Maternal Consumption of Non-Nutritive Sweeteners during Pregnancy Is Associated with Alterations in the Colostrum Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Ethical Considerations
2.2. Selection and Evaluation of Patients
2.3. Non-Nutritive Sweeteners Consumption Assessment
2.4. Sample Collection and Processing
2.5. DNA Extraction
2.6. Preparation of the 16S rDNA Library and High-Throughput Sequencing
2.7. High-Throughput DNA Sequencing
2.8. Taxonomic Assignment and Bacterial Diversity
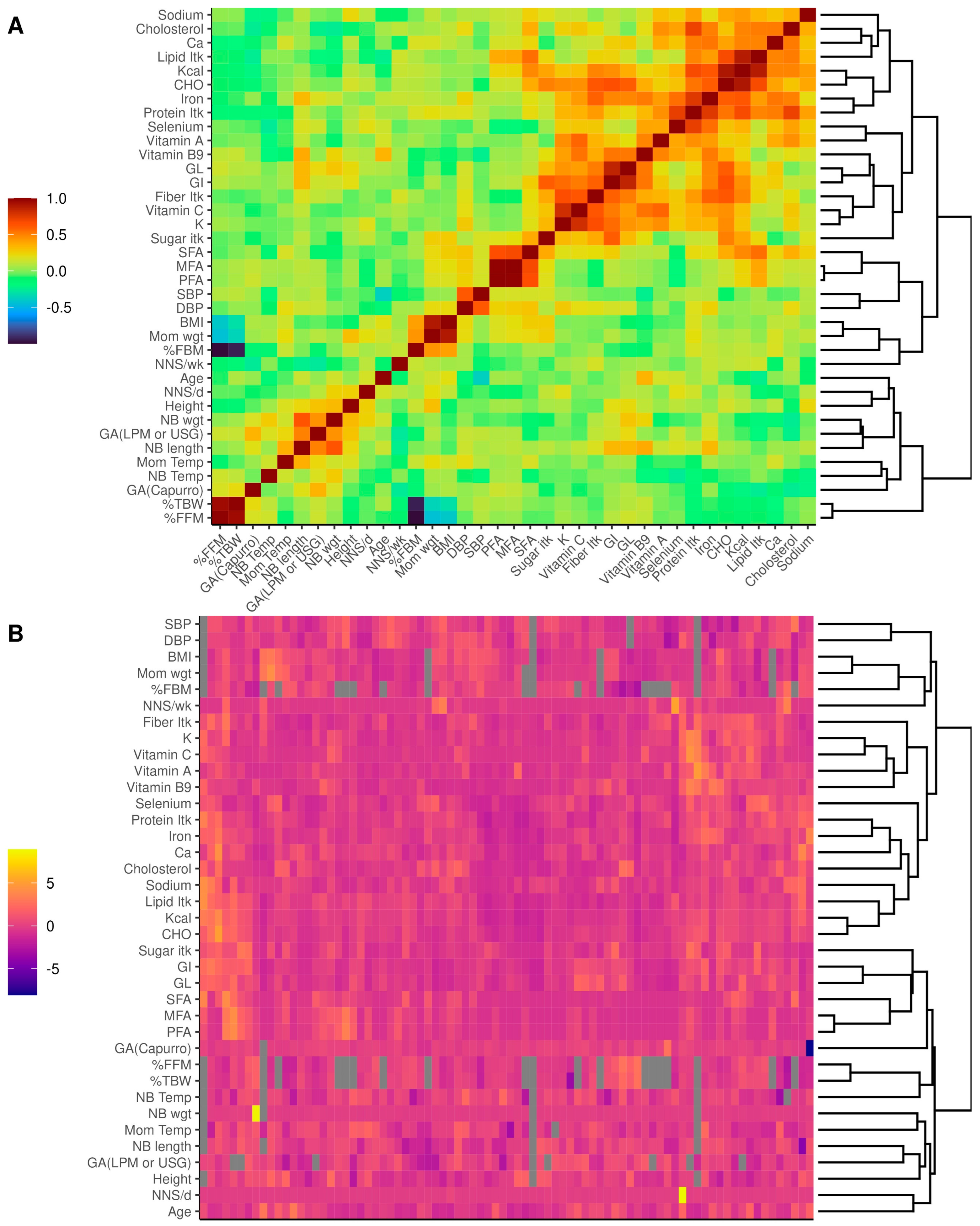
2.9. Statistical Analysis
3. Results
3.1. Study Population
3.1.1. Descriptive Statistics
3.1.2. Diet Characteristics
3.2. Colostrum Microbiota
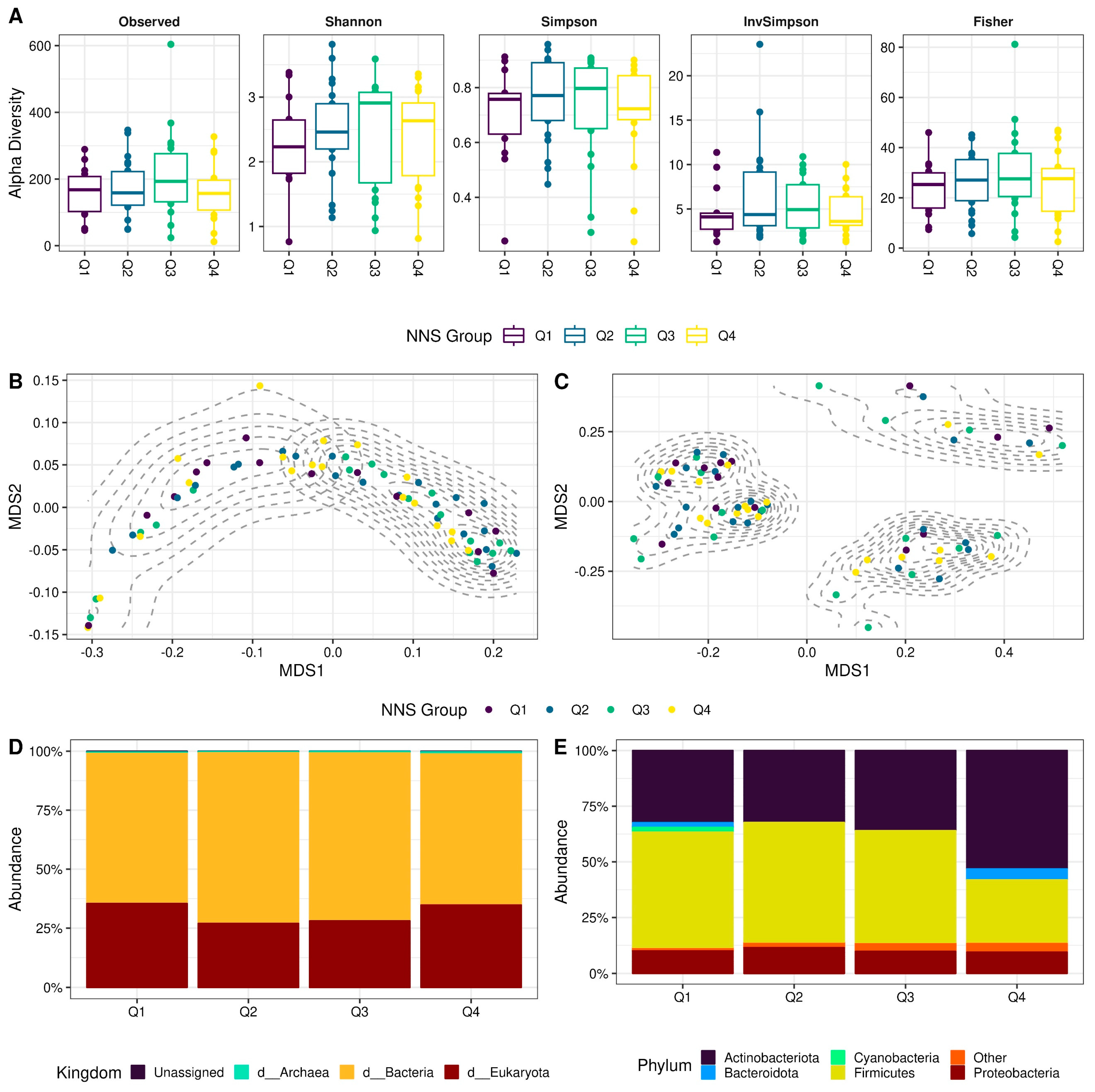
3.2.1. Alpha and Beta Diversity of Colostrum Microbiota Are Not Related to the Consumption of Non-Nutritive Sweeteners
3.2.2. The Composition of the Colostrum Microbiota Appears Unrelated to the Consumption of Non-Nutritive Sweeteners at the Kingdom and Phylum Levels
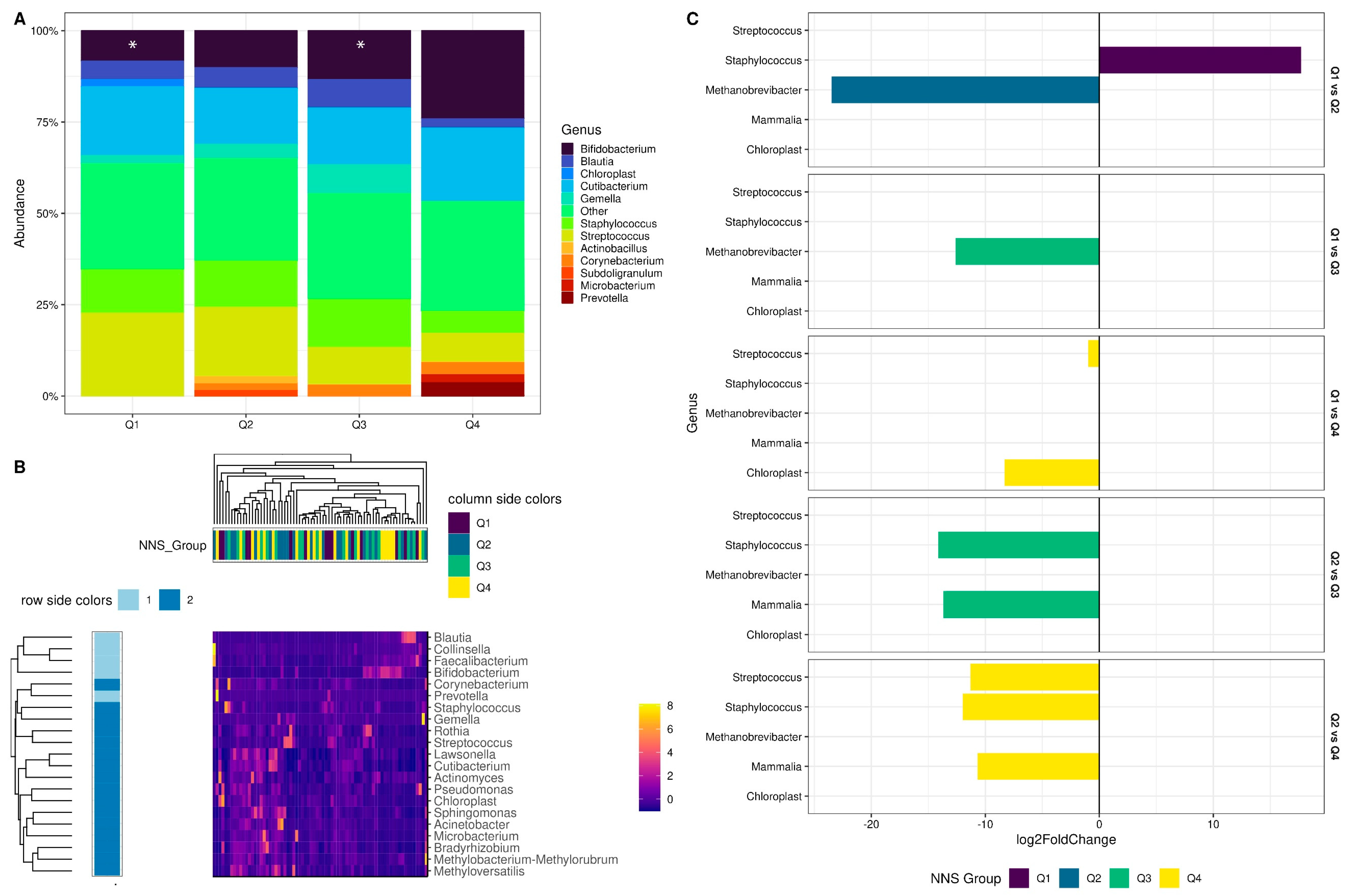
3.2.3. Non-Nutritive Sweetener Consumption during Pregnancy Associated with Changes in Specific Genera of the Colostrum Microbiota
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stanhope, K.L. Sugar Consumption, Metabolic Disease and Obesity: The State of the Controversy. Crit. Rev. Clin. Lab. Sci. 2016, 53, 52–67. [Google Scholar] [CrossRef]
- Shankar, P.; Ahuja, S.; Sriram, K. Non-Nutritive Sweeteners: Review and Update. Nutrition 2013, 29, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Fitch, C.; Keim, K.S. Position of the Academy of Nutrition and Dietetics: Use of Nutritive and Nonnutritive Sweeteners. J. Acad. Nutr. Diet. 2012, 112, 739–758. [Google Scholar] [CrossRef] [PubMed]
