Novel Functional Food Properties of Forest Onion (Eleutherine bulbosa Merr.) Phytochemicals for Treating Metabolic Syndrome: New Insights from a Combined Computational and In Vitro Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Extraction of Forest Onion
2.2. Identification of Metabolite–Peptide Profile via Untargeted Metabolomic Profiling
2.3. In Silico Study Assessment
2.3.1. Prediction of Bioactive Compound Activities, Toxicity Analysis, and Drug-Likeness
2.3.2. Target Protein Identification and Analysis
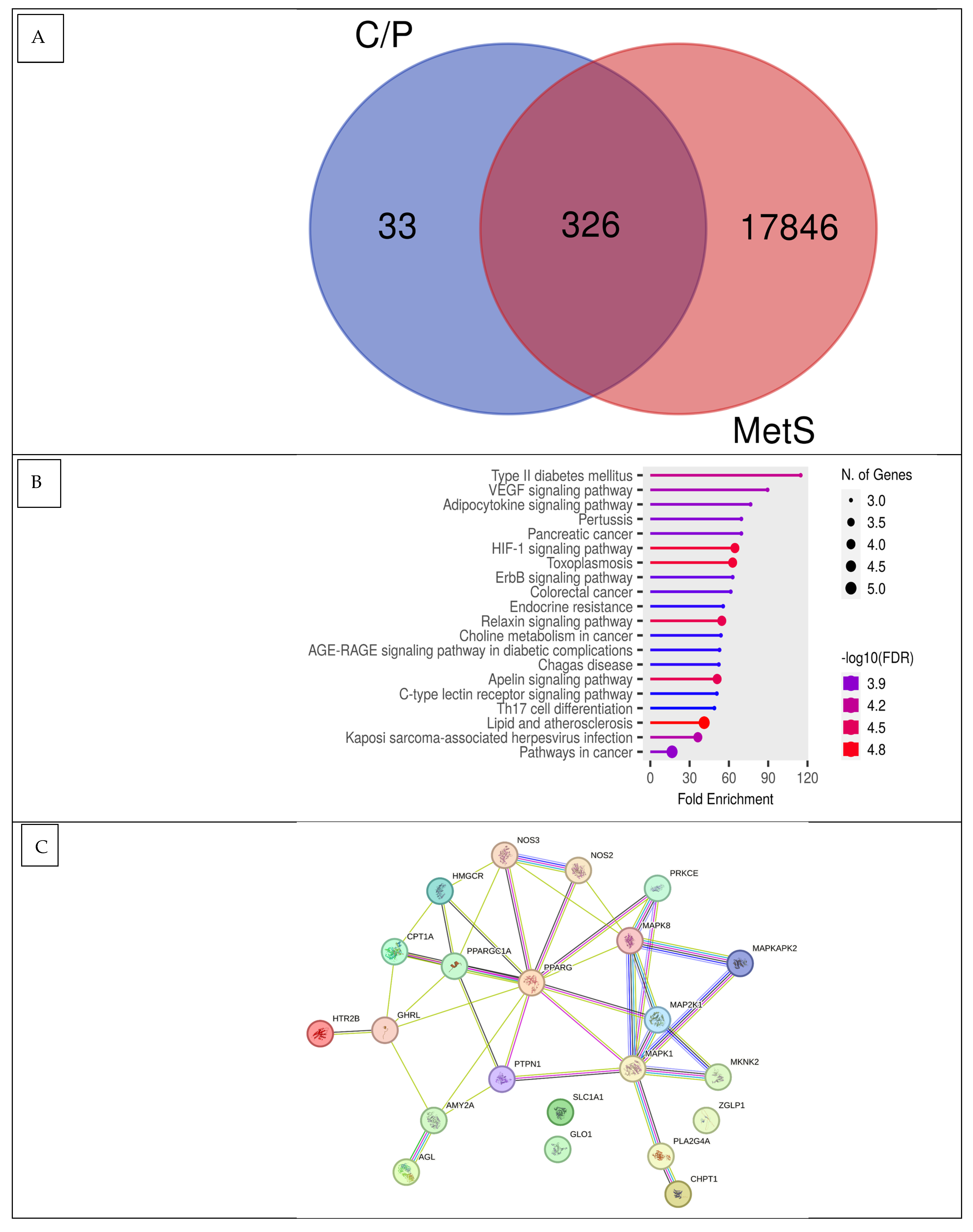
2.3.3. Network Pharmacology Analysis
2.3.4. Molecular Docking Simulation
2.4. In Vitro Study Assessments
2.4.1. Antiobesity Assessment via Pancreatic Lipase Inhibition
2.4.2. Antidiabetic Assessment via α-Glucosidase and α-Amylase Inhibition
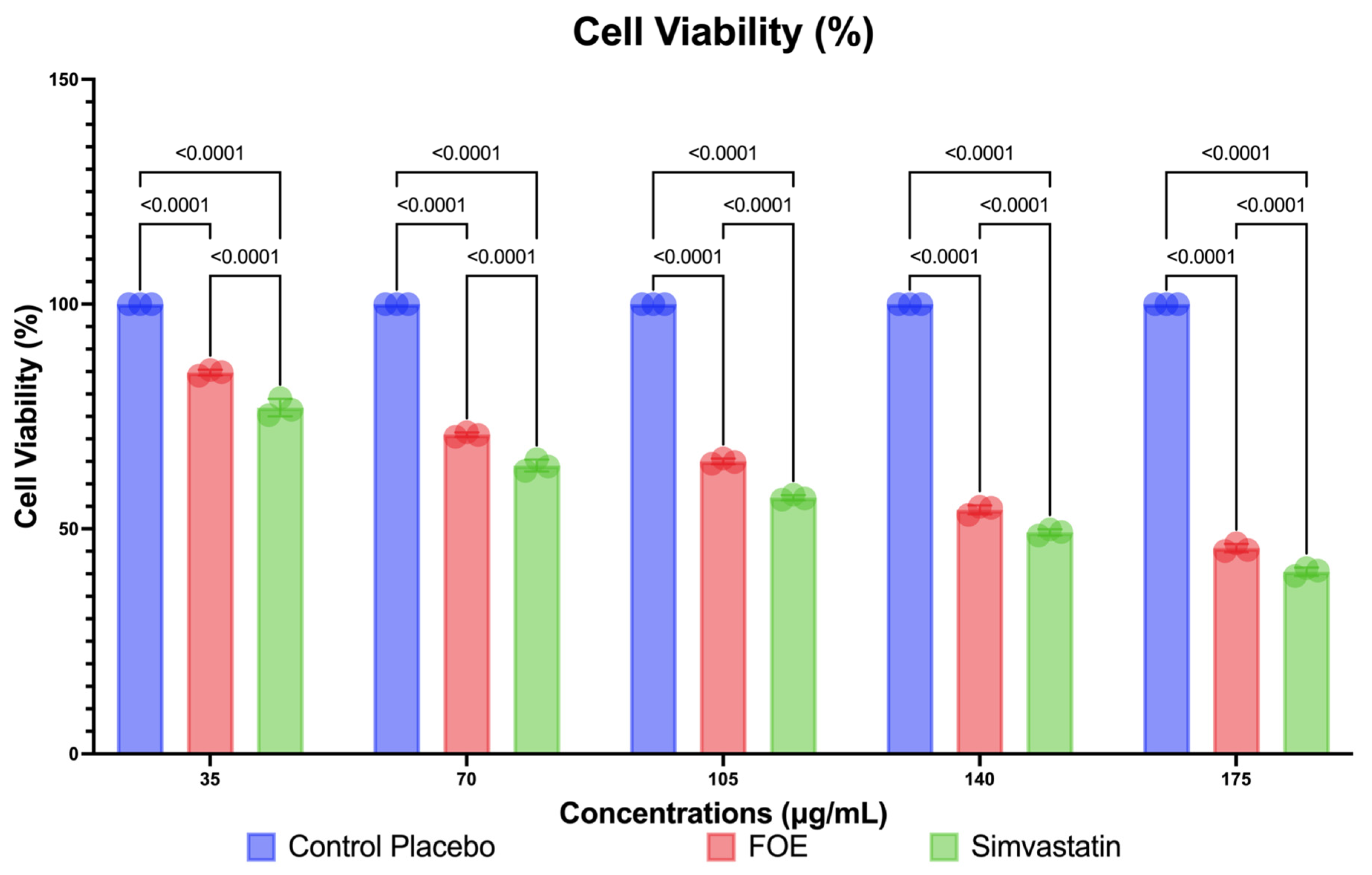
2.4.3. Cell Culture and Cell Viability of 3T3-L1 Mouse Cells
In Vitro Assessment of MAPK8, PPARG, HMGCR, CPT-1, and GLP1 Expression on Preadipocyte 3T3-L1 Mouse Cells
2.5. Data Analytics and Management
3. Results
3.1. In Silico Study Results
3.1.1. List of Compounds and Peptides after Metabolomic Profiling
3.1.2. Pa Score, Toxicity Prediction, Drug-Likeness, and Network Pharmacology Analysis
3.1.3. Docking Potency of Compound Found in Eleutherine bulbosa
3.2. In Vitro Study Results
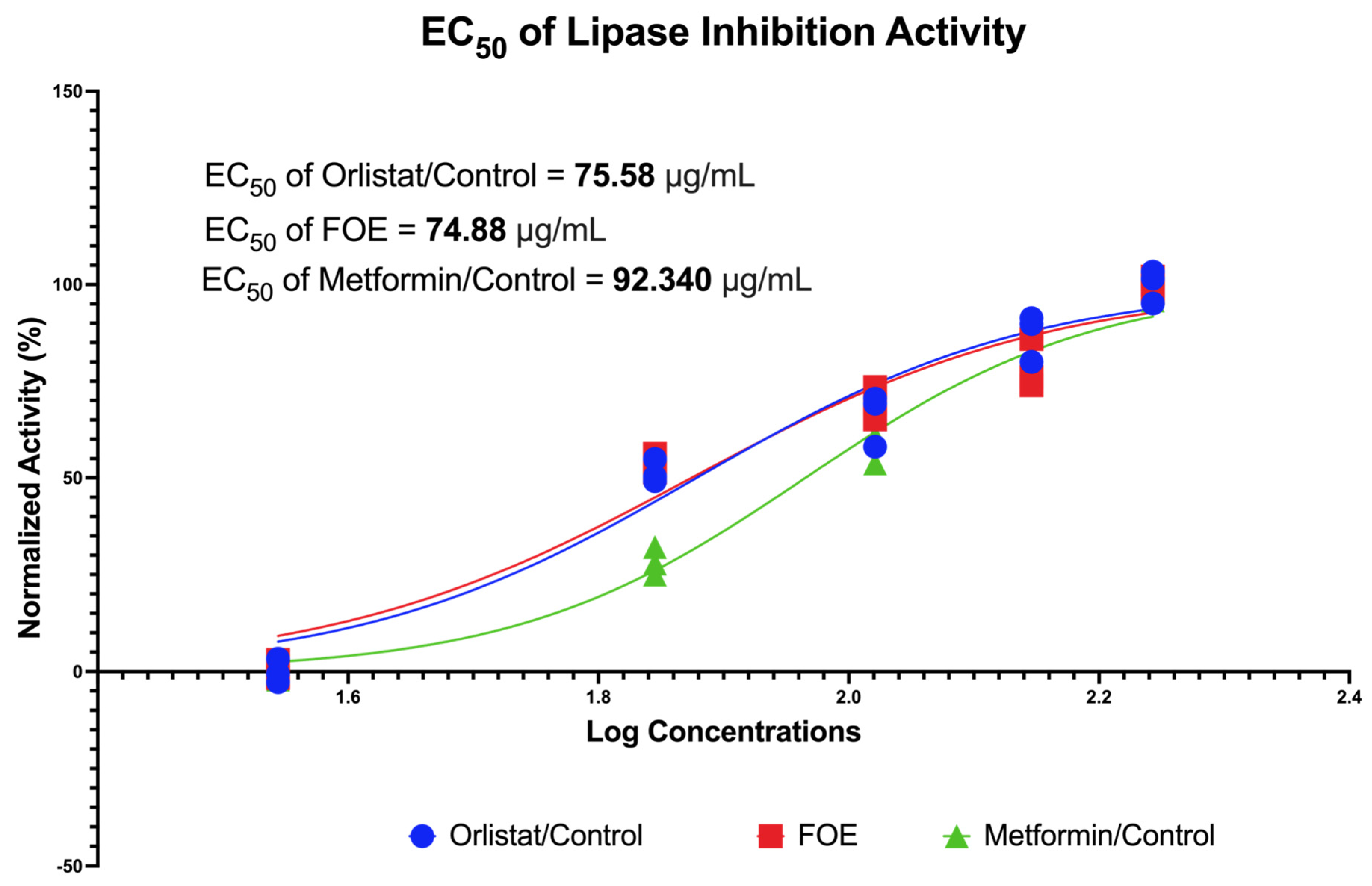
3.2.1. Lipase Inhibition Potential of FOE
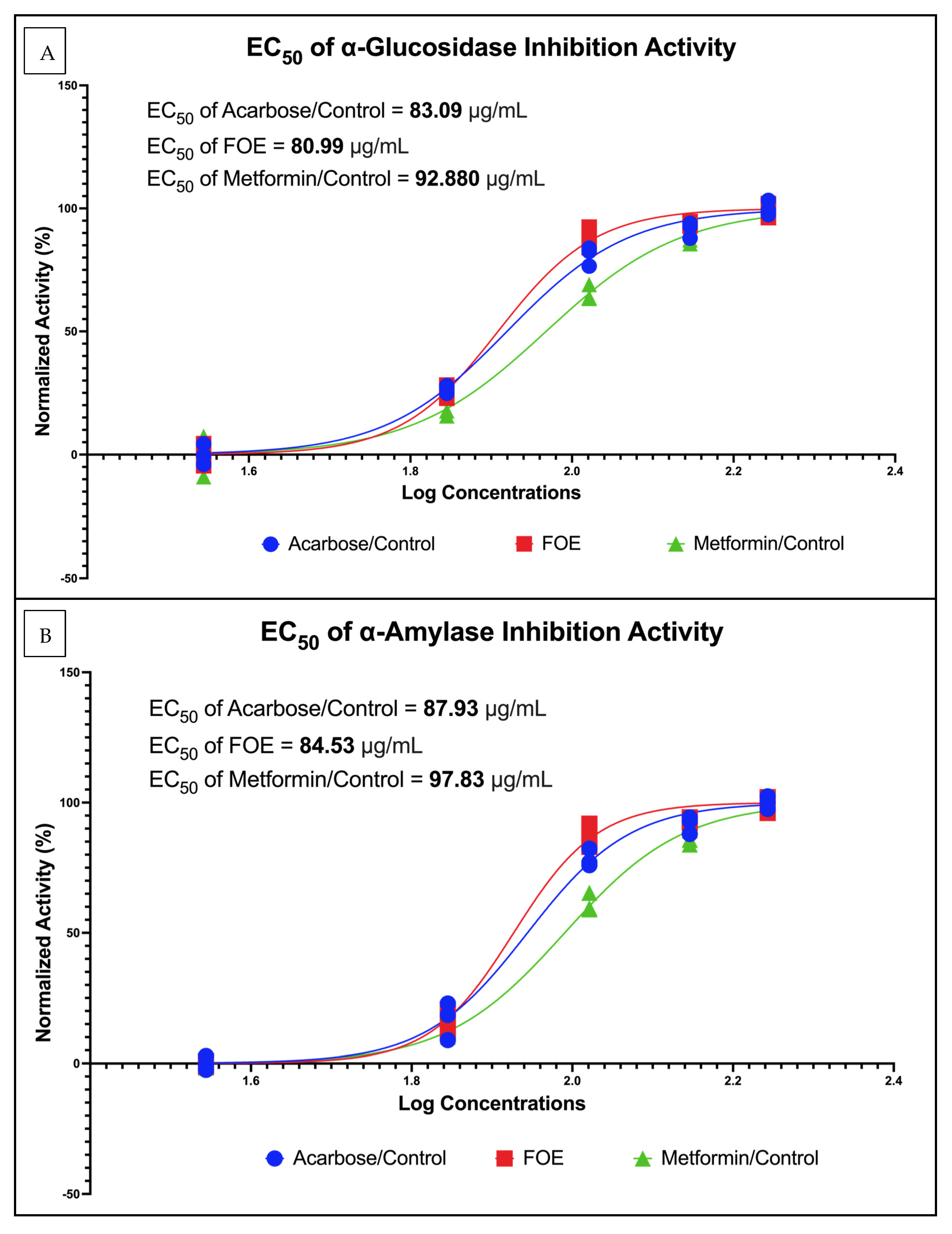
3.2.2. α-Glucosidase and α-Amylase Potential of FOE
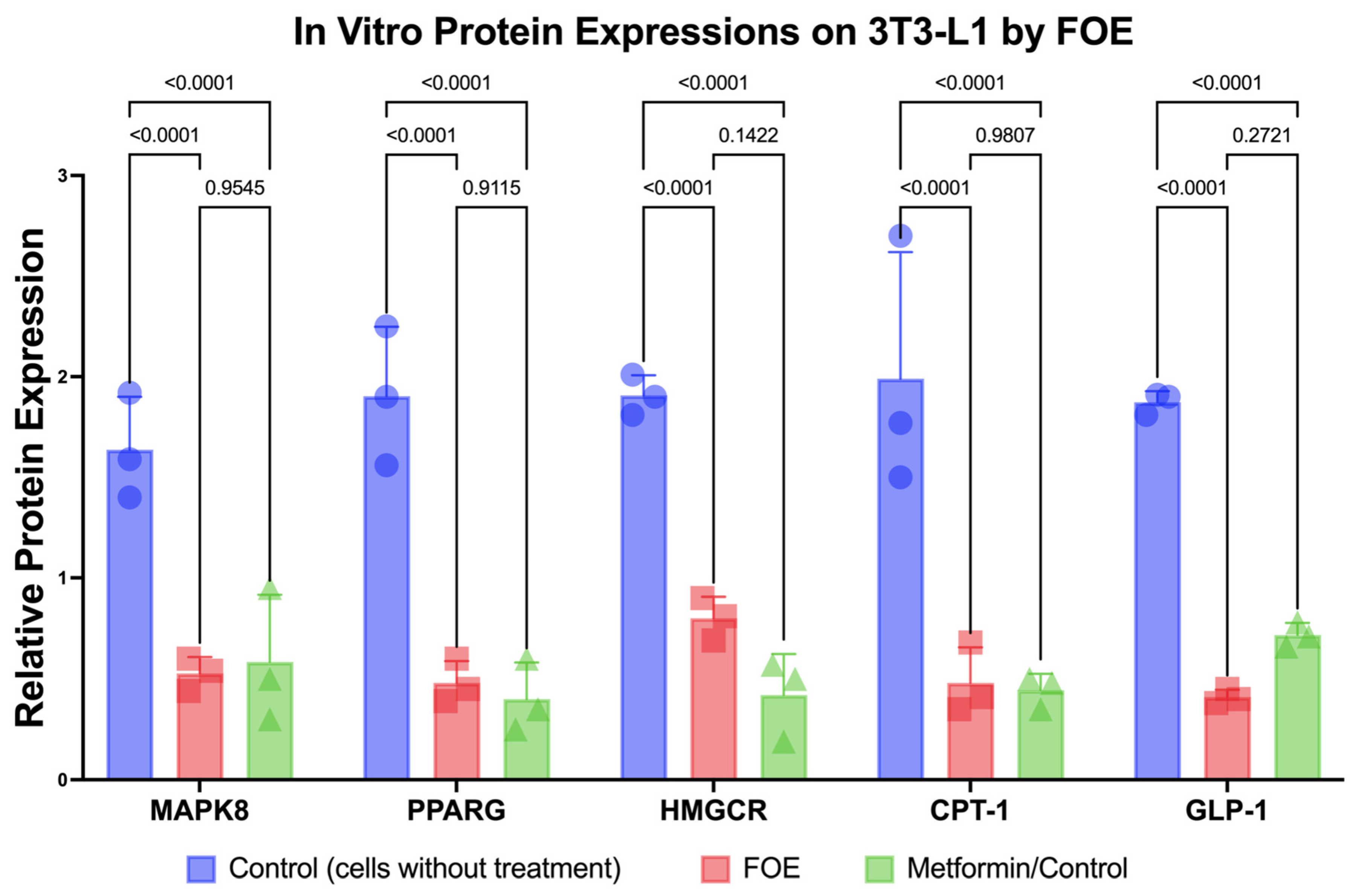
3.2.3. Downregulation of Protein Expression and Reduction in 3T3-L1 Mouse Cells by FOE
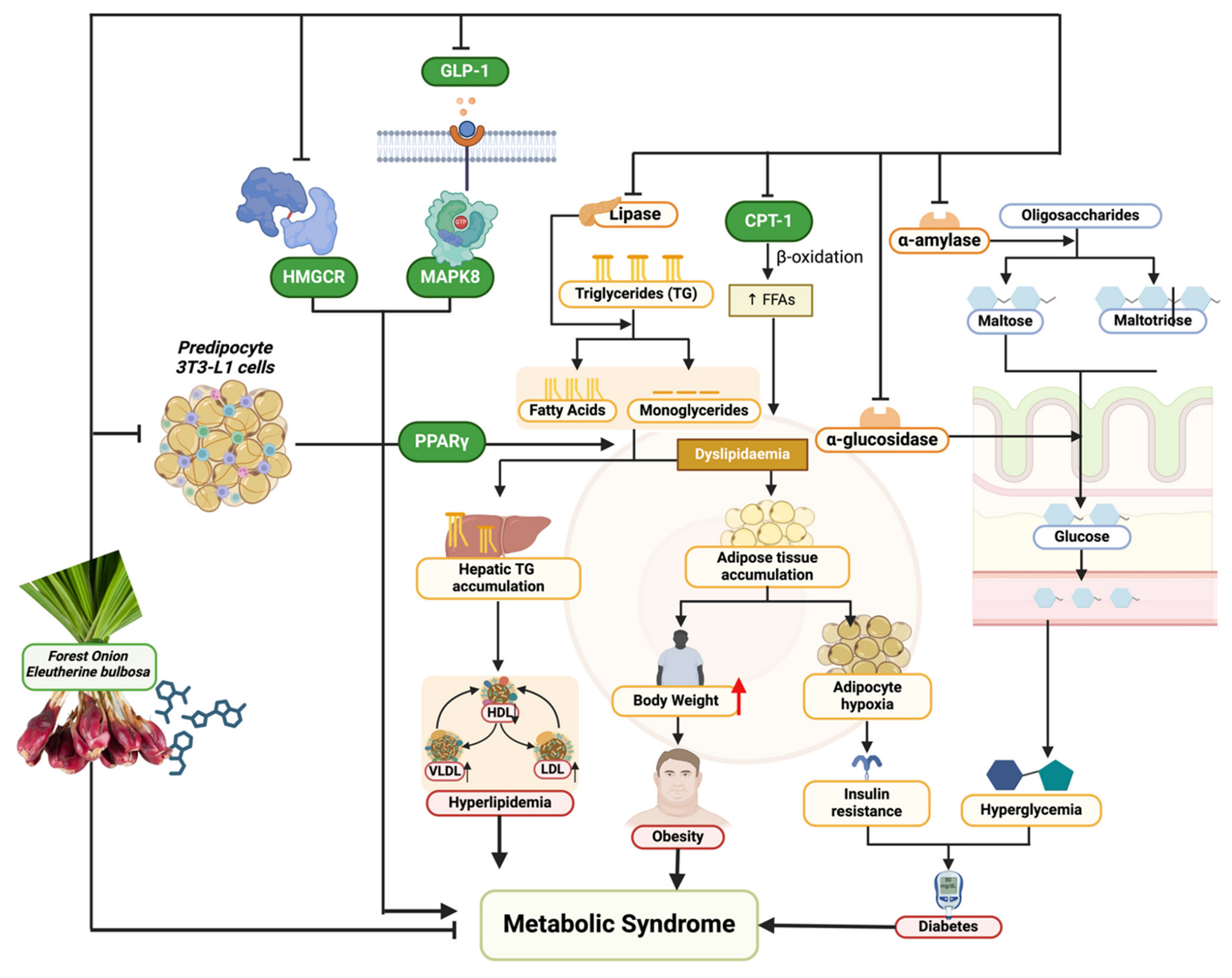
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Engin, A. The Definition and Prevalence of Obesity and Metabolic Syndrome. In Obesity and Lipotoxicity; Engin, A.B., Engin, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–17. ISBN 978-3-319-48382-5. [Google Scholar]
- Nolan, P.B.; Carrick-Ranson, G.; Stinear, J.W.; Reading, S.A.; Dalleck, L.C. Prevalence of Metabolic Syndrome and Metabolic Syndrome Components in Young Adults: A Pooled Analysis. Prev. Med. Rep. 2017, 7, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Mathangasinghe, Y.; Jayawardena, R.; Hills, A.P.; Misra, A. Prevalence and Trends of Metabolic Syndrome among Adults in the Asia-Pacific Region: A Systematic Review. BMC Public Health 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, P.M.; Tuomilehto, J.; Rydén, L. The Metabolic Syndrome—What Is It and How Should It Be Managed? Eur. J. Prev. Cardiol. 