (10.6–17.7), muscle mass % = 34.4 (27.6–44.9)) | Dominant phylum (CTRL/LA): Firmicutes (73.9%/75.6%), Bacteroidetes (18.6%/14.4%), Proteobacteria (0.5%/1.5%). F/B ratio + α-diversity: no statistical difference between 2 groups. Family level: LA → ↑ Ruminococcaceae, ↓ Bacteroidaceae, Clostridiales Incertae Sedis XI, Cytophagia. Genus level: LA → ↑ Prevotella, Intestimonas, Subdoligranulum, Pseudobutyrivibrio, Marvinbryantia, Vallitalea, Porphyromonas, Anaerovorax, ↓ Bacteroides, Anaerosporobacter, Phascolarctobacterium, Bacteroides/Prevotella ratio. |
| Tamura et al., 2017 [47] | 56 | M and F | 72.1 ± 0.6 (65–84) | 23.1 ± 0.4 | NR | Most abundant families: Lachnospiraceae (25.4% ± 1.3%), Ruminococcaceae (13.5% ± 1.0%), Bifidobacteriaceae (9.9% ± 1.2%), Streptococcaceae (6.0% ± 1.2%), Bacteroidaceae (5.9% ± 0.7%), Eubacteriaceae (4.9% ± 0.4%), Coriobacteriaceae (4.3% ± 0.5%), Peptostreptococcaceae (2.8% ± 0.5%), Enterobacteriaceae (2.0% ± 0.5%), Erysipelotrichaceae (1.7% ± 0.4%), Clostridiaceae (1.5% ± 0.3%), Lactobacillaceae (1.0% ± 0.2%), Porphyromonadaceae (0.8% ± 0.1%), Rikenellaceae (0.7% ± 0.1%), Prevotellaceae (0.6% ± 0.2%). Correlations between BMI and fecal microbiota: Negative correlations → Porphyromonadaceae (r = −0.342), Rikenellaceae (r = −0.299), Christensenellaceae (r = −0.341), Oxalobacteraceae (r = −0.329)—Positive correlations → Aerococcaceae (r = 0.32). |
| Tavella et al., 2021 [48] | 201 | M and F | 71.2 ± 3.8 (65–79) | G1: 27.04 ± 3.60 G2: 24.68 ± 3.25 G3: 28.48 ± 4.18 | G1 (waist circumference cm = 93.12 ± 11.63, hip circumference cm = 1014.3 ± 7.75, waist/hip ratio = 0.92 ± 0.09) G2 (waist circumference cm = 84.75 ± 9.31, hip circumference cm = 97.58 ± 7.36, waist/hip ratio = 0.86 ± 0.07) G3 (waist circumference cm = 95.79 ± 11.05, hip circumference cm = 104.75 ± 7.04, waist/hip ratio = 0.91 ± 0.08) | Overall: Most abundant phylum → Firmicutes (80%), Bacteroidetes (8.9%), Actinobacteria (7.4%). Most abundant family → Ruminococcaceae (37.5%), Lachnospiraceae (27.6%)—both belonging to Firmicutes). Most abundant genus → Subdoligranulum (12.5%), Faecalibacterium (7.8%), Bifidobacterium (4.6%). 3 groups: G1, G2, G3. α-diversity: ↑ G2, G3. G1 → enriched in Lachnospiraceae (Eubacterium rectale group, Fusitanetibacter, Blautia: negatively correlated with SMI—positively correlated with DXA variables, especially those related to fat mass distribution—FM, FMI, AF/AL, AF/GF, VAT) G2 (significantly ↓ anthropometric and body composition values) → enriched in Christensellaceae, Porphyromonadaceae, Rikenellaceae (Christensellaceae R7 group, Parabacteroides, Alistipes: inversely associated with DXA variables—visceral adipose tissue) G3 → enriched in Ruminococcaceae (Ruminococcaceae UCG 014, 002, 005: negatively correlated with most adiposity-related DXA variables, directly correlated with SMI and Faecalibacterium, Subdoligranulum, Ruminococcus: positively correlated with most adiposity-related DXA variables, negatively correlated with SMI). |
AE = Aerobic Exercise Training; BMI = Body Mass Index; CTRL = Control; F = Female; LA: Lifetime Elderly Endurance Athletes; M = Male; NR = Not Reported; TM = Trunk Muscle Training.

Table 5.
Characteristics of studies investigating gut microbiome composition regardless of age.
Table 5.
Characteristics of studies investigating gut microbiome composition regardless of age.
| Author(s), Date | N | Sex | Age (Years) | BMI Category (kg/m2) | Body Composition | Results |
|---|---|---|---|---|---|---|
| Kulecka et al., 2020 [49] | 71 | M and F | 14–72 | NR | FMR (TBW lt = 30.9 ± 4.4, BF kg = 8.2 ± 1.1, FFM kg = 42.2 ± 5.9, MM kg = 23.4 ± 3.25) FCCS (TBW lt = 36.5 ± 2.7, BF kg = 9.3 ± 1.8, FFM kg = 50 ± 3.9, MM kg = 28.3 ± 2.3) MMR (TBW lt = 43.2 ± 3.6, BF kg = 5.9 ± 2.7, FFM kg = 59.8 ± 5.1, MM kg = 38.5 ± 10.1) MCCS (TBW lt = 49 ± 3.4, BF kg = 4.9 ± 1, FFM kg = 67 ± 4.74, MM kg = 39.3 ± 2.9) | Both athlete groups (MR, CCS) compared with healthy controls: ↓ Bacteroides, ↑ Prevotella, microbial diversity, and richness. F/B ratio: ↓ in healthy controls compared with CCS (p = 0.043), no statistically significant difference between healthy controls and MR. |
| La-Ongkham et al., 2020 [50] | 120 | M and F | Adult: 34.60 ± 3.19, elderly: 69.53 ± 3.44 | Adult: 22.39 ± 3.33, elderly: 24.30 ± 2.68 | NR | Phylum: >96% belonged to Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria. Statistically significant differences only in Bacteroidetes and Actinobacteria. Elderly → ↑ Bacteroidetes (phylum) (p = 0.019)—Bacteroidaceae (family) (p = 0.001)—Bacteroides (genus) (p = 0.001)—species: Bacteroides uniformis, Bacteroides ovatus, Bacteroides caccae, Bacteroides thetaiotaomicron, Parabacteroides (genus) (p = 0.02), ↓ Actinobacteria (phylum) (p = 0.001)—Bifidobacteriaceae (family) (p = 0.001)—Bifidobacterium (genus) (p = 0.001)—species: Bifidobacterium adolescentis, Bifidobacterium longum, Bifidobacterium pseudocatenulatum, Dorea (genus) (p = 0.01), F/B ratio (p = 0.01). ↑ age → ↓ Bifidobacterium, ↑ Bacteroides. |
| Latorre-Pérez et al., 2021 [51] | 528 | M and F | 18.3–71 | 17.26–36.33 | NR | All participants: Dominant phylum → Firmicutes (53.9%), Bacteroidetes (37.2%), Proteobacteria (5%), Verrucomicrobia (1.8%), Actinobacteria (0.9%). Dominant genera → Bacteroides (18.4%), Faecalibacterium (12.5%) (12.5%), Prevotella (6.7%), Alistipes (3.4%), Oscillospiraceae taxa (2.3%). ↑ BMI → positive correlation with Roseburia (genus), proteobacteria (phylum)—negative association with Marvinbryantia (genus) and Christensenellaceae (family). ↑ Age → ↓ Faecalibacterium, Bifidobacterium, ↑ alpha diversity—no significant associations with Akkermansia and Bacteroides |
| Martínez-Cuesta et al., 2021 [52] | 26 | M and F | 18+ | Normo-weight (N): 18–25, obese (O): >30 | NR | Richness and diversity: Obese → ↓ Chao1 index (α diversity), no other statistical differences. Phylum: No statistical differences in Firmicutes, Bacteroidetes, F/B ratio. Family: Obese → ↓ Ruminococcaceae, Rikenellaceae, Peptostreptococcaceae, Clostridiales. Genus: Obese → ↑ Collisnella, Clostridium XIVa, Catenibacterium, ↓ Alistipes, Clostridium sensu stricto, Romboutsia, Oscilibacter. |
| Oki et al., 2016 [53] | 516 | M and F | 52.4 ± 13.4 (21–88) | Lean: <25, obese: >30 | NR | Predominant bacterial families: Bacteroidaceae (33.1 ± 19.0%), Lachnospiraceae (17.6 ± 10.1%), Ruminococcaceae (15.8 ± 9.3%), Prevotellaceae (9.1 ± 18.0%). Obese: ↓ Christensenellaceae, Mogibacteriaceae, Rikenellaceae (p < 0.05). |
| Schwiertz et al., 2010 [54] | 98 | M and F | 47 ± 13 (14–74) | Lean: 18.5–24.9, overweight: 25.0–29.9, obese: ≥30.0 | NR | Most abundant bacterial groups in all groups: Clostridium leptum group, Clostridium coccoides group, Bacteroides spp. → all belonged to Firmicutes and Bacteroidetes phyla. Differences between groups: Overweight/obese compared with lean → ↓ Firmicutes (p = 0.001, p = 0.002), F/B ratio (p = 0.001, p = 0.005), Ruminococcus flacefaciens subgroup (phylum: Firmicutes; p = 0.006, p = 0.011), ↑ Bacteroidetes (p = 0.001, p = 0.006). Overweight compared with lean → ↑ Bacteroides (p = 0.002). Obese compared with lean → ↓ Clostridium leptum group (p = 0.07), Bifidobacterium (p = 0.02), Methanobrevibacter (p = 0.017). |
BF = Body Fat; BMI = Body Mass Index; CCS = Cross-Country Skiers; F/B Ratio = Firmicutes to Bacteroidetes Ratio; F = Female; FCCS = Female Cross-Country Skiers; FFM = Fat-Free Mass; FMR = Female Marathon Runners; M = Male; MCCS = Male Cross-Country Skiers; MM = Muscle Mass; MMR = Male Marathon Runners; MR = Marathon Runners; NR = Not Reported; TBW = Total Body Water.