- Debras, C.; Chazelas, E.; Srour, B.; Druesne-Pecollo, N.; Esseddik, Y.; de Edelenyi, F.S.; Agaësse, C.; De Sa, A.; Lutchia, R.; Gigandet, S.; et al. Artificial Sweeteners and Cancer Risk: Results from the NutriNet-Santé Population-Based Cohort Study. PLoS Med. 2022, 19, e1003950. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.; Wylie-Rosett, J.; Gidding, S.S.; Steffen, F.L.M.; Johnson, F.R.K.; Reader, D.; Lichtenstein, A.H. Nonnutritive Sweeteners: Current Use and Health Perspectives: A Scientific Statement from the American Heart Association and the American Diabetes Association. Diabetes Care 2012, 35, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Cohen, Y.; Valdés-Mas, R.; Mor, U.; Dori-Bachash, M.; Federici, S.; Zmora, N.; Leshem, A.; Heinemann, M.; Linevsky, R.; et al. Personalized Microbiome-Driven Effects of Non-Nutritive Sweeteners on Human Glucose Tolerance. Cell 2022, 185, 3307–3328. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A.; et al. Artificial Sweeteners Induce Glucose Intolerance by Altering the Gut Microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef]
- Bueno-Hernández, N.; Esquivel-Velázquez, M.; Alcántara-Suárez, R.; Gómez-Arauz, A.; de León-Barrera, K.; Mendoza-Martínez, V.; Sánchez Medina, G.; Ruiz-Barranco, A.; Escobedo, G.; Meléndez, G. Chronic Sucralose Consumption Induces Elevation of Serum Insulin in Young Healthy Adults: A Randomized, Double Blind, Controlled Trial. Nutr. J. 2020, 19, 32. [Google Scholar] [CrossRef]
