Bacteroides salyersiae Is a Candidate Probiotic Species with Potential Anti-Colitis Properties in the Human Colon: First Evidence from an In Vivo Mouse Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animal Experiment
2.3. 16S rRNA Gene Amplicon High-Throughput Sequencing and Bioinformatics Analysis
2.4. Metabolome Analysis
2.5. Statistical Analyses
3. Results
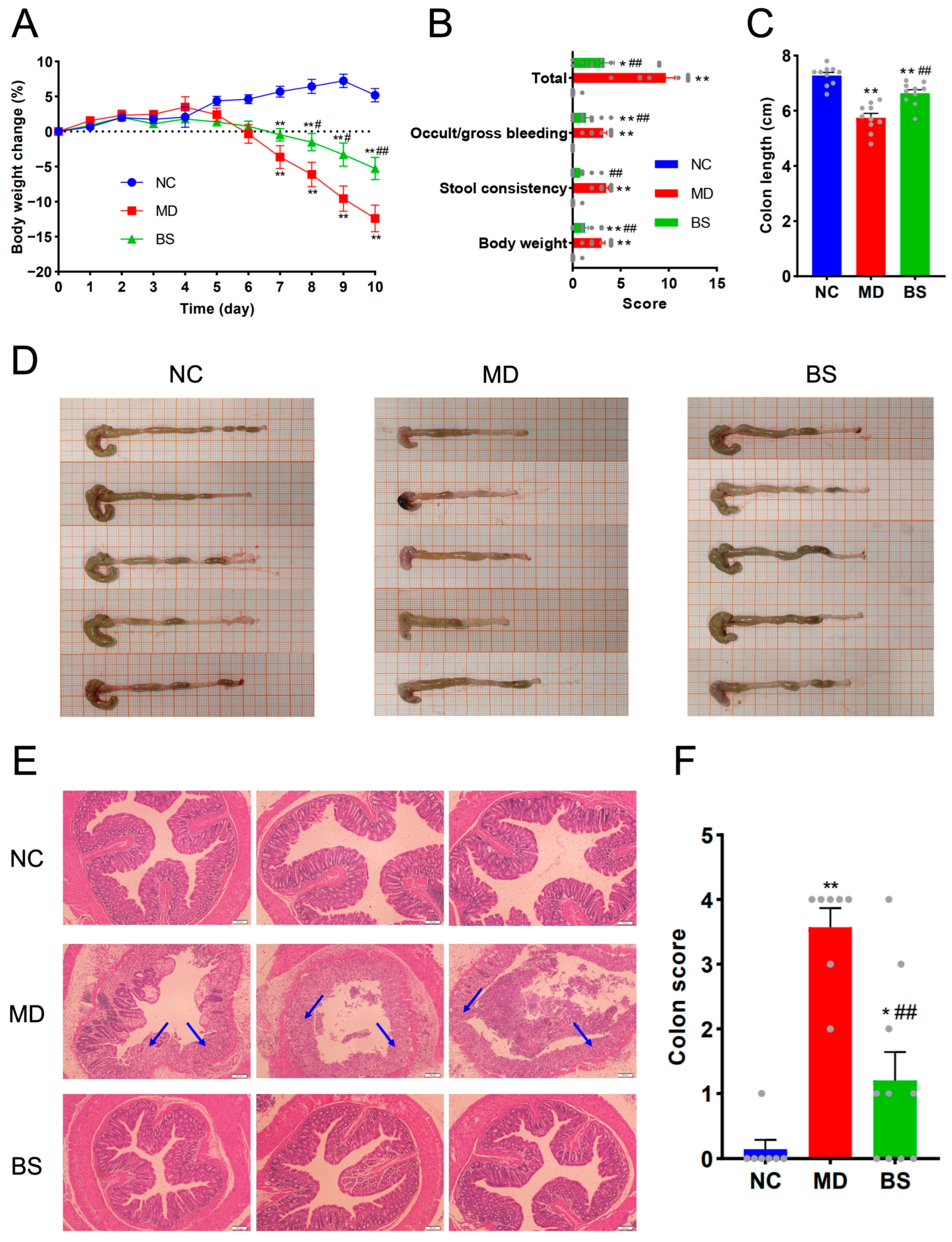
3.1. Oral Intake of B. salyersiae CSP6 Protected against DSS-Induced Colitis in C57BL/6J Mice
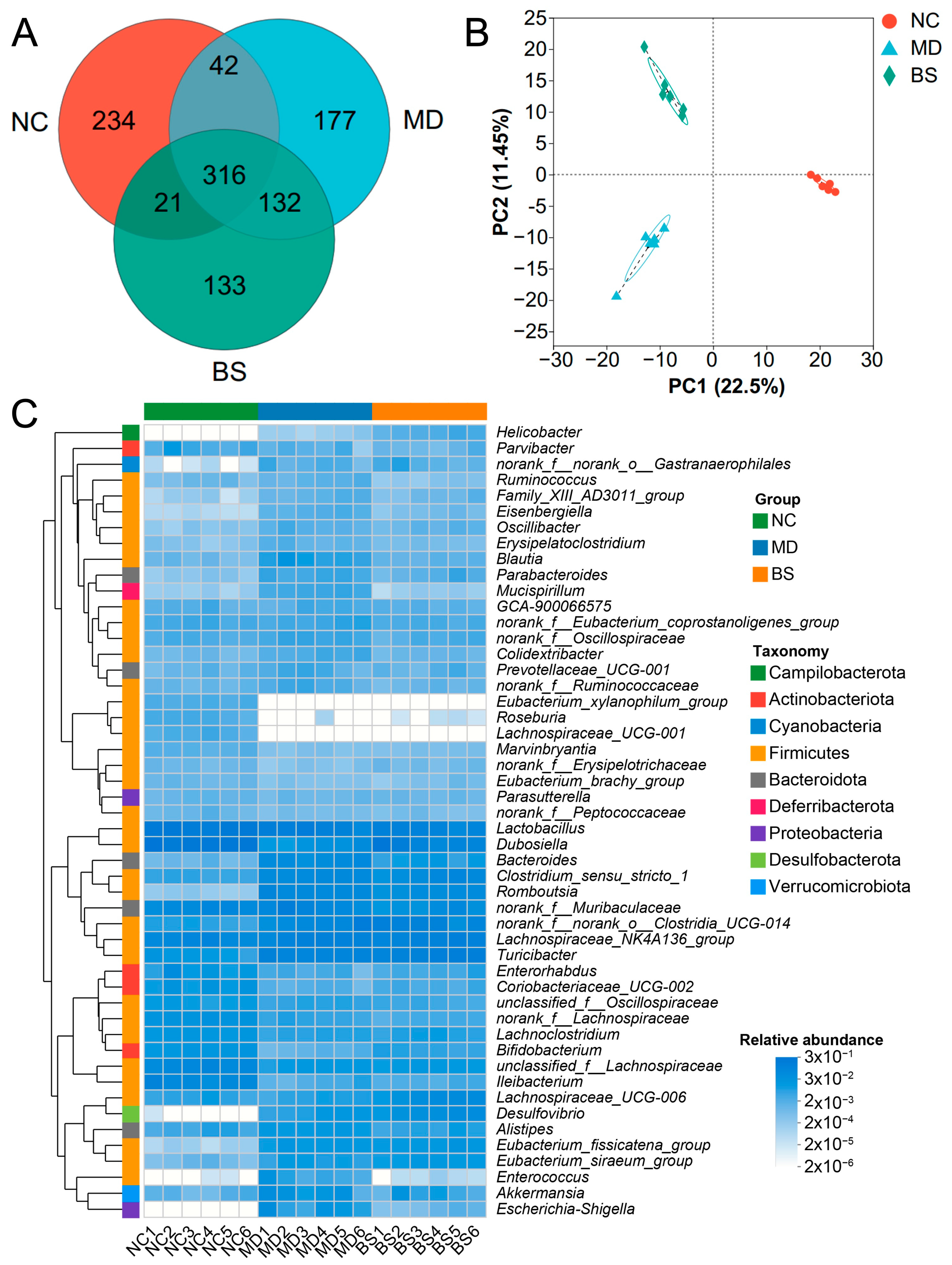
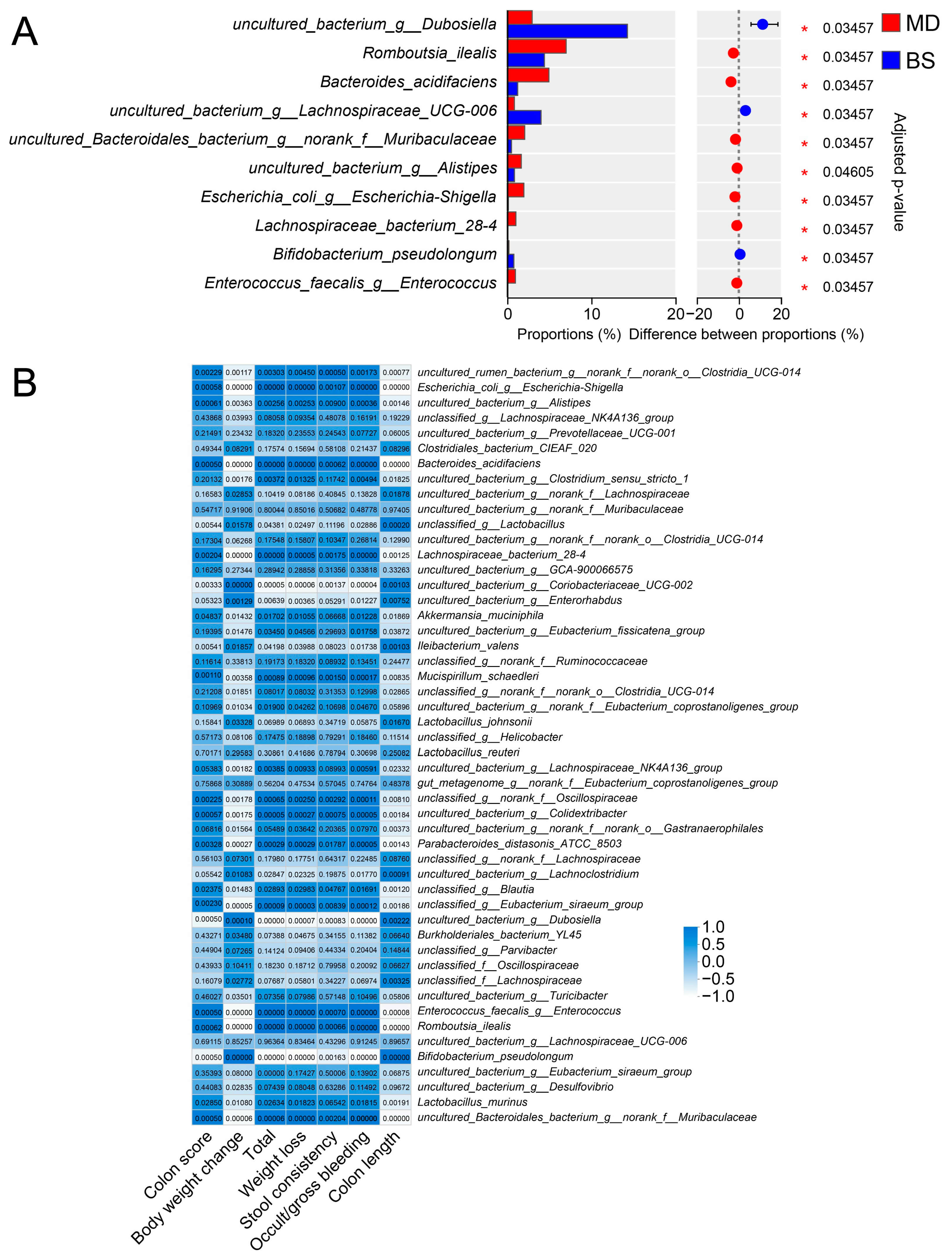
3.2. B. salyersiae CSP6 Changed the Structure of the Intestinal Microbiota and Attenuated Gut Dysbiosis in DSS-Fed Mice
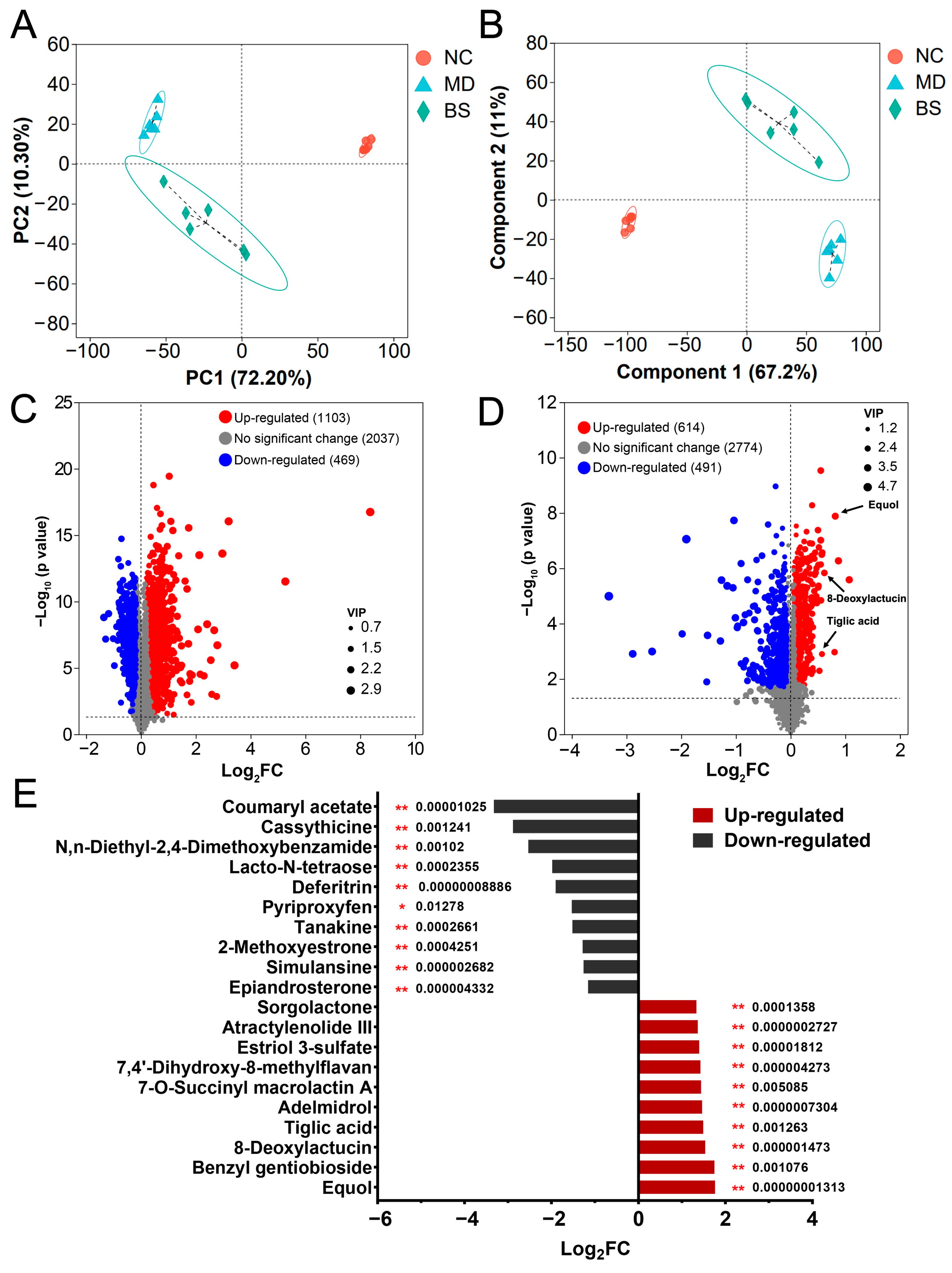
3.3. B. salyersiae CSP6 Modulated the Composition of Intestinal Metabolites in UC Mice and Increased the Fecal Concentrations of Anti-Inflammatory Equol, 8-Deoxylactucin, and Tiglic Acid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Le Berre, C.; Honap, S.; Peyrin-Biroulet, L. Ulcerative colitis. Lancet 2023, 402, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.F. Ulcerative colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef] [PubMed]
- Gros, B.; Kaplan, G.G. Ulcerative colitis in adults: A review. JAMA 2023, 330, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Ekstedt, N.; Jamioł-Milc, D.; Pieczyńska, J. Importance of gut microbiota in patients with inflammatory bowel disease. Nutrients 2024, 16, 2092. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Baumgartner, M.; Zirnbauer, R.; Schlager, S.; Mertens, D.; Gasche, N.; Sladek, B.; Herbold, C.; Bochkareva, O.; Emelianenko, V.; Vogelsang, H.; et al. Atypical enteropathogenic E. coli are associated with disease activity in ulcerative colitis. Gut Microbes 2022, 14, 2143218. [Google Scholar] [CrossRef]