2019, 26, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Bressan, J.; de Carvalho Vidigal, F.; Hermsdorff, H.H.M. Social Components of the Obesity Epidemic. Curr. Obes. Rep. 2013, 2, 32–41. [Google Scholar] [CrossRef][Green Version]
- Mendrick, D.L.; Diehl, A.M.; Topor, L.S.; Dietert, R.R.; Will, Y.; La Merrill, M.A.; Bouret, S.; Varma, V.; Hastings, K.L.; Schug, T.T.; et al. Metabolic Syndrome and Associated Diseases: From the Bench to the Clinic. Toxicol. Sci. 2018, 162, 36–42. [Google Scholar] [CrossRef]
- McCracken, E.; Monaghan, M.; Sreenivasan, S. Pathophysiology of the Metabolic Syndrome. Clin. Dermatol. 2018, 36, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Mexitalia, M.; Dewi, Y.O.; Pramono, A.; Anam, M.S. Effect of Tuberculosis Treatment on Leptin Levels, Weight Gain, and Percentage Body Fat in Indonesian Children. Korean J. Pediatr. 2017, 60, 118. [Google Scholar] [CrossRef]
- Aguilar-Salinas, C.A.; Viveros-Ruiz, T. Recent Advances in Managing/Understanding the Metabolic Syndrome. F1000Research 2019, 8, 370. [Google Scholar] [CrossRef]
- Sherling, D.H.; Perumareddi, P.; Hennekens, C.H. Metabolic Syndrome. J. Cardiovasc. Pharmacol. Ther. 2017, 22, 365–367. [Google Scholar] [CrossRef]
- Rask Larsen, J.; Dima, L.; Correll, C.U.; Manu, P. The Pharmacological Management of Metabolic Syndrome. Expert. Rev. Clin. Pharmacol. 2018, 11, 397–410. [Google Scholar] [CrossRef]
- Munaeni, W.; Widanarni, W.; Yuhana, M.; Setiawati, M.; Wahyudi, A. The potential of Buton forest onion Eleutherine bulbosa (Mill.) Urb. extract as a prebiotic and an antioxidant. J. Microbiol. Biotechnol. Food Sci. 2020, 10, 107–111. [Google Scholar] [CrossRef]
- Insanu, M.; Kusmardiyani, S.; Hartati, R. Recent Studies on Phytochemicals and Pharmacological Effects of Eleutherine americana Merr. Procedia Chem. 2014, 13, 221–228. [Google Scholar] [CrossRef]
- da Silva, R.M.G.; Alves, C.P.; Barbosa, F.C.; Santos, H.H.; Adão, K.M.; Granero, F.O.; Figueiredo, C.C.M.; Figueiredo, C.R.; Nicolau-Junior, N.; Silva, L.P. Antioxidant, Antitumoral, Antimetastatic Effect and Inhibition of Collagenase Enzyme Activity of Eleutherine bulbosa (Dayak Onion) Extract: In Vitro, in Vivo and in Silico Approaches. J. Ethnopharmacol. 2024, 318, 117005. [Google Scholar] [CrossRef] [PubMed]