Table 6.
Main differences in gut microbiome composition in all BMI categories and age groups.
Table 6.
Main differences in gut microbiome composition in all BMI categories and age groups.
| α-diversity | Phyla | Genera (Phylum) | Species | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteroidetes | Firmicutes | Firmicutes/ Bacteroidetes Ratio | Akkermansia (Verrucomicrobia) | Alistipes (Bacteroidetes) | Bacteroides (Bacteroidetes) | Bifidobacterium (Actinobacteria) | Dorea (Firmicutes) | Eubacterium (Firmicutes) | Faecalibacterium (Firmicutes) | Intestimonas (Firmicutes) | Lactobacillus (Firmicutes) | Megasphaera (Firmicutes) | Oscilibacter (Firmicutes) | Streptococcus (Firmicutes) | Faecalibacterium Prausnitzii | Lactobacillus Plantarum | Akkermansia Muciniphila | Roseburia spp. | |||
| Children | Normo-weight | ↑ | ↑ | ↓ | ↓ | ↑ | – | ↑ | ↑ | ↓ | ↓ | – | – | – | – | ↑ | – | ↓ | ↑ | ↑ | – |
| Overweight | ↓ | ↓ | ↑ | ↑ | ↓ | – | ↓ | ↓ | ↑ | ↑ | – | – | – | – | ↓ | – | ↑ | ↓ | ↓ | – | |
| Obese | ↓ | ↓ | ↑ | ↑ | ↓ | – | ↓ | ↓ | ↑ | ↑ | – | – | – | – | ↓ | – | ↑ | ↓ | ↓ | – | |
| Athletes | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| Adults | Normo-weight | ↑ | ↑ | ↓ | ↓ | – | ↑ | ↑ | ↑ | ↓ | ↓ | ↑ | ↑ | ↓ | ↓ | ↑ | ↓ | ↑ | ↑ | ↑ | – |
| Overweight | ↓ | ↓ | ↑ | ↑ | – | ↓ | ↓ | ↓ | ↑ | ↑ | ↓ | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | ↓ | ↓ | – | |
| Obese | ↓ | ↓ | ↑ | ↑ | – | ↓ | ↓ | ↓ | ↑ | ↑ | ↓ | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | ↓ | ↓ | – | |
| Athletes | ↑↑ | ↓ | – | – | ↑ | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ↑ | |
| Older Adults | Normo-weight | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Overweight | – | – | – | – | – | – | ↑ | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| Obese | – | – | – | – | – | – | ↑ | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| Athletes | ↑↓ | – | – | ↑↓ | – | – | ↓ | – | – | – | – | ↓ | – | – | – | – | – | – | – | – | |
↑ = increased, ↓ = decreased, ↑↓ = contradictory, – = data not available.
3.3. Children
Most of the results from the studies in this category were presented comparing normal-weight with underweight or overweight children (Table 2). Regarding α-diversity, seven studies took it into consideration [23,24,26,27,28,29,57]; however, only two studies reported statistically significant differences (p < 0.05) in relation to body composition [23,28]. The first study identified three groups according to muscle mass [23], and the second one showed less α-diversity in obese children compared with normal-weight children [28]. Only one study included β-diversity, with statistically significant differences between obese and normal-weight children (p < 0.05) [28].
The dominant phyla identified throughout the studies, both in normal-weight and obese children, were, in descending order, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia [24,26,27,29,30,32]. Riva and colleagues identified the dominant families and genera. The dominant families were Ruminococcaceae, Lachnospiraceae, Bacteroidaceae, Veillonellaceae, Bifidobacteriaceae, Prevotellaceae, Verrucomicrobiaceae, Rikenellaceae, and Christensellaceae, while the dominant genera were Bacteroides, Subdoligranulum, Faecalibacterium, Dialister, Bifidobacterium, Pseudobutyrivibrio, and Blautia [29].
Correlations between the gut microbiome composition and BMI were observed in 15 studies [24,25,26,27,28,29,30,55,56,57,58,59,60,69,75] and can be summarized into six classification categories: phylum, class, order, family, genus, and species. At the phylum level, the composition of the gut microbiome in obese children comprised decreased Bacteroidetes and increased Firmicutes, Actinobacteria, and Firmicutes/Bacteroidetes ratios (F/B ratio), in comparison to normal-weight children [24,26,29,30,60,69]. At the class level, no study showed any results, while, at the order level, high levels of Pasteurellales were observed [32]. At the family level, obese children’s microbiomes were characterized by increased levels of Lactobacillaceae, Enterobacteriaceae, and Lachnospiraceae and decreased levels of Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, Desulfovibrio, Christensenellaceae, and Ruminococcaceae [28,29,55,57]. At the genus level, the studies showed increased levels of Blautia, Dorea, Eubacterium, Fusitanetibacter, and Bifidobacterium, and decreased levels of Bacteroides, Oscilibacter, Parabacteroidetes, Ruminococcus, Akkermansia, and Haemophilus in obese children [24,26,29,57,69,75]. Finally, at the species level, increased levels occurred in Faecalibacterium prausnitzii, Bacteroides fragilis group, Lactobacillus spp., Bacteroides eggerthii, Lachnospira, and Prevotella member and decreased levels occurred in Bifidobacterium spp., Akkermansia muciniphila, Bacteroides plebeius, and species from Christensenellaceae, Alistipes, and the Lactobacillus gasseri subgroup [25,27,28,56,57,59].
Other body composition parameters, besides BMI were correlated with the gut microbiome composition in five studies [23,31,55,58,69]. More specifically, a positive correlation was observed between the phylum Firmicutes and the circumferences of the waist and head [58], while correlations were also observed between Bacteroidetes (p = 0.031; p = 0.012; p = 0.003), the F/B ratio (p = 0.075; p = 0.032; p = 0.002), Actinobacteria (p = 0.039; p = 0.053; p = 0.078), and visceral, subcutaneous, and hepatic fat [69]. A positive correlation was observed between the family Lactobacillaceae and visceral fat [55], while a correlation was also observed between the family Ruminococcaceae and the fat-free mass index (FFMI) Z-score in boys (p = 0.027) [31]. At the genus level, Faecalibacterium and Lachnospira were positively correlated with at least one of the following: ratio of total body lean soft tissue mass (TSM) to weight (TSMR), appendicular skeletal mass (ASM), ratio of appendicular skeletal mass to height (ASMI), and ASMI z-score. They were negatively correlated with at least one of the following: ratio of TSMR, total body lean soft tissue mass/total body fat (TSM/TBF), appendicular skeletal mass to weight (ASMR), appendicular skeletal mass/appendicular fat mass (ASM/AFM), and ASMR z-score [23]. The genera Actinomyces, Bifidobacterium, Streptococcus, and Blautia were positively correlated with body fat storage, while, in contrast, the genera Odoribacter, Oscillospira, Bacteroides, and Faecalibacterium were negatively correlated with fat [69].