- Mills, R.H.; Dulai, P.S.; Vázquez-Baeza, Y.; Sauceda, C.; Daniel, N.; Gerner, R.R.; Batachari, L.E.; Malfavon, M.; Zhu, Q.; Weldon, K.; et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 2022, 7, 262–276. [Google Scholar] [CrossRef]
- Kudo, T.; Aoyagi, Y.; Fujii, T.; Ohtsuka, Y.; Nagata, S.; Shimizu, T. Development of Candida albicans colitis in a child undergoing steroid therapy for ulcerative colitis. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 96–99. [Google Scholar] [CrossRef]
- Martín, R.; Chain, F.; Miquel, S.; Lu, J.; Gratadoux, J.J.; Sokol, H.; Verdu, E.F.; Bercik, P.; Bermúdez-Humarán, L.G.; Langella, P. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm. Bowel Dis. 2014, 20, 417–430. [Google Scholar] [CrossRef]
- Guo, W.; Mao, B.; Cui, S.; Tang, X.; Zhang, Q.; Zhao, J.; Zhang, H. Protective effects of a novel probiotic Bifidobacterium pseudolongum on the intestinal barrier of colitis mice via modulating the Pparγ/STAT3 pathway and intestinal microbiota. Foods 2022, 11, 1551. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.-M.; Guo, H.-X.; Cai, J.-W.; Meng, X.-C. Bifidobacterium breve alleviates DSS-induced colitis in mice by maintaining the mucosal and epithelial barriers and modulating gut microbes. Nutrients 2022, 14, 3671. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.; Williams, M.A.; O’Callaghan, A.A.; Dempsey, E.; Cabrera-Rubio, R.; Raverdeau, M.; Crispie, F.; Cotter, P.D.; Corr, S.C. Lactobacillus salivarius UCC118™ dampens inflammation and promotes microbiota recovery to provide therapeutic benefit in a DSS-induced colitis model. Microorganisms 2022, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Ahl, D.; Liu, H.; Schreiber, O.; Roos, S.; Phillipson, M.; Holm, L. Lactobacillus reuteri increases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol. 2016, 217, 300–310. [Google Scholar] [CrossRef]
- Delday, M.; Mulder, I.; Logan, E.T.; Grant, G. Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of Crohn’s disease. Inflamm. Bowel Dis. 2019, 25, 85–96. [Google Scholar] [CrossRef]
- Ihekweazu, F.D.; Fofanova, T.Y.; Queliza, K.; Nagy-Szakal, D.; Stewart, C.J.; Engevik, M.A.; Hulten, K.G.; Tatevian, N.; Graham, D.Y.; Versalovic, J.; et al. Bacteroides ovatus ATCC 8483 monotherapy is superior to traditional fecal transplant and multi-strain bacteriotherapy in a murine colitis model. Gut Microbes 2019, 10, 504–520. [Google Scholar] [CrossRef]
- Fu, T.; Wang, Y.; Ma, M.; Dai, W.; Pan, L.; Shang, Q.; Yu, G. Isolation of alginate-degrading bacteria from the human gut microbiota and discovery of Bacteroides xylanisolvens AY11-1 as a novel anti-colitis probiotic bacterium. Nutrients 2023, 15, 1352. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, J.; Chen, L.; Yu, J.; Zhang, J.; Yin, H.; Shang, Q.; Yu, G. Discovery of Bacteroides uniformis F18-22 as a safe and novel probiotic bacterium for the treatment of ulcerative colitis from the healthy human colon. Int. J. Mol. Sci. 2023, 24, 14669. [Google