- Alamsyah Lubis, I.; Ichwan, M.F.; Mustofa, M.; Satria, D. Anticancer Activity of Eleutherine bulbosa (Mill.) Urb. Extract on WiDr Cell Line In Vitro. In Proceedings of the 2nd Public Health International Conference (PHICo 2017), Surakarta, Indonesia, 6–7 September 2017; Atlantis Press: Paris, France, 2018. [Google Scholar]
- Shi, P.; Du, W.; Wang, Y.; Teng, X.; Chen, X.; Ye, L. Total Phenolic, Flavonoid Content, and Antioxidant Activity of Bulbs, Leaves, and Flowers Made from Eleutherine bulbosa (Mill.) Urb. Food Sci. Nutr. 2019, 7, 148–154. [Google Scholar] [CrossRef]
- Herman, H.; Ibrahim, A.; Junaidin, J.; Arifuddin, M.; Hikmawan, B.D.; Siska, S.; Bariroh, T.; Purwoko, R.Y.; Febrina, L.; Faisal, M.; et al. Pharmacognostic Profile and Antidiabetic Activity of Eleutherine bulbosa Mills. Bulbs from East Kalimantan, Indonesia. Pharmacogn. J. 2024, 16, 118–125. [Google Scholar] [CrossRef]
- Kamarudin, A.A.; Esa, N.M.; Saad, N.; Sayuti, N.H.; Razak, N.A.A. Heat Assisted Extraction of Phenolic Compounds from Eleutherine bulbosa (Mill.) Bulb and Its Bioactive Profiles Using Response Surface Methodology. Ind. Crops Prod. 2020, 144, 112064. [Google Scholar] [CrossRef]
- Simbala, H.E.I.; Nurkolis, F.; Mayulu, N.; Rotty, L.W.A. New Discovery of Covid-19 Natural-Based Potential Antivirus Herbal Supplement Products from Pinang Yaki (Areca Vestiaria) Extract: A Preliminary Study by Untargeted Metabolomic Profiling. F1000Research 2022, 10, 1021. [Google Scholar] [CrossRef]
- Druzhilovskiy, D.S.; Rudik, A.V.; Filimonov, D.A.; Gloriozova, T.A.; Lagunin, A.A.; Dmitriev, A.V.; Pogodin, P.V.; Dubovskaya, V.I.; Ivanov, S.M.; Tarasova, O.A.; et al. Computational Platform Way2Drug: From the Prediction of Biological Activity to Drug Repurposing. Russ. Chem. Bull. 2017, 66, 1832–1841. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Norinder, U.; Bergström, C.A.S. Prediction of ADMET Properties. ChemMedChem 2006, 1, 920–937. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A Platform for Systematic ADMET Evaluation Based on a Comprehensively Collected ADMET Database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Dunkel, M.; Gunther, S.; Ahmed, J.; Wittig, B.; Preissner, R. SuperPred: Drug Classification and Target Prediction. Nucleic Acids Res. 2008, 36, W55–W59. [Google Scholar] [CrossRef] [PubMed]