- Méndez-García, L.A.; Bueno-Hernández, N.; Cid-Soto, M.A.; De León, K.L.; Mendoza-Martínez, V.M.; Espinosa-Flores, A.J.; Carrero-Aguirre, M.; Esquivel-Velázquez, M.; León-Hernández, M.; Viurcos-Sanabria, R.; et al. Ten-Week Sucralose Consumption Induces Gut Dysbiosis and Altered Glucose and Insulin Levels in Healthy Young Adults. Microorganisms 2022, 10, 434. [Google Scholar] [CrossRef]
- Azad, M.B.; Abou-Setta, A.M.; Chauhan, B.F.; Rabbani, R.; Lys, J.; Copstein, L.; Mann, A.; Jeyaraman, M.M.; Reid, A.E.; Fiander, M.; et al. Nonnutritive Sweeteners and Cardiometabolic Health: A Systematic Review and Meta-Analysis of Randomized Controlled Trials and Prospective Cohort Studies. CMAJ 2017, 189, E929–E939. [Google Scholar] [CrossRef]
- Azad, M.B.; Sharma, A.K.; De Souza, R.J.; Dolinsky, V.W.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Lefebvre, D.L.; Sears, M.R.; et al. Association between Artificially Sweetened Beverage Consumption during Pregnancy and Infant Body Mass Index. JAMA Pediatr. 2016, 170, 662–670. [Google Scholar] [CrossRef]
- Archibald, A.J.; Dolinsky, V.W.; Azad, M.B. Early-Life Exposure to Non-Nutritive Sweeteners and the Developmental Origins of Childhood Obesity: Global Evidence from Human and Rodent Studies. Nutrients 2018, 10, 194. [Google Scholar] [CrossRef]
- Olivier-Van Stichelen, S.; Rother, K.I.; Hanover, J.A. Maternal Exposure to Non-Nutritive Sweeteners Impacts Progeny’s Metabolism and Microbiome. Front. Microbiol. 2019, 10, 1360. [Google Scholar] [CrossRef]