Scholar] [CrossRef]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In vitro characterization of gut microbiota-derived commensal strains: Selection of Parabacteroides distasonis strains alleviating TNBS-induced colitis in mice. Cells 2020, 9, 2104. [Google Scholar] [CrossRef]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Barua, N.; Ip, M. Mucin-degrading gut commensals isolated from healthy faecal donor suppress intestinal epithelial inflammation and regulate tight junction barrier function. Front. Immunol. 2022, 13, 1021094. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, M.; Dai, W.; Shang, Q.; Yu, G. Bacteroides salyersiae is a potent chondroitin sulfate-degrading species in the human gut microbiota. Microbiome 2024, 12, 41. [Google Scholar] [CrossRef]
- Yang, C.; Merlin, D. Unveiling colitis: A journey through the dextran sodium sulfate-induced model. Inflamm. Bowel Dis. 2024, 30, 844–853. [Google Scholar] [CrossRef]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, S.; Neufert, C.; Weigmann, B.; Neurath, M.F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2007, 2, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.; Corr, S.C. Gut microbial metabolite-mediated regulation of the intestinal barrier in the pathogenesis of inflammatory bowel disease. Nutrients 2021, 13, 4259. [Google Scholar] [CrossRef]
- Cai, J.; Sun, L.L.; Gonzalez, F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef]
- Márquez-Flores, Y.K.; Martínez-Galero, E.; Correa-Basurto, J.; Sixto-López, Y.; Villegas, I.; Rosillo, M.Á.; Cárdeno, A.; Alarcón-de-la-Lastra, C. Daidzein and equol: Ex vivo and in silico approaches targeting COX-2, iNOS, and the canonical inflammasome signaling pathway. Pharmaceuticals 2024, 17, 647. [Google Scholar] [CrossRef]