3.4. Adults
The results from the studies in the adult category were presented by comparing normal-weight adults with either athletes or overweight/obese adults (Table 3). Five studies showed statistically significant differences in α-diversity (p < 0.05) [34,40,71,73,80], while two studies showed no differences (p > 0.05) [33,79]. More specifically, α-diversity was significantly lower in overweight/obese individuals, compared with the normal-weight control group (p < 0.05) [40,71,73], although Kasai and his colleagues reported the opposite result [80]. Clarke et al. [34] compared elite athletes with two groups of non-athletes, including both low and high BMI levels. Elite athletes showed statistically significantly higher levels of α-diversity compared with both groups, while no differences between the control groups were observed. Statistically significant differences between normal-weight and overweight individuals for β-diversity (p < 0.05) were observed in two studies [33,40].
The dominant phyla in all BMI adult groups in descending order were Firmicutes, Bacteroidetes, Proteobacteria, Verrucomicrobia, Fusobacteria, and Actinobacteria [36,41,42,43,63,68,70,78,79]. The three dominant families were Bacteroidaceae, Ruminococcaceae, and Lachnospiraceae [36]. The dominant genera were Bacteroides, Clostridium, Dialister, Blautia, Faecalibacterium, and Ruminococcus, all of which belong to the Bacteroidetes and Firmicutes phyla [36,42,68,70].
Correlations between the gut microbiome composition and BMI were observed from 20 studies [16,33,34,35,37,38,40,41,44,61,62,65,66,67,71,72,77,79,80,81], some of which did not show statistically significant differences [44,65]. The results were summarized into six classification categories, beginning with the phylum level. The composition of the gut microbiome in obese adults comprised increased levels of Firmicutes, increased F/B ratios, increased P/B ratios, and decreased levels of Bacteroidetes [38,40,65,67,80]. At the family level, Ruminococcaceae, Succinivibrionaceae, and Akkermansia were observed to be lower in obese adults, while Microbacteriaceae was higher [38]. At the genus level, obese adults had statistically significantly higher levels of Mogibacterium, Mitsuokella, Megamonas, Howardella, Anaerovibrio, Allisonella, Adlercreutzia, Abiotrophia, Pseudobutyrivibrio, Adlercreutzia, Selemonas, Megasphaera, Streptococcus, Lachnobacterium, Jannaschia, Dialister, Eubacterium, and Actinobacterium and lower levels of Victivallis, Succinivibrio, Rothia, Parvimonas, Intestimonas, Haemophilus, Faecalibacterium, Anaerococcus, Paraprevotella, and Desulfovibrio [33,34,35,40,41,71,79,81]. The results in the genera Bacteroides [33,40,81] and Dorea [33,34,79] were conflicting between studies. The Bacteroides levels [40,81] were decreased in obese adults, while, on the other hand, the Dorea levels [34,79] were elevated in the majority. Finally, at the species level, conflicting results were also observed for Clostridium leptum [62,66]. Otherwise, obese adults’ gut microbiomes were composed of increased levels of Bacteroides thetaiotaomicron, Blautia hydrogenotorophica, Coprococcus catus, Eubacterium rentriosum, Ruminococcus bromii, and Lactobacillus reuteri, most of which belong to the Firmicutes phylum, and decreased levels of Lactococcus lactis, Flavonifractor plautii, Faecalibacterium prausnitzii, Lactobacillus plantarum, Akkermansia muciniphila, Bifidobacterium genus, Bifidobacterium longum, Bifidobacterium animalis, Clostridium coccoides, Clostridium perfringens, Escherichia coli, Bacillus spp., Erysipelothrix spp., Holdemania spp., and Methanobrevibacter smithii [16,33,37,38,40,62,66,67,71,80,81].
Other body composition parameters, apart from BMI, were correlated with the gut microbiome composition in seven studies [34,40,41,45,65,66,70]. Obesity parameters, examples of which include the body fat percentage, visceral fat, waist circumference, and waist/hip ratio, were positively correlated with Firmicutes, the Firmicutes taxa, and the F/B ratio and negatively correlated with Bacteroidetes and the Bacteroidetes taxa, both in males and females (p < 0.05) [40,70]. A positive correlation was also observed in women between the body fat percentage and Eubacterium rectale and Clostridium coccoides [45]. On the other hand, another study showed a negative correlation between the waist circumference and Clostridium leptum (p < 0.05), as well as between body fat and Bifidobacterium, Clostridium leptum, and Lactobacillus plantarum in women (p < 0.05) [66]. A study conducted with a male sample showed a negative correlation between body fat and Faecalibacterium (p < 0.05) [41]. Both lean body mass and fat mass were negatively correlated with the Firmicutes taxa in males and females and positively correlated with Faecalibacerium in males (p < 0.05) [40,41]. Clarke et al. [34] compared elite male athletes with a control group of men with high BMI levels, who were not statistically significantly different from athletes. The two groups differed in the body fat percentage, lean body mass, and waist/hip ratio. Statistically significantly higher levels of the Akkermansiaceae family (p = 0.049) and the Akkermansia genus (p = 0.035) and lower levels of the Bacteroidetes phylum (p = 0.022) were observed in athletes [34]. Finally, one study did not show any statistically significant correlation between the gut microbiome composition and lean body mass or fat mass (p > 0.05) [65].
3.5. Older Adults
The results from the studies in the older adult category were presented by comparing older adults with different BMIs and long-term athletes versus sedentary control groups (Table 4). Two studies investigated α-diversity [46,48]; one did not show any statistically significant difference between the athlete group and the sedentary control group (p > 0.05) [46], while the other study categorized its sample into three groups according to different gut microbiome compositions and did show significant differences between groups (p < 0.05) [48]. None of the studies investigated β-diversity.
The dominant phyla in older adults were, in descending order, Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria [46,48]. The three dominant families were Lachnospiraceae, Ruminococcaceae, and Bifidobacteriaceae [47,48]. The results on the genus level differed between studies. The dominant genera in older adults with lower BMI values (18.8–23.1 kg/m2) were Bacteroides, Clostridium subcluster XIVa, Bifidobacterium, and Clostridium cluster IV [74]. The dominant genera in older adults with higher BMI ranges were Subdoligranulum, Faecalibacterium, and Bifidobacterium [48].
Two out of four studies observed correlations between the gut microbiome composition and BMI [46,47]. Tamura et al. [47] showed a negative correlation between BMI and the families Porphyromonadaceae (r = −0.342), Rikenellaceae (r = −0.299), Christensellaceae (r = −0.341), and Oxalobacteraceae (r = −0.329) and a positive correlation between BMI and the family Aerococcaceae (r = 0.32). On the other hand, Soltys et al. [46] and her colleagues compared long-term athletes with a sedentary control group that had statistically significantly higher BMI values (p < 0.05). At the phylum level, the F/B ratio was not different between groups, while, at the family level, athletes had higher levels of Ruminococcaceae and lower levels of the Bacteroidaceae, Clostridiales Incertae Sedis XI, and Cytophagia families. Moreover, athletes had higher levels of the genera Prevotella, Intestimonas, Subdoligranulum, Pseudobutyrivibrio, Marvinbryantia, Vallitalea, Porphyromonas, and Anaerovorax and lower levels of Bacteroides, Anaerosporobacter, Phascolarctobacterium, and the Bacteroides/Prevotella ratio (p < 0.05).