- Gallo, K.; Goede, A.; Preissner, R.; Gohlke, B.-O. SuperPred 3.0: Drug Classification and Target Prediction—A Machine Learning Approach. Nucleic Acids Res. 2022, 50, W726–W731. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A Web Server for Target Prediction of Bioactive Small Molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, A.; Ghorbani, N.; Dastan, K. Identification of Druggable Hub Genes and Key Pathways Associated with Cervical Cancer by Protein-Protein Interaction Analysis: An in Silico Study. Int. J. Reprod. BioMed. (IJRM) 2023, 21, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Yang, Y.; Cheng, H.; Fu, S.; Liu, Y.; Qiu, Y.; Chen, H.; Zhang, J.; Zhou, H.; Shi, L.; et al. Integrated Analysis of Long Non-Coding RNA Expression Profiles in Glaesserella Parasuis-Induced Meningitis: New Insight into Pathogenesis. Microbiol. Res. 2023, 14, 1427–1441. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Gan, J.; Xiao, Z.-X.; Cao, Y. FitDock: Protein–Ligand Docking by Template Fitting. Brief. Bioinform. 2022, 23, bbac087. [Google Scholar] [CrossRef]
- Nurkolis, F.; Taslim, N.A.; Subali, D.; Kurniawan, R.; Hardinsyah, H.; Gunawan, W.B.; Kusuma, R.J.; Yusuf, V.M.; Pramono, A.; Kang, S.; et al. Dietary Supplementation of Caulerpa Racemosa Ameliorates Cardiometabolic Syndrome via Regulation of PRMT-1/DDAH/ADMA Pathway and Gut Microbiome in Mice. Nutrients 2023, 15, 909. [Google Scholar] [CrossRef]
- Kurniawan, R.; Nurkolis, F.; Taslim, N.A.; Subali, D.; Surya, R.; Gunawan, W.B.; Alisaputra, D.; Mayulu, N.; Salindeho, N.; Kim, B. Carotenoids Composition of Green Algae Caulerpa Racemosa and Their Antidiabetic, Anti-Obesity, Antioxidant, and Anti-Inflammatory Properties. Molecules 2023, 28, 3267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, J.; Mao, X.; Qi, P.; Zhang, X. Separation and Lipid Inhibition Effects of a Novel Decapeptide from Chlorella Pyenoidose. Molecules 2019, 24, 3527. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-U.; Park, Y.-J. Ergosterol Peroxide from the Medicinal Mushroom Ganoderma Lucidum Inhibits Differentiation and Lipid Accumulation of 3T3-L1 Adipocytes. Int. J. Mol. Sci. 2020, 21, 460. [Google Scholar] [CrossRef] [PubMed]