- Sekikawa, A.; Wharton, W.; Butts, B.; Veliky, C.V.; Garfein, J.; Li, J.; Goon, S.; Fort, A.; Li, M.; Hughes, T.M. Potential protective mechanisms of s-equol, a metabolite of soy isoflavone by the gut microbiome, on cognitive decline and dementia. Int. J. Mol. Sci. 2022, 23, 11921. [Google Scholar] [CrossRef]
- Cankar, K.; Hakkert, J.C.; Sevenier, R.; Papastolopoulou, C.; Schipper, B.; Baixinho, J.P.; Fernández, N.; Matos, M.S.; Serra, A.T.; Santos, C.N.; et al. Lactucin synthase inactivation boosts the accumulation of anti-inflammatory 8-deoxylactucin and its derivatives in chicory (Cichorium intybus L.). J. Agric. Food Chem. 2023, 71, 6061–6072. [Google Scholar] [CrossRef] [PubMed]
- Baixinho, J.P.; Anastácio, J.D.; Ivasiv, V.; Cankar, K.; Bosch, D.; Menezes, R.; de Roode, M.; dos Santos, C.N.; Matias, A.A.; Fernández, N. Supercritical CO2 extraction as a tool to isolate anti-inflammatory sesquiterpene lactones from Cichorium intybus L. roots. Molecules 2021, 26, 2583. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Ma, S.G.; Lin, M.B.; Hou, Q.; Ma, M.; Yu, S.S. Hydroxylated ethacrylic and tiglic acid derivatives from the stems and branches of Enkianthus chinensis and their potential anti-inflammatory activities. J. Nat. Prod. 2020, 83, 2867–2876. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 2024, 22, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.O.; Ridgway, K.; Scovell, L.; Kemsley, E.K.; Lund, E.K.; Jamieson, C.; Johnson, I.T.; Narbad, A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010, 10, 134. [Google Scholar] [CrossRef]