In addition to BMI, other body composition parameters were correlated with the gut microbiome composition in two studies [46,48]. Soltys et al. [46], as described before, reported statistically significant differences between athletes and control groups in terms of the body fat percentage, visceral fat, and muscle mass percentage; these differences may have been responsible for the gut microbiome differences between the groups. The results of the second study were categorized into three groups according to the composition of the gut microbiome. The first group (G1) was enriched in the Lachnospiraceae family. The Eubacterium rectale group, Fusitanetibacter, and Blautia were negatively correlated with the skeletal muscle index (SMI) and positively correlated with the body fat distribution parameters (fat mass (FM), fat mass index (FMI), ratio of android fat mass/android lean mass (AF/AL), ratio of android fat mass/gynoid fat mass (AF/GF), visceral adipose tissue (VAT)). The second group (G2), with the significantly lowest anthropometric measurements, was enriched in the Christensellaceae, Porphyromonadaceae, and Rikenellaceae families. In the Christensellaceae R7 group, Parabacteroides and Alistipes were negatively correlated with visceral fat. The last group (G3) was enriched in the Ruminococcaceae family. Ruminococcaceae UCG 014, 002, and 005 were negatively correlated with body composition parameters referring to fat and positively correlated with the SMI. Faecalibacterium, Subdoligranulum, and Ruminococcus showed a reverse pattern compared to the above, with a positive correlation with body fat parameters and a negative correlation with the SMI (p < 0.05) [48].
3.6. Whole Age Range
The results from the studies in the whole age range category were presented by comparing people with different body composition measurements, regardless of age (Table 5). Three studies showed statistically significant differences for α-diversity (p < 0.05) [49,51,52]. The α-diversity was higher in athletes compared with non-athletes (p < 0.05) [49], older adults compared with adults (p < 0.05) [51], and normo-weight compared to obese individuals (p < 0.05) [52]. None of the studies investigated β-diversity.
The dominant phyla in all age groups were Firmicutes, Bacteroidetes, Proteobacteria, Verrucomicrobia, and Actinobacteria [50,51]. However, increasing age was observed to cause an increase in the Bacteroides and Bacteroides taxa and a decrease in the Actinobacteria and Actinobacteria taxa [50]. The dominant families were Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Prevotellaceae [53]. The dominant genera were the Bacteroides, Faecalibacterium, Prevotella, Alistipes, and Oscillosperaceae taxa [51]. Finally, Schwiertz et al. [54] identified the most abundant bacterial groups, which were the Clostridium leptum group, Clostridium coccoides group, and Bacteroides spp., all belonging to the Firmicutes and Bacteroidetes phyla.
Three out of six studies described correlations between the gut microbiome and BMI [51,52,54]. The BMI was positively correlated with the Roseburia genus, while a negative correlation was found in the Marvinbryantia genus and Christensellaceae family [51]. Moreover, Martinez-Cuesta et al. [52] compared normo-weight with obese individuals. At the phylum level, no statistically significant correlation was observed in Firmicutes, Bacteroidetes, and the F/B ratio (p > 0.05). On the other hand, obese people had lower levels of the families Ruminococcaceae, Rikenellaceae, Peptostreptococcaceae, and Clostridiales and the genera Alistipes, Clostridium sensu stricto, Romboutsia, and Oscilibacter and higher levels of the genera Collisnella, Clostridium XIVa, and Catenibacterium (p < 0.05). Schwiertz et al. [54] compared normo-weight, overweight, and obese individuals. The gut microbiomes of overweight and obese individuals were found to have lower Firmicutes levels (p = 0.001, p = 0.002), F/B ratios (p = 0.001, p = 0.005), and Ruminococcus flacefaciens subgroup levels (p = 0.006, p = 0.011) and higher levels of Bacteroidetes (p = 0.001, p = 0.006). Overweight people had higher levels of Bacteroides (p = 0.002) and obese people had lower levels of the Clostridium leptum group (p = 0.07), Bifidobacterium (p = 0.02), and Methanobrevibacter (p = 0.017) compared with normal-weight individuals.
Correlations of the gut microbiome with other body composition parameters, besides BMI, were found only by Kulecka et al. [49]. The sample was categorized into three groups, marathon runners, skier athletes, and a sedentary control group. The body composition parameters, like body fat, lean body mass, and muscle mass, differed between the two athlete groups and the control group (p < 0.05). The results showed reduced levels of Bacteroides and increased levels of Prevotella in both athlete groups compared to the control group (p < 0.05). Increased levels of the F/B ratio were also observed in skiers compared with the control group (p = 0.043), while no statistically significant difference was observed in marathon runners (p > 0.05).
The main differences in the gut microbiome composition in all BMI categories in all age groups are presented in Table 6. Figure 2 and Figure 3 show a comparative representation of the gut microbiome’s formation across the human lifespan. Children, adults, and older adults are categorized according to BMI into (i) normo-weight, (ii) overweight, (iii) obese, and (iv) athletes and are compared in terms of the gut microbiome composition regarding α-diversity and the most commonly found phyla, genera, and species.
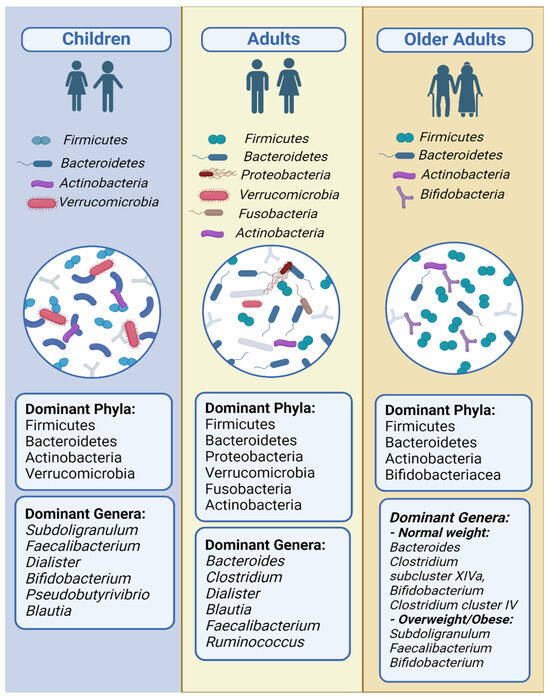
Figure 2.
Predominant bacterial phyla and genera across distinct age groups.
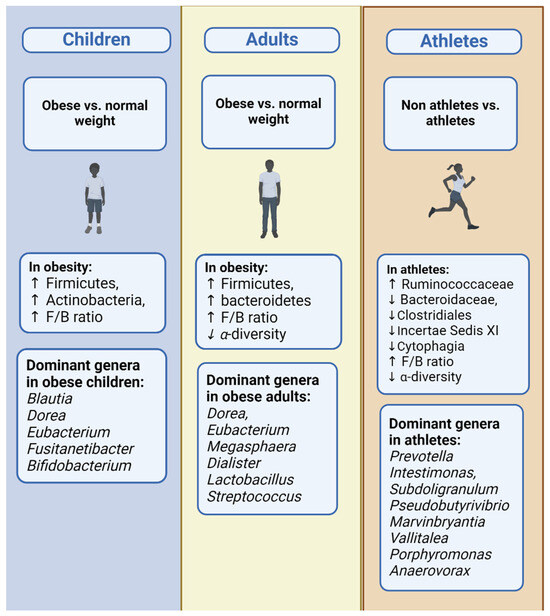
Figure 3.
Bacterial phyla and general dynamics: contrasts between obesity and normal weight and across age groups including children, adults, and athletes.
4. Discussion
The present systematic review aimed to identify different gut microbiome profiles in healthy individuals, from children to older adults, and to correlate them with body composition formation. It was found that there are significant differences in the gut microbiome composition in individuals with excess weight or athletes across different age groups.
It was observed that the gut microbiome composition of overweight and obese participants was characterized by decreased α-diversity, mostly in adults compared to children, where only two [23,28] out of the seven studies [23,24,26,27,28,29,57] showed statistically significant differences. In addition, decreased levels of the Bacteroidetes phylum and its taxa and increased levels of the Firmicutes phylum, its taxa, and the F/B ratio were observed in comparison to normal-weight participants. Other body composition parameters, apart from the BMI, followed similar correlations. More specifically, a positive correlation between the Firmicutes phylum, its taxa, and obesity parameters, examples of which include the body fat mass and waist circumference, was observed, while a negative correlation was observed between the Bacteroidetes phylum, its taxa, and obesity parameters. On the other hand, the Bacteroidetes phylum and its taxa were also positively correlated with the lean body mass and muscle mass. These outcomes appeared to be more significant in athletes, even compared to normal-weight individuals.