- Nurkolis, F.; Purnomo, A.F.; Alisaputra, D.; Gunawan, W.B.; Qhabibi, F.R.; Park, W.; Moon, M.; Taslim, N.A.; Park, M.N.; Kim, B. In Silico and In Vitro Studies Reveal a Synergistic Potential Source of Novel Anti-Ageing from Two Indonesian Green Algae. J. Funct. Foods 2023, 104, 105555. [Google Scholar] [CrossRef]
- Kamarudin, A.A.; Sayuti, N.H.; Saad, N.; Razak, N.A.A.; Esa, N.M. Eleutherine bulbosa (Mill.) Urb. Bulb: Review of the Pharmacological Activities and Its Prospects for Application. Int. J. Mol. Sci. 2021, 22, 6747. [Google Scholar] [CrossRef] [PubMed]
- Fahed, G.; Aoun, L.; Bou Zerdan, M.; Allam, S.; Bou Zerdan, M.; Bouferraa, Y.; Assi, H.I. Metabolic Syndrome: Updates on Pathophysiology and Management in 2021. Int. J. Mol. Sci. 2022, 23, 786. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-N.; Shin, M.-R.; Shin, S.H.; Lee, A.R.; Lee, J.Y.; Seo, B.-I.; Kim, M.Y.; Kim, T.H.; Noh, J.S.; Rhee, M.H.; et al. Study of Antiobesity Effect through Inhibition of Pancreatic Lipase Activity of Diospyros kaki Fruit and Citrus unshiu Peel. Biomed. Res. Int. 2016, 2016, 1723042. [Google Scholar] [CrossRef]
- Kashtoh, H.; Baek, K.-H. Recent Updates on Phytoconstituent Alpha-Glucosidase Inhibitors: An Approach towards the Treatment of Type Two Diabetes. Plants 2022, 11, 2722. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; An, X.; Yang, C.; Sun, W.; Ji, H.; Lian, F. The Crucial Role and Mechanism of Insulin Resistance in Metabolic Disease. Front. Endocrinol. 2023, 14, 1149239. [Google Scholar] [CrossRef]
- Sun, C.; Mao, S.; Chen, S.; Zhang, W.; Liu, C. PPARs-Orchestrated Metabolic Homeostasis in the Adipose Tissue. Int. J. Mol. Sci. 2021, 22, 8974. [Google Scholar] [CrossRef]
- Pangestika, I.; Oksal, E.; Tengku Muhammad, T.S.; Amir, H.; Syamsumir, D.F.; Wahid, M.E.A.; Andriani, Y. Inhibitory Effects of Tangeretin and Trans-Ethyl Caffeate on the HMG-CoA Reductase Activity: Potential Agents for Reducing Cholesterol Levels. Saudi J. Biol. Sci. 2020, 27, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Liang, K. Mitochondrial CPT1A: Insights into Structure, Function, and Basis for Drug Development. Front. Pharmacol. 2023, 14, 1160440. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Vella, A. Effects of GLP-1 on Appetite and Weight. Rev. Endocr. Metab. Disord. 2014, 15, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-Q.; Xiao, H.-T.; Yang, F.; Wang, Y.-D.; Li, P.; Zheng, Z.-G. Advancing Targeted Protein Degradation for Metabolic Diseases Therapy. Pharmacol. Res. 2023, 188, 106627. [Google Scholar] [CrossRef] [PubMed]