- Palatnik, A.; Moosreiner, A.; Olivier-Van Stichelen, S. Consumption of Non-Nutritive Sweeteners during Pregnancy. Am. J. Obstet. Gynecol. 2020, 223, 211–218. [Google Scholar] [CrossRef]
- Rother, K.I.; Sylvetsky, A.C.; Schiffman, S.S. Non- Nutritive Sweeteners in Breast Milk: Perspective on Potential Implications of Recent Findings. Arch. Toxicol. 2015, 89, 2169–2171. [Google Scholar] [CrossRef] [PubMed]
- Sylvetsky, A.C.; Gardner, A.L.; Bauman, V.; Blau, J.E.; Garraffo, H.M.; Walter, P.J.; Rother, K.I. Nonnutritive Sweeteners in Breast Milk. J. Toxicol. Environ. Health A 2015, 78, 1029–1032. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical Mother-Neonate Transfer of Maternal Gut Bacteria via Breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The Human Milk Microbiome Changes over Lactation and Is Shaped by Maternal Weight and Mode of Delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Taylor, R.; Keane, D.; Borrego, P.; Arcaro, K. Effect of Maternal Diet on Maternal Milk and Breastfed Infant Gut Microbiomes: A Scoping Review. Nutrients 2023, 15, 1420. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human Milk Microbiota Profiles in Relation to Birthing Method, Gestation and Infant Gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef]
- Moossavi, S.; Azad, M.B. Origins of Human Milk Microbiota: New Evidence and Arising Questions. Gut Microbes 2019, 12, 1667722. [Google Scholar] [CrossRef]
- Capurro, H.; Konichezky, S.; Fonseca, D.; Caldeyro-Barcia, R. A Simplified Method for Diagnosis of Gestational Age in the Newborn Infant. J. Pediatr. 1978, 93, 120–122. [Google Scholar] [CrossRef]