The relationship between the gut microbiome composition and body weight has recently been discovered and continues to be studied widely, especially during the last decade [10,82]. Studies conducted in mice observed an alteration in body weight after a fecal transplant intervention from obese mice to mice without any microbiome; such an observation is responsible for the expanding studies conducted in humans [83]. The three main mechanisms through which the gut microbiome contributes to body weight are well known and have already been described in the Introduction of the current systematic review. Briefly, the first mechanism involves LPS promoting underlying inflammation, a common sign of obesity. The second mechanism involves the SCFAs that metabolize undigested food components like fiber, resulting in 10% more energy intake, while, in contrast, they contribute to other metabolic pathways, activating the secretion of anorexic hormones. The last mechanism involves bile acids, through which energy expenditure and the secretion of anorexigenic GLP-1 are promoted [8]. Despite the fact that the above mechanisms are well studied, the responsible bacteria are not yet fully identified [84].
According to the existing literature, the results for α-diversity between individuals with normal and excess weight are controversial. A meta-analysis conducted by Walters et al. [85] in 2014 did not show any statistically significant difference between normo-weight and overweight adults. In contrast, two more recent meta-analyses confirmed the reduced α-diversity in obesity observed in the current systematic review, although only two of the ten studies in Sze and Schloss’s meta-analysis showed statistically significant differences (p < 0.05) [86,87]. It is noteworthy that α-diversity is related to the better functionality of the gut microbiome; thus, a reduced α-diversity can lead to the disruption of the gut microbiome’s functioning and, ultimately, host dysbiosis [88]. Two recent systematic reviews examined the impact of exercise on α-diversity, confirming a positive association between α-diversity and individuals with high levels of fitness or cardiorespiratory fitness, as well as individuals with lower fitness levels after the impact of exercise [89,90].
At the phylum level, the F/B ratio, in the majority of the studies, was observed to be higher in obese compared with normo-weight individuals, in all age groups. However, two meta-analyses were in disagreement with our results, showing that the F/B ratio did not display statistically significant differences (p > 0.05) [15,91]. Thus, this measure cannot be considered a strong indicator for the separation of individuals based on BMI [87,92]. The phyla Firmicutes and Bacteroidetes are well known as the dominant phyla of the gut microbiome, making up over 90% of its composition [93]. The increased levels of the Firmicutes and decreased levels of Bacteroidetes observed in obese participants in the present systematic review are in agreement with a number of studies confirming the respective relationship [14,15,92]. More specifically, the phylum Firmicutes is positively correlated with parameters related to obesity, such as the body fat percentage, and negatively correlated with the lean body mass. In contrast, the phylum Bacteroidetes is negatively correlated with obesity parameters, a result that is also consistent with the present findings [19,85,94]. The observed relationship between Firmicutes and obesity parameters seems to be explained by the fact that many enzymes involved in carbohydrate metabolism belong to this phylum. The exact mechanism that promotes obesity is probably the one involving the production of SCFAs, as a positive correlation has been observed between the phylum Firmicutes and SCFAs in feces. This indicates that obese individuals prevail in the fermentation of undigested nutrients in the large intestine and, by extension, in the 10% excess energy production and in body weight gain [92,95,96,97,98]. Moreover, a second mechanism concerning SCFAs can explain the observed positive correlation between the phylum Firmicutes and body fat. The fermentation of fiber by SCFAs can also lead to the promotion of hepatic lipogenesis, increasing the storage and accumulation of fatty acids and triglycerides in the adipose tissue. Acetic acid is considered to be the main culprit responsible for this process and is mainly produced by bacteria belonging to Firmicutes [19,93].
Recent meta-analyses that investigated the gut microbiome’s composition in normo-weight and obese individuals confirm the results of the current systematic review at the genus level. Some commonly detected genera in obese individuals are increased levels of Dorea, Eubacterium, Megasphaera, Dialister, Lactobacillus, and Streptococcus (phylum Firmicutes) and decreased levels of Bacteroides, Alistipes (phylum Bacteroidetes), Bifidobacterium (phylum Actinobacteria), Faecalibacterium, and Oscilibacter (phylum Firmicutes). However, it is obvious that the relationship between the phylum level and obesity does not necessarily expand at the genus level. For instance, the genera Faecalibacterium and Oscilibacter are reduced in obese people, while the expected observation would be increased levels due to belonging to the phylum Firmicutes [15,85,99,100]. The exact mechanism through which some bacteria affect body weight has already been discovered. The bacteria Lactobacillus plantarum, Faecalibacterium prausnitzii, and Akkermansia muciniphila appear to be reduced in obese compared to normo-weight people, a correlation that was also found in the present study. The genus Lactobacillus, as a member of the phylum Firmicutes, has been associated with obesity and is found to be increased in those with excess weight. Some specific species, like Lactobacillus plantarum, have been shown to prevent dysbiosis through the production of bacteriocins that prevent the growth of pathogenic microorganisms [101]. Faecalibacterium prausnitzii causes the production of butyric acid from the fermentation of undigested nutrients and is also characterized by its anti-inflammatory role, explaining its protective role against obesity [102]. Akkermansia muciniphila participates in mucus metabolism and the maintenance of intestinal barrier integrity in the host, while it prevents the colonization of pathogenic microorganisms and dysbiosis [103].
As in every research study, there are some issues that need to be considered when interpreting the data of this review. Firstly, the majority of the studies included were cross-sectional; hence, their results do not reflect a cause–effect relationship. It is important to note that the prospective studies and clinical trials reported in this review included baseline data, before any intervention took place. Moreover, the heterogeneity between studies should also be considered, not only regarding the definition of obesity, which differs by country and age, but also regarding the level of bacterial taxonomy investigated by each study, making the comparison of the results difficult.
5. Conclusions
To conclude, the composition of the gut microbiome is evidently different in overweight individuals or athletes in all age groups. The composition of the gut microbiome in obese people comprises decreased α-diversity, decreased levels of the phylum Bacteroidetes and its taxa, and increased levels of the phylum Firmicutes, its taxa, and the F/B ratio. Besides the BMI, obesity parameters, like body fat mass, are positively correlated with the Firmicutes taxa and negatively correlated with the Bacteroidetes taxa, and lean fat mass and muscle mass are positively correlated with the Bacteroidetes taxa. Additional studies are needed to confirm the above results, including those with healthy older adults.