- Al-Ishaq, R.K.; Abotaleb, M.; Kubatka, P.; Kajo, K.; Büsselberg, D. Flavonoids and Their Anti-Diabetic Effects: Cellular Mechanisms and Effects to Improve Blood Sugar Levels. Biomolecules 2019, 9, 430. [Google Scholar] [CrossRef] [PubMed]
- Samota, M.K.; Sharma, M.; Kaur, K.; Sarita; Yadav, D.K.; Pandey, A.K.; Tak, Y.; Rawat, M.; Thakur, J.; Rani, H. Onion Anthocyanins: Extraction, Stability, Bioavailability, Dietary Effect, and Health Implications. Front. Nutr. 2022, 9, 917617. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Badshah, S.L.; Khan, N.; Ghani, L.; Poulson, B.G.; Emwas, A.-H.; Jaremko, M. Important Flavonoids and Their Role as a Therapeutic Agent. Molecules 2020, 25, 5243. [Google Scholar] [CrossRef]
| No. | Observed Compounds | Molecular Formula | Structural Class | RT (Min) | Observed MW (m/z) | PubChem ID or Substance ID |
|---|---|---|---|---|---|---|
| C1 | p-Cresol | C7H8O | Aromatic Alcohol | 6.993 | 108.05783 | 2879 |
| C2 | 4-Fluorophenol | C6H5FO | Aromatic Halide | 0.678 | 112.03258 | 9732 |
| C3 | Guvacine | C6H9NO2 | Amino Acid | 0.832 | 127.06334 | 3532 |
| C4 | Furaneol | C6H8O3 | Ester | 0.854 | 128.04738 | 19309 |
| C5 | p-cymene | C10H14 | Bicyclic Hydrocarbon | 5.345 | 134.10948 | 7463 |
| C6 | Elaeokanine C | C12H21NO2 | Amine | 11.249 | 211.15705 | 442855 |
| C7 | Flavokawain A | C18H18O5 | Anthraquinone | 7.357 | 314.11498 | 5355469 |
| C8 | 3-Hydroxy-3,4-bis[(4-hydroxy-3-methoxyphenyl)methyl]oxolan-2-one | C20H22O7 | Polyphenol | 4.171 | 374.13597 | 321311 |
| C9 | Euparin | C13H12O3 | Phenolic Acid | 6.218 | 216.0783 | 119039 |
| C10 | Eleutherol | C14H12O4 | Phenolic Aldehyde | 8.492 | 244.07336 | 120697 |
| Type | Observed Peptides | Molecular Formula | RT (Min) | Observed MW (m/z) | PubChem ID or Substance ID |
|---|---|---|---|---|---|
| P1 | Gly-Leu | C8H16N2O3 | 0.872 | 188.11589 | 92843 |
| P2 | Ala-Leu | C9H18N2O3 | 2.847 | 202.13164 | 96801 |
| P3 | Val-Ser | C8H16N2O4 | 0.874 | 204.11071 | 139506 |
| P4 | Gly-Phe | C11H14N2O3 | 3.223 | 222.10013 | 92953 |
| P5 | Ala-phe | C12H16N2O3 | 2.809 | 236.11589 | 96814 |