- Bueno-Hernández, N.; Melendez-Mier, G.; Alcántara-Suarez, R.; Pérez-Castañeda, M.; A Hernández-León, Y.; Ruiz-Barranco, A.; Escobedo, G.; Islas-Andrade, S.; Meléndez, G. Content Validity and Reliability of a Food Frequency Questionnaire with Intense Sweeteners (FFQIS) in a Hispanic Population. J. Nutr. Food Sci. 2018, 8, 716. [Google Scholar] [CrossRef]
- Denova-Gutiérrez, E.; Ramírez-Silva, I.; Rodríguez-Ramírez, S.; Jiménez-Aguilar, A.; Shamah-Levy, T.; Rivera-Dommarco, J.A. Validity of a Food Frequency Questionnaire to Assess Food Intake in Mexican Adolescent and Adult Population. Salud Publica Mex. 2016, 58, 617–628. [Google Scholar] [CrossRef]
- Corona-Cervantes, K.; García-González, I.; Villalobos-Flores, L.E.; Hernández-Quiroz, F.; Piña-Escobedo, A.; Hoyo-Vadillo, C.; Rangel-Calvillo, M.N.; García-Mena, J. Human Milk Microbiota Associated with Early Colonization of the Neonatal Gut in Mexican Newborns. PeerJ 2020, 8, e9205. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible Sequence Taxonomy Reference Database Management for the Masses. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software; PBC: Boston, MA, USA, 2023. [Google Scholar]
- Bisanz, J.E. Qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. Version 0.99.6, 2018; GitHub. Available online: https://github.com/jbisanz/qiime2R (accessed on 15 May 2023).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, Version 2.6-4, GitHub. Available online: https://github.com/vegandevs/vegan (accessed on 15 May 2023).