Author Contributions
I.K., C.D.G. and E.A. designed the study; I.K. and A.V. conducted the literature search; I.K., A.V. and C.D.G. analyzed the data; I.K., A.V., C.D.G., M.P., M.C., K.F. and E.A. drafted and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gomaa, E.Z. Human Gut Microbiota/Microbiome in Health and Diseases: A Review. Antonie Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef] [PubMed]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the Human Microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [PubMed]
- Baranowski, T.; Motil, K.J. Simple Energy Balance or Microbiome for Childhood Obesity Prevention? Nutrients 2021, 13, 2730. [Google Scholar] [CrossRef]
- Appleton, J. The Gut-Brain Axis: Influence of Microbiota on Mood and Mental Health. Integr. Med. 2018, 17, 28–32. [Google Scholar]
- Heiss, C.N.; Olofsson, L.E. Gut Microbiota-Dependent Modulation of Energy Metabolism. J. Innate Immun. 2018, 10, 163–171. [Google Scholar] [CrossRef]
- de Clercq, N.C.; Groen, A.K.; Romijn, J.A.; Nieuwdorp, M. Gut Microbiota in Obesity and Undernutrition. Adv. Nutr. 2016, 7, 1080–1089. [Google Scholar] [CrossRef]
- Cornejo-Pareja, I.; Muñoz-Garach, A.; Clemente-Postigo, M.; Tinahones, F.J. Importance of Gut Microbiota in Obesity. Eur. J. Clin. Nutr. 2019, 72, 26–37. [Google Scholar] [CrossRef]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The Human Gut Microbiota: Metabolism and Perspective in Obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef] [PubMed]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S.F. Microbial Dysbiosis-Induced Obesity: Role of Gut Microbiota in Homoeostasis of Energy Metabolism. Br. J. Nutr. 2020, 123, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Bakker, G.J.; Zhao, J.; Herrema, H.; Nieuwdorp, M. Gut Microbiota and Energy Expenditure in Health and Obesity. J. Clin. Gastroenterol. 2015, 49, S13–S19. [Google Scholar] [CrossRef]
- Bliss, E.S.; Whiteside, E. The Gut-Brain Axis, the Human Gut Microbiota and Their Integration in the Development of Obesity. Front. Physiol. 2018, 9, 900. [Google Scholar] [CrossRef]
- Duca, F.A.; Lam, T.K.T. Gut Microbiota, Nutrient Sensing and Energy Balance. Diabetes Obes. Metab. 2014, 16, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Companys, J.; Gosalbes, M.J.; Pla-Pagà, L.; Calderón-Pérez, L.; Llauradó, E.; Pedret, A.; Valls, R.M.; Jiménez-Hernández, N.; Sandoval-Ramirez, B.A.; del Bas, J.M.; et al. Gut Microbiota Profile and Its Association with Clinical Variables and Dietary Intake in Overweight/Obese and Lean Subjects: A Cross-Sectional Study. Nutrients 2021, 13, 2032. [Google Scholar] [CrossRef]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the Gut Microbiota of Adults with Obesity: A Systematic Review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Angelakis, E.; Maraninchi, M.; Henry, M.; Giorgi, R.; Valero, R.; Vialettes, B.; Raoult, D. Correlation between Body Mass Index and Gut Concentrations of Lactobacillus Reuteri, Bifidobacterium Animalis, Methanobrevibacter Smithii and Escherichia Coli. Int. J. Obes. 2013, 37, 1460–1466. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, G.; Zhao, Y. Gut Microbiota and Alimentary Tract Injury; Springer: Berlin/Heidelberg, Germany, 2020; pp. 11–22. [Google Scholar]
- Huang, K.; Wu, L.; Yang, Y. Gut Microbiota: An Emerging Biological Diagnostic and Treatment Approach for Gastrointestinal Diseases. JGH Open 2021, 5, 973–975. [Google Scholar] [CrossRef]
- Davis, C.D. The Gut Microbiome and Its Role in Obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef]
- Maruvada, P.; Leone, V.; Kaplan, L.M.; Chang, E.B. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe 2017, 22, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Radua, J. PRISMA 2020—An Updated Checklist for Systematic Reviews and Meta-Analyses. Neurosci. Biobehav. Rev. 2021, 124, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Wells, G.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Available online: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 15 June 2022).
- Chen, F.; Li, Q.; Chen, Y.; Wei, Y.; Liang, J.; Song, Y.; Shi, L.; Wang, J.; Mao, L.; Zhang, B.; et al. Association of the Gut Microbiota and Fecal Short-chain Fatty Acids with Skeletal Muscle Mass and Strength in Children. FASEB J. 2022, 36, e22109. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.Y. Lifestyle Modifications Result in Alterations in the Gut Microbiota in Obese Children. BMC Microbiol. 2021, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Correlation between Body Mass Index and Faecal Microbiota from Children. Clin. Microbiol. Infect. 2016, 22, 258.e1–258.e8. [Google Scholar] [CrossRef] [PubMed]
- Karvonen, A.M.; Sordillo, J.E.; Gold, D.R.; Bacharier, L.B.; O’Connor, G.T.; Zeiger, R.S.; Beigelman, A.; Weiss, S.T.; Litonjua, A.A. Gut Microbiota and Overweight in 3-Year Old Children. Int. J. Obes. 2019, 43, 713–723. [Google Scholar] [CrossRef]
- López-Contreras, B.E.; Morán-Ramos, S.; Villarruel-Vázquez, R.; Macías-Kauffer, L.; Villamil-Ramírez, H.; León-Mimila, P.; Vega-Badillo, J.; Sánchez-Muñoz, F.; Llanos-Moreno, L.E.; Canizalez-Román, A.; et al. Composition of Gut Microbiota in Obese and Normal-Weight Mexican School-Age Children and Its Association with Metabolic Traits. Pediatr. Obes. 2018, 13, 381–388. [Google Scholar] [CrossRef]
- McCann, J.R.; Bihlmeyer, N.A.; Roche, K.; Catherine, C.; Jawahar, J.; Kwee, L.C.; Younge, N.E.; Silverman, J.; Ilkayeva, O.; Sarria, C.; et al. The Pediatric Obesity Microbiome and Metabolism Study (POMMS): Methods, Baseline Data, and Early Insights. Obesity 2021, 29, 569–578. [Google Scholar] [CrossRef]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. Pediatric Obesity Is Associated with an Altered Gut Microbiota and Discordant Shifts in F. irmicutes Populations. Environ. Microbiol. 2017, 19, 95–105. [Google Scholar] [CrossRef]
- Ruiz, A.; Cerdó, T.; Jáuregui, R.; Pieper, D.H.; Marcos, A.; Clemente, A.; García, F.; Margolles, A.; Ferrer, M.; Campoy, C.; et al. One-Year Calorie Restriction Impacts Gut Microbial Composition but Not Its Metabolic Performance in Obese Adolescents. Environ. Microbiol. 2017, 19, 1536–1551. [Google Scholar] [CrossRef] [PubMed]
- Smith-Brown, P.; Morrison, M.; Krause, L.; Davies, P.S.W. Male-Specific Association Between Fat-Free Mass Index and Fecal Microbiota in 2- to 3-Year-Old Australian Children. J. Pediatr. Gastroenterol. Nutr. 2018, 66, 147–151. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, R.; Zhang, Y.; Lin, X.; Yang, X. Gut Microbiota: Effect of Pubertal Status. BMC Microbiol. 2020, 20, 334. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Cuevas-Sierra, A.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Comprehensive Analysis Reveals Novel Interactions between Circulating MicroRNAs and Gut Microbiota Composition in Human Obesity. Int. J. Mol. Sci. 2020, 21, 9509. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and Associated Dietary Extremes Impact on Gut Microbial Diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Dekker Nitert, M.; Mousa, A.; Barrett, H.L.; Naderpoor, N.; de Courten, B. Altered Gut Microbiota Composition Is Associated with Back Pain in Overweight and Obese Individuals. Front. Endocrinol. 2020, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Henning, S.M.; Yang, J.; Woo, S.L.; Lee, R.-P.; Huang, J.; Rasmusen, A.; Carpenter, C.L.; Thames, G.; Gilbuena, I.; Tseng, C.-H.; et al. Hass Avocado Inclusion in a Weight-Loss Diet Supported Weight Loss and Altered Gut Microbiota: A 12-Week Randomized, Parallel-Controlled Trial. Curr. Dev. Nutr. 2019, 3, nzz068. [Google Scholar] [CrossRef] [PubMed]