| P6 | Asp-Leu | C10H18N2O5 | 2.946 | 246.1213 | 332962 |
| P7 | Val-Met | C10H20N2O3S | 2.62 | 248.1192 | 292427 |
| P8 | Ala-Tyr | C12H16N2O4 | 1.948 | 252.11066 | 92946 |
| P9 | Lys-Leu | C12H25N3O3 | 1.391 | 259.18932 | 7016103 |
| P10 | Arg-Leu | C12H25N5O3 | 1.508 | 287.19524 | 6992563 |
| P11 | Arg-Glu | C11H21N5O5 | 0.804 | 303.15392 | 6995004 |
| Compounds/Peptides | Pa Score | Toxicity Model Computation Analysis | Drug-Likeness | |||
|---|---|---|---|---|---|---|
| Insulin Promoter | Predicted LD50 (mg/kg) | Toxicity Class | Lipinski Rule | Pfizer Rule | GSK | |
| C1 | 0.605 | 160 | 3 | Accepted | Accepted | Accepted |
| C2 | 0.433 | 270 | 3 | Accepted | Accepted | Accepted |
| C3 | 0.457 | 1000 | 4 | Accepted | Accepted | Accepted |
| C5 | 0.751 | 3 | 1 | Accepted | Rejected | Accepted |
| C6 | 0.559 | 338 | 4 | Accepted | Accepted | Accepted |
| P1 | 0.563 | 6838 | 6 | Accepted | Accepted | Accepted |
| P2 | 0.608 | 5000 | 5 | Accepted | Accepted | Accepted |
| P3 | 0.659 | 5000 | 5 | Accepted | Accepted | Accepted |
| P4 | 0.678 | 1000 | 4 | Accepted | Accepted | Accepted |
| P5 | 0.724 | 1000 | 4 | Accepted | Accepted | Accepted |
| P6 | 0.634 | 6836 | 6 | Accepted | Accepted | Accepted |
| P8 | 0.596 | 1000 | 4 | Accepted | Accepted | Accepted |
| P9 | 0.405 | 5000 | 5 | Accepted | Accepted | Accepted |
| Name | Degree | Betweenness Centrality | Closeness Centrality | Overall Score | Pathway |
|---|---|---|---|---|---|
| PPARG | 10 | 0.32564103 | 0.8125 | 11.138141 | Peroxisomal beta-oxidation pathway of fatty acids; tissue-specific adipocyte P2 (aP2) enhancer |
| MAPK8 | 7 | 0.25769231 | 0.68421053 | 7.94190283 | Protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway; MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK8/JNK |
| HMGCR | 4 | 0.01495726 | 0.54166667 | 4.55662393 | Cholesterol biosynthesis; obesity |
| GLP-1 | 4 | 0.01068376 | 0.54166667 | 4.55235043 | Adenylyl cyclase is activated and intracellular cAMP levels are raised as a result of ligand binding activating a signaling cascade; diabetes and insulin |
| Compounds/Peptides and Control as Ligands | MAPK8 (3ELJ) | PPARG (8BF1) | HMGCR (2R4F) | CPT-1 (1NDB) | GLP-1 (4ZGM) |
|---|---|---|---|---|---|
| Control Metformin (4901) | −4.5 | −4.9 | −5.6 | −5.4 | −4.5 |
| Control Orlistat (3034010) | −6.6 | −6.4 | |||