- Hennig, C. Fpc: Flexible Procedures for Clustering, Version 2.2-10; Unibo. Available online: https://www.unibo.it/sitoweb/christian.hennig/en/ (accessed on 15 May 2023).
- Lahti, L.; Shetty, S. Microbiome R Package, Version 1.22.0. GitHub. Available online: http://microbiome.github.io/microbiome (accessed on 15 May 2023).
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R Package for Creating Interactive Cluster Heatmaps for Online Publishing. Bioinformatics 2017, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Ahlmann-Eltze, C.; Forbes, K.; Anders, S.; Huber, W. DESeq2: Differential Gene Expression Analysis Based on the Negative Binomial Distribution. Version 1.40.2. GitHub. Available online: https://github.com/thelovelab/DESeq2 (accessed on 17 August 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots. Version 0.6.0. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 31 August 2023).
- Wickham, H.; Seidel, D. Scales: Scale Functions for Visualization. Version 1.2.1. Available online: https://github.com/r-lib/scales (accessed on 18 August 2023).
- Akoglu, H. User’s Guide to Correlation Coefficients. Turk. J. Emerg. Med. 2018, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- INEGI Censo de Población y Vivienda. 2020. Available online: https://www.inegi.org.mx/programas/ccpv/2020/default.html#Resultados_generales (accessed on 21 September 2023).
- Institto Nacional de Geografía y Estadística (INEGI). COMUNICADO DE PRENSA NÚM 98/21. 2021. Available online: https://www.inegi.org.mx/contenidos/saladeprensa/boletines/2021/EstSociodemo/ResultCenso2020_CdMx.pdf (accessed on 1 September 2023).