- Hjorth, M.F.; Blædel, T.; Bendtsen, L.Q.; Lorenzen, J.K.; Holm, J.B.; Kiilerich, P.; Roager, H.M.; Kristiansen, K.; Larsen, L.H.; Astrup, A. Prevotella-to-Bacteroides Ratio Predicts Body Weight and Fat Loss Success on 24-Week Diets Varying in Macronutrient Composition and Dietary Fiber: Results from a Post-Hoc Analysis. Int. J. Obes. 2019, 43, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Osaki, T.; Oikawa, S. Use of T-RFLP and Seven Restriction Enzymes to Compare the Faecal Microbiota of Obese and Lean Japanese Healthy Men. Benef. Microbes 2015, 6, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, N.; Burke, L.; Vlahovich, N.; Charlesson, B.; O’Neill, H.; Ross, M.; Campbell, K.; Krause, L.; Morrison, M. The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers. Nutrients 2019, 11, 261. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut Microbiota Markers Associated with Obesity and Overweight in Italian Adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef]
- Resende, A.S.; Leite, G.S.F.; Lancha Junior, A.H. Changes in the Gut Bacteria Composition of Healthy Men with the Same Nutritional Profile Undergoing 10-Week Aerobic Exercise Training: A Randomized Controlled Trial. Nutrients 2021, 13, 2839. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, I.N.; Aljutaily, T.; Walton, G.; Huarte, E. Effects of Synbiotic Supplement on Human Gut Microbiota, Body Composition and Weight Loss in Obesity. Nutrients 2020, 12, 222. [Google Scholar] [CrossRef]
- Valeriani, F.; Gallè, F.; Cattaruzza, M.S.; Antinozzi, M.; Gianfranceschi, G.; Postiglione, N.; Romano Spica, V.; Liguori, G. Are Nutrition and Physical Activity Associated with Gut Microbiota? A Pilot Study on a Sample of Healthy Young Adults. Ann. Ig. 2020, 32, 521–527. [Google Scholar] [CrossRef]
- Whisner, C.M.; Maldonado, J.; Dente, B.; Krajmalnik-Brown, R.; Bruening, M. Diet, Physical Activity and Screen Time but Not Body Mass Index Are Associated with the Gut Microbiome of a Diverse Cohort of College Students Living in University Housing: A Cross-Sectional Study. BMC Microbiol. 2018, 18, 210. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, Y.; Wiklund, P.; Tan, X.; Wu, N.; Zhang, X.; Tikkanen, O.; Zhang, C.; Munukka, E.; Cheng, S. The Association between Cardiorespiratory Fitness and Gut Microbiota Composition in Premenopausal Women. Nutrients 2017, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Šoltys, K.; Lendvorský, L.; Hric, I.; Baranovičová, E.; Penesová, A.; Mikula, I.; Bohmer, M.; Budiš, J.; Vávrová, S.; Grones, J.; et al. Strenuous Physical Training, Physical Fitness, Body Composition and Bacteroides to Prevotella Ratio in the Gut of Elderly Athletes. Front. Physiol. 2021, 12, 670989. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Hoshi, C.; Kobori, M.; Takahashi, S.; Tomita, J.; Nishimura, M.; Nishihira, J. Quercetin Metabolism by Fecal Microbiota from Healthy Elderly Human Subjects. PLoS ONE 2017, 12, e0188271. [Google Scholar] [CrossRef] [PubMed]
- Tavella, T.; Rampelli, S.; Guidarelli, G.; Bazzocchi, A.; Gasperini, C.; Pujos-Guillot, E.; Comte, B.; Barone, M.; Biagi, E.; Candela, M.; et al. Elevated Gut Microbiome Abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae Is Associated with Reduced Visceral Adipose Tissue and Healthier Metabolic Profile in Italian Elderly. Gut Microbes 2021, 13, 1880221. [Google Scholar] [CrossRef] [PubMed]
- Kulecka, M.; Fraczek, B.; Mikula, M.; Zeber-Lubecka, N.; Karczmarski, J.; Paziewska, A.; Ambrozkiewicz, F.; Jagusztyn-Krynicka, K.; Cieszczyk, P.; Ostrowski, J. The Composition and Richness of the Gut Microbiota Differentiate the Top Polish Endurance Athletes from Sedentary Controls. Gut Microbes 2020, 11, 1374–1384. [Google Scholar] [CrossRef]
- La-ongkham, O.; Nakphaichit, M.; Nakayama, J.; Keawsompong, S.; Nitisinprasert, S. Age-Related Changes in the Gut Microbiota and the Core Gut Microbiome of Healthy Thai Humans. 3 Biotech 2020, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Pérez, A.; Hernández, M.; Iglesias, J.R.; Morán, J.; Pascual, J.; Porcar, M.; Vilanova, C.; Collado, L. The Spanish Gut Microbiome Reveals Links between Microorganisms and Mediterranean Diet. Sci. Rep. 2021, 11, 21602. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cuesta, M.C.; del Campo, R.; Garriga-García, M.; Peláez, C.; Requena, T. Taxonomic Characterization and Short-Chain Fatty Acids Production of the Obese Microbiota. Front. Cell. Infect. Microbiol. 2021, 11, 598093. [Google Scholar] [CrossRef]
- Oki, K.; Toyama, M.; Banno, T.; Chonan, O.; Benno, Y.; Watanabe, K. Comprehensive Analysis of the Fecal Microbiota of Healthy Japanese Adults Reveals a New Bacterial Lineage Associated with a Phenotype Characterized by a High Frequency of Bowel Movements and a Lean Body Type. BMC Microbiol. 2016, 16, 284. [Google Scholar] [CrossRef] [PubMed]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in Lean and Overweight Healthy Subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, T.; Nava, G.M.; Olvera-Ramírez, A.M.; Ronquillo, D.; Camacho, M.; Zavala, G.A.; Caamaño, M.C.; Acevedo-Whitehouse, K.; Rosado, J.L.; García, O.P. Gut Bacterial Families Are Associated with Body Composition and Metabolic Risk Markers in School-Aged Children in Rural Mexico. Child. Obes. 2020, 16, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, R.; George, G.; Kabeerdoss, J.; Hepsiba, J.; Chandragunasekaran, A.M.S.; Ramakrishna, B.S. Quantitative Differences in Intestinal Faecalibacterium prausnitzii in Obese Indian Children. Br. J. Nutr. 2010, 103, 335–338. [Google Scholar] [CrossRef]
- Karlsson, C.L.J.; Önnerfält, J.; Xu, J.; Molin, G.; Ahrné, S.; Thorngren-Jerneck, K. The Microbiota of the Gut in Preschool Children With Normal and Excessive Body Weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Miranda, V.P.N.; dos Santos Amorim, P.R.; Bastos, R.R.; de Faria, E.R.; de Castro Moreira, M.E.; do Carmo Castro Franceschini, S.; do Carmo Gouveia Peluzio, M.; de Luces Fortes Ferreira, C.L.; Priore, S.E. Abundance of Gut Microbiota, Concentration of Short-Chain Fatty Acids, and Inflammatory Markers Associated with Elevated Body Fat, Overweight, and Obesity in Female Adolescents. Mediat. Inflamm. 2019, 2019, 7346863. [Google Scholar] [CrossRef]
- Nagata, S.; Chiba, Y.; Wang, C.; Yamashiro, Y. The Effects of the Lactobacillus casei Strain on Obesity in Children: A Pilot Study. Benef. Microbes 2017, 8, 535–543. [Google Scholar] [CrossRef]
- Xu, P.; Li, M.; Zhang, J.; Zhang, T. Correlation of Intestinal Microbiota with Overweight and Obesity in Kazakh School Children. BMC Microbiol. 2012, 12, 283. [Google Scholar] [CrossRef]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sport. Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.C.; Kim, H.; Fang, C.; Bennett, W.; Nemec, M.; Sirven, M.A.; Suchodolski, J.S.; Deutz, N.; Britton, R.A.; Mertens-Talcott, S.U.; et al. Body Mass Index as a Determinant of Systemic Exposure to Gallotannin Metabolites during 6-Week Consumption of Mango (Mangifera indica L.) and Modulation of Intestinal Microbiota in Lean and Obese Individuals. Mol. Nutr. Food Res. 2019, 63, 1800512. [Google Scholar] [CrossRef] [PubMed]
- Basciani, S.; Camajani, E.; Contini, S.; Persichetti, A.; Risi, R.; Bertoldi, L.; Strigari, L.; Prossomariti, G.; Watanabe, M.; Mariani, S.; et al. Very-Low-Calorie Ketogenic Diets With Whey, Vegetable, or Animal Protein in Patients with Obesity: A Randomized Pilot Study. J. Clin. Endocrinol. Metab. 2020, 105, 2939–2949. [Google Scholar] [CrossRef] [PubMed]
- Bielik, V.; Hric, I.; Baláž, V.; Penesová, A.; Vávrová, S.; Grones, J.; Bokor, B.; Budiš, J.; Bohmer, M.; Minárik, G.; et al. Gut Microbiota Diversity in Lean Athletes Is Associated with Positive Energy Balance. Ann. Nutr. Metab. 2020, 76, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Durk, R.P.; Castillo, E.; Márquez-Magaña, L.; Grosicki, G.J.; Bolter, N.D.; Lee, C.M.; Bagley, J.R. Gut Microbiota Composition Is Related to Cardiorespiratory Fitness in Healthy Young Adults. Int. J. Sport Nutr. Exerc. Metab. 2019, 29, 249–253. [Google Scholar] [CrossRef]