- Nomura, K.; Ishikawa, D.; Okahara, K.; Ito, S.; Haga, K.; Takahashi, M.; Arakawa, A.; Shibuya, T.; Osada, T.; Kuwahara-Arai, K.; et al. Bacteroidetes species are correlated with disease activity in ulcerative colitis. J. Clin. Med. 2021, 10, 1749. [Google Scholar] [CrossRef]
- Tan, H.; Zhai, Q.; Chen, W. Investigations of Bacteroides spp. towards next-generation probiotics. Food Res. Int. 2019, 116, 637. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Fang, Z.; Ma, M.; Wang, Y.; Dai, W.; Shang, Q.; Yu, G. Degradation and fermentation of hyaluronic acid by Bacteroides spp. from the human gut microbiota. Carbohydr. Polym. 2024, 334, 122074. [Google Scholar] [CrossRef]
- La Rosa, S.L.; Ostrowski, M.P.; Vera-Ponce de León, A.; McKee, L.S.; Larsbrink, J.; Eijsink, V.G.; Lowe, E.C.; Martens, E.C.; Pope, P.B. Glycan processing in gut microbiomes. Curr. Opin. Microbiol. 2022, 67, 102143. [Google Scholar] [CrossRef]
- McKee, L.S.; La Rosa, S.L.; Westereng, B.; Eijsink, V.G.; Pope, P.B.; Larsbrink, J. Polysaccharide degradation by the Bacteroidetes: Mechanisms and nomenclature. Environ. Microbiol. Rep. 2021, 13, 559–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tu, S.; Ji, X.; Wu, J.; Meng, J.; Gao, J.; Shao, X.; Shi, S.; Wang, G.; Qiu, J.; et al. Dubosiella newyorkensis modulates immune tolerance in colitis via the L-lysine-activated AhR-IDO1-Kyn pathway. Nat. Commun. 2024, 15, 1333. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.H.; Wang, J.; Zhang, C.Y.; Zhao, L.; Sheng, Y.Y.; Tao, G.S.; Xue, Y.Z. Gut microbial characteristical comparison reveals potential anti-aging function of Dubosiella newyorkensis in mice. Front. Endocrinol. 2023, 14, 1133167. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Han, X.; Sun, L.; Liu, X.; Zhang, W.; Hao, J. Indole-3-acetic acid alleviates DSS-induced colitis by promoting the production of R-equol from Bifidobacterium pseudolongum. Gut Microbes 2024, 16, 2329147. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, W.; Lv, Y.; Quan, M.; Ma, M.; Shang, Q.; Yu, G. Bacteroides salyersiae Is a Candidate Probiotic Species with Potential Anti-Colitis Properties in the Human Colon: First Evidence from an In Vivo Mouse Model. Nutrients 2024, 16, 2918. https://doi.org/10.3390/nu16172918
Dai W, Lv Y, Quan M, Ma M, Shang Q, Yu G. Bacteroides salyersiae Is a Candidate Probiotic Species with Potential Anti-Colitis Properties in the Human Colon: First Evidence from an In Vivo Mouse Model. Nutrients. 2024; 16(17):2918. https://doi.org/10.3390/nu16172918
Chicago/Turabian StyleDai, Wei, Youjing Lv, Min Quan, Mingfeng Ma, Qingsen Shang, and Guangli Yu. 2024. "Bacteroides salyersiae Is a Candidate Probiotic Species with Potential Anti-Colitis Properties in the Human Colon: First Evidence from an In Vivo Mouse Model" Nutrients 16, no. 17: 2918. https://doi.org/10.3390/nu16172918
APA StyleDai, W., Lv, Y., Quan, M., Ma, M., Shang, Q., & Yu, G. (2024). Bacteroides salyersiae Is a Candidate Probiotic Species with Potential Anti-Colitis Properties in the Human Colon: First Evidence from an In Vivo Mouse Model. Nutrients, 16(17), 2918. https://doi.org/10.3390/nu16172918