- De Koning, L.; Malik, V.S.; Rimm, E.B.; Willett, W.C.; Hu, F.B. Sugar-Sweetened and Artificially Sweetened Beverage Consumption and Risk of Type 2 Diabetes in Men. Am. J. Clin. Nutr. 2011, 93, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Djoussé, L.; Gaziano, J.M. Dietary Cholesterol and Coronary Artery Disease: A Systematic Review. Curr. Atheroscler. Rep. 2009, 11, 418–422. [Google Scholar] [CrossRef]
- Oh, C.; Keats, E.C.; Bhutta, Z.A. Vitamin and Mineral Supplementation during Pregnancy on Maternal, Birth, Child Health and Development Outcomes in Low-and Middle-Income Countries: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 491. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Impact of Human Milk Bacteria and Oligosaccharides on Neonatal Gut Microbiota Establishment and Gut Health. Nutr. Rev. 2015, 73, 426–437. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut Microbiota Markers Associated with Obesity and Overweight in Italian Adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of Inflammation-Driven Bacterial Dysbiosis in the Gut. Mucosal Immunol. 2017, 10, 18. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of Chloroplast Contamination in 16S RRNA Gene Pyrosequencing of Insect Herbivore Bacterial Communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef]
- Beck, D.L.; Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schu, U.M.E.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A.; Schütte, U.M.E.; et al. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Ojima, M.N.; Jiang, L.; Arzamasov, A.A.; Yoshida, K.; Odamaki, T.; Xiao, J.; Nakajima, A.; Kitaoka, M.; Hirose, J.; Urashima, T.; et al. Priority Effects Shape the Structure of Infant-Type Bifidobacterium Communities on Human Milk Oligosaccharides. ISME J. 2022, 16, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Bottacini, F.; Van Sinderen, D.; Ventura, M. Omics of Bifidobacteria: Research and Insights into Their Health-Promoting Activities. Biochem. J. 2017, 474, 4137–4152. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Gerasimidis, K.; Bryden, K.; Chen, X.; Papachristou, E.; Verney, A.; Roig, M.; Hansen, R.; Nichols, B.; Papadopoulou, R.; Parrett, A. The Impact of Food Additives, Artificial Sweeteners and Domestic Hygiene Products on the Human Gut Microbiome and Its Fibre Fermentation Capacity. Eur. J. Nutr. 2020, 59, 3213–3230. [Google Scholar] [CrossRef]
- Abou-Donia, M.B.; El-Masry, E.M.; Abdel-Rahman, A.A.; McLendon, R.E.; Schiffman, S.S. Splenda Alters Gut Microflora and Increases Intestinal P-Glycoprotein and Cytochrome p-450 in Male Rats. J. Toxicol. Environ. Health A 2008, 71, 1415–1429. [Google Scholar] [CrossRef]
- Martín, R.; Jiménez, E.; Heilig, H.; Fernández, L.; Marín, M.L.; Zoetendal, E.G.; Rodríguez, J.M. Isolation of Bifidobacteria from Breast Milk and Assessment of the Bifidobacterial Population by PCR-Denaturing Gradient Gel Electrophoresis and Quantitative Real-Time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Shil, A.; Olusanya, O.; Ghufoor, Z.; Forson, B.; Marks, J.; Chichger, H. Artificial Sweeteners Disrupt Tight Junctions and Barrier Function in the Intestinal Epithelium through Activation of the Sweet Taste Receptor, T1R3. Nutrients 2020, 12, 1862. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Fernández, L.; Verhasselt, V. The Gut–Breast Axis: Programming Health for Life. Nutrients 2021, 13, 606. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and Bifidobacteria in Human Breast Milk: Influence of Antibiotherapy and Other Host and Clinical Factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef]
- Togo, A.H.; Grine, G.; Khelaifia, S.; des Robert, C.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 18653. [Google Scholar] [CrossRef]