- Teixeira, T.F.S.; Grześkowiak, Ł.M.; Salminen, S.; Laitinen, K.; Bressan, J.; do Carmo Gouveia Peluzio, M. Faecal Levels of Bifidobacterium and Clostridium Coccoides but Not Plasma Lipopolysaccharide Are Inversely Related to Insulin and HOMA Index in Women. Clin. Nutr. 2013, 32, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.; Su, W.; Rahat-Rozenbloom, S.; Wolever, T.M.S.; Comelli, E.M. Adiposity, Gut Microbiota and Faecal Short Chain Fatty Acids Are Linked in Adult Humans. Nutr. Diabetes 2014, 4, e121. [Google Scholar] [CrossRef]
- Most, J.; Penders, J.; Lucchesi, M.; Goossens, G.H.; Blaak, E.E. Gut Microbiota Composition in Relation to the Metabolic Response to 12-Week Combined Polyphenol Supplementation in Overweight Men and Women. Eur. J. Clin. Nutr. 2017, 71, 1040–1045. [Google Scholar] [CrossRef]
- Goffredo, M.; Mass, K.; Parks, E.J.; Wagner, D.A.; McClure, E.A.; Graf, J.; Savoye, M.; Pierpont, B.; Cline, G.; Santoro, N. Role of Gut Microbiota and Short Chain Fatty Acids in Modulating Energy Harvest and Fat Partitioning in Youth. J. Clin. Endocrinol. Metab. 2016, 101, 4367–4376. [Google Scholar] [CrossRef]
- Bezek, K.; Petelin, A.; Pražnikar, J.; Nova, E.; Redondo, N.; Marcos, A.; Jenko Pražnikar, Z. Obesity Measures and Dietary Parameters as Predictors of Gut Microbiota Phyla in Healthy Individuals. Nutrients 2020, 12, 2695. [Google Scholar] [CrossRef]
- Borgo, F.; Garbossa, S.; Riva, A.; Severgnini, M.; Luigiano, C.; Benetti, A.; Pontiroli, A.E.; Morace, G.; Borghi, E. Body Mass Index and Sex Affect Diverse Microbial Niches within the Gut. Front. Microbiol. 2018, 9, 213. [Google Scholar] [CrossRef]
- Brignardello, J.; Morales, P.; Diaz, E.; Romero, J.; Brunser, O.; Gotteland, M. Pilot Study: Alterations of Intestinal Microbiota in Obese Humans Are Not Associated with Colonic Inflammation or Disturbances of Barrier Function. Aliment. Pharmacol. Ther. 2010, 32, 1307–1314. [Google Scholar] [CrossRef]
- Janssens, P.L.H.R.; Penders, J.; Hursel, R.; Budding, A.E.; Savelkoul, P.H.M.; Westerterp-Plantenga, M.S. Long-Term Green Tea Supplementation Does Not Change the Human Gut Microbiota. PLoS ONE 2016, 11, e0153134. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Yokoyama, H.; Imai, D.; Takeda, R.; Ota, A.; Kawai, E.; Hisada, T.; Emoto, M.; Suzuki, Y.; Okazaki, K. Aerobic Exercise Training with Brisk Walking Increases Intestinal Bacteroides in Healthy Elderly Women. Nutrients 2019, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Leong, C.; Haszard, J.J.; Heath, A.-L.M.; Tannock, G.W.; Lawley, B.; Cameron, S.L.; Szymlek-Gay, E.A.; Gray, A.R.; Taylor, B.J.; Galland, B.C.; et al. Using Compositional Principal Component Analysis to Describe Children’s Gut Microbiota in Relation to Diet and Body Composition. Am. J. Clin. Nutr. 2020, 111, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Joller, P.; Cabaset, S.; Maurer, S. Influence of a Food Supplement on the Gut Microbiome in Healthy Overweight Women. Funct. Foods Health Dis. 2020, 10, 428. [Google Scholar] [CrossRef]
- Koliada, A.; Moseiko, V.; Romanenko, M.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Vaiserman, A. Sex Differences in the Phylum-level Human Gut Microbiota Composition. BMC Microbiol. 2021, 21, 131. [Google Scholar] [CrossRef] [PubMed]
- Bloemendaal, M.; Szopinska-Tokov, J.; Belzer, C.; Boverhoff, D.; Papalini, S.; Michels, F.; van Hemert, S.; Arias Vasquez, A.; Aarts, E. Probiotics-Induced Changes in Gut Microbial Composition and Its Effects on Cognitive Performance after Stress: Exploratory Analyses. Transl. Psychiatry 2021, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Gallè, F.; Valeriani, F.; Cattaruzza, M.S.; Gianfranceschi, G.; Liguori, R.; Antinozzi, M.; Mederer, B.; Liguori, G.; Romano Spica, V. Mediterranean Diet, Physical Activity and Gut Microbiome Composition: A Cross-Sectional Study among Healthy Young Italian Adults. Nutrients 2020, 12, 2164. [Google Scholar] [CrossRef]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the Gut Microbiota Composition between Obese and Non-Obese Individuals in a Japanese Population, as Analyzed by Terminal Restriction Fragment Length Polymorphism and next-Generation Sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Zuo, H.-J. Gut Bacteria Alteration in Obese People and Its Relationship with Gene Polymorphism. World J. Gastroenterol. 2011, 17, 1076. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Kassam, Z.; Mullish, B.H.; Chiang, A.; Carrellas, M.; Hurtado, J.; Marchesi, J.R.; McDonald, J.A.K.; Pechlivanis, A.; Barker, G.F.; et al. Effects of Fecal Microbiota Transplantation with Oral Capsules in Obese Patients. Clin. Gastroenterol. Hepatol. 2020, 18, 855–863.e2. [Google Scholar] [CrossRef]
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J. Diabetes Res. 2018, 2018, 3462092. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-Analyses of Human Gut Microbes Associated with Obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; Debojyoti, D. Meta-Analysis Reveals Obesity Associated Gut Microbial Alteration Patterns and Reproducible Contributors of Functional Shift. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio 2016, 7, e01018-16. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Ortiz-Alvarez, L.; Xu, H.; Martinez-Tellez, B. Influence of Exercise on the Human Gut Microbiota of Healthy Adults: A Systematic Review. Clin. Transl. Gastroenterol. 2020, 11, e00126. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, X.; Tian, X.Y.; Ma, A.C.; Wong, S.H. Does the Gut Microbiota Contribute to the Antiobesity Effect of Exercise? A Systematic Review and Meta-analysis. Obesity 2022, 30, 407–423. [Google Scholar] [CrossRef] [PubMed]
- Castaner, O.; Goday, A.; Park, Y.-M.; Lee, S.-H.; Magkos, F.; Shiow, S.-A.T.E.; Schröder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.; Gasbarrini, A.; Mele, M. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity Alters Gut Microbial Ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-Chain Fatty Acids in Control of Body Weight and Insulin Sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Ibrahim, M.; Anishetty, S. A Meta-Metabolome Network of Carbohydrate Metabolism: Interactions between Gut Microbiota and Host. Biochem. Biophys. Res. Commun. 2012, 428, 278–284. [Google Scholar] [CrossRef]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.J.; Soininen, P.; Wang, Z.; Ala-Korpela, M.; Hazen, S.L.; et al. Relationships between Gut Microbiota, Plasma Metabolites, and Metabolic Syndrome Traits in the METSIM Cohort. Genome Biol. 2017, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Rahat-Rozenbloom, S.; Fernandes, J.; Gloor, G.B.; Wolever, T.M.S. Evidence for Greater Production of Colonic Short-Chain Fatty Acids in Overweight than Lean Humans. Int. J. Obes. 2014, 38, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-Analysis of Gut Microbiome Studies Identifies Disease-Specific and Shared Responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef] [PubMed]
- Pinart, M.; Dötsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut Microbiome Composition in Obese and Non-Obese Persons: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef]
- Drissi, F.; Merhej, V.; Angelakis, E.; El Kaoutari, A.; Carrière, F.; Henrissat, B.; Raoult, D. Comparative Genomics Analysis of Lactobacillus Species Associated with Weight Gain or Weight Protection. Nutr. Diabetes 2014, 4, e109. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium Prausnitzii: From Microbiology to Diagnostics and Prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Belzer, C.; de Vos, W.M. Akkermansia Muciniphila and Its Role in Regulating Host Functions. Microb. Pathog. 2017, 106, 171–181. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).