| Control Simvastatin (54454) | −7.7 | ||||
| C3 | −4.9 | −5.1 | −6.0 | −5.1 | −4.6 |
| C6 | −6.0 | −6.4 | −6.1 | −6.2 | −5.3 |
| P1 | −5.4 | −5.3 | −6.0 | −6.5 | −5.1 |
| P2 | −5.4 | −5.8 | −6.4 | −6.5 | −5.2 |
| P3 | −5.6 | −5.4 | −5.9 | −6.1 | −4.8 |
| P4 | −7.5 | −6.3 | −6.6 | −7.9 | −6.1 |
| P5 | −7.0 | −6.3 | −6.9 | −7.9 | −6.2 |
| P6 | −6.3 | −6.0 | −6.1 | −6.7 | −5.8 |
| P8 | −7.1 | −6.8 | −7.2 | −7.8 | −6.2 |
| P9 | −5.5 | −5.5 | −5.8 | −6.6 | −5.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Permatasari, H.K.; Abshori, N.F.; Syahputra, R.A.; Harahap, U.; Amalia, N.; Kumalawati, D.A.; Mayulu, N.; Taslim, N.A.; Tallei, T.E.; Tjandrawinata, R.R.; et al. Novel Functional Food Properties of Forest Onion (Eleutherine bulbosa Merr.) Phytochemicals for Treating Metabolic Syndrome: New Insights from a Combined Computational and In Vitro Approach. Nutrients 2024, 16, 1441. https://doi.org/10.3390/nu16101441
Permatasari HK, Abshori NF, Syahputra RA, Harahap U, Amalia N, Kumalawati DA, Mayulu N, Taslim NA, Tallei TE, Tjandrawinata RR, et al. Novel Functional Food Properties of Forest Onion (Eleutherine bulbosa Merr.) Phytochemicals for Treating Metabolic Syndrome: New Insights from a Combined Computational and In Vitro Approach. Nutrients. 2024; 16(10):1441. https://doi.org/10.3390/nu16101441
Chicago/Turabian StylePermatasari, Happy Kurnia, Nuril Farid Abshori, Rony Abdi Syahputra, Urip Harahap, Nurlinah Amalia, Dian Aruni Kumalawati, Nelly Mayulu, Nurpudji Astuti Taslim, Trina Ekawati Tallei, Raymond Rubianto Tjandrawinata, and et al. 2024. "Novel Functional Food Properties of Forest Onion (Eleutherine bulbosa Merr.) Phytochemicals for Treating Metabolic Syndrome: New Insights from a Combined Computational and In Vitro Approach" Nutrients 16, no. 10: 1441. https://doi.org/10.3390/nu16101441
APA StylePermatasari, H. K., Abshori, N. F., Syahputra, R. A., Harahap, U., Amalia, N., Kumalawati, D. A., Mayulu, N., Taslim, N. A., Tallei, T. E., Tjandrawinata, R. R., Wiyarta, E., Pramono, A., Kim, B., Tsopmo, A., Serra-Majem, L., & Nurkolis, F. (2024). Novel Functional Food Properties of Forest Onion (Eleutherine bulbosa Merr.) Phytochemicals for Treating Metabolic Syndrome: New Insights from a Combined Computational and In Vitro Approach. Nutrients, 16(10), 1441. https://doi.org/10.3390/nu16101441