- Samuel, B.S.; Gordon, J.I. A Humanized Gnotobiotic Mouse Model of Host-Archaeal-Bacterial Mutualism. Proc. Natl. Acad. Sci. USA 2006, 103, 10011–10016. [Google Scholar] [CrossRef] [PubMed]
- Mbakwa, C.A.; Penders, J.; Savelkoul, P.H.; Thijs, C.; Dagnelie, P.C.; Mommers, M.; Arts, I.C.W. Gut Colonization with Methanobrevibacter Smithii Is Associated with Childhood Weight Development. Obesity 2015, 23, 2508–2516. [Google Scholar] [CrossRef]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The Gut Microbiome of Mexican Children Affected by Obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Araújo, J.R.; Martel, F.; Keating, E. Exposure to Non-Nutritive Sweeteners during Pregnancy and Lactation: Impact in Programming of Metabolic Diseases in the Progeny Later in Life. Reprod. Toxicol. 2014, 49, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Englund-Ögge, L.; Brantsæter, A.L.; Haugen, M.; Sengpiel, V.; Khatibi, A.; Myhre, R.; Myking, S.; Meltzer, H.M.; Kacerovsky, M.; Nilsen, R.M.; et al. Association between Intake of Artificially Sweetened and Sugar-Sweetened Beverages and Preterm Delivery: A Large Prospective Cohort Study. Am. J. Clin. Nutr. 2012, 96, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, T.I.; Strøm, M.; Petersen, S.B.; Olsen, S.F. Intake of Artificially Sweetened Soft Drinks and Risk of Preterm Delivery: A Prospective Cohort Study in 59,334 Danish Pregnant Women. Am. J. Clin. Nutr. 2010, 92, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Petherick, E.S.; Goran, M.I.; Wright, J. Relationship between Artificially Sweetened and Sugar-Sweetened Cola Beverage Consumption during Pregnancy and Preterm Delivery in a Multi-Ethnic Cohort: Analysis of the Born in Bradford Cohort Study. Eur. J. Clin. Nutr. 2014, 68, 404–407. [Google Scholar] [CrossRef]
- Maslova, E.; Strøm, M.; Olsen, S.F.; Halldorsson, T.I. Consumption of Artificially-Sweetened Soft Drinks in Pregnancy and Risk of Child Asthma and Allergic Rhinitis. PLoS ONE 2013, 8, e57261. [Google Scholar] [CrossRef]
- Shamah-Levy, T.; Cuevas-Nasu, L.; Gaona-Pineda, E.B.; Valenzuela-Bravo, D.G.; Méndez Gómez-Humarán, I.; Ávila-Arcos, M.A. Childhood Obesity in Mexico: Influencing Factors and Prevention Strategies. Front. Public. Health 2022, 10, 949893. [Google Scholar] [CrossRef]
- Weihrauch-Blüher, S.; Schwarz, P.; Klusmann, J.H. Childhood Obesity: Increased Risk for Cardiometabolic Disease and Cancer in Adulthood. Metabolism 2019, 92, 147–152. [Google Scholar] [CrossRef]
- Weihrauch-Blüher, S.; Wiegand, S. Risk Factors and Implications of Childhood Obesity. Curr. Obes. Rep. 2018, 7, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles across Specific Geographic Locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef]
- Molina-Torres, R.; Nolasco-Jáuregui, O.; Rodriguez-Torres, E.E.; Itzá-Ortiz, B.A.; Quezada-Téllez, L.A. A Comparative Analysis of Urban Development, Economic Level, and COVID-19 Cases in Mexico City. J. Urban Manag. 2021, 10, 265–274. [Google Scholar] [CrossRef]
- Walker, S.P.; Barrett, M.; Hogan, G.; Flores Bueso, Y.; Claesson, M.J.; Tangney, M. Non-Specific Amplification of Human DNA Is a Major Challenge for 16S RRNA Gene Sequence Analysis. Sci. Rep. 2020, 10, 16356. [Google Scholar] [CrossRef] [PubMed]
- Witkowska-Zimny, M.; Kaminska-El-Hassan, E. Cells of Human Breast Milk. Cell Mol. Biol. Lett. 2017, 22, 11. [Google Scholar] [CrossRef]
- Toews, I.; Lohner, S.; Küllenberg de Gaudry, D.; Sommer, H.; Meerpohl, J. Association between Intake of Non-Sugar Sweeteners and Health Outcomes: Systematic Review and Meta-Analyses of Randomised and Non-Randomised Controlled Trials and Observational Studies. BMJ 2019, 364, l156. [Google Scholar] [CrossRef]
- Soria-Contreras, D.C.; Trejo-Valdivia, B.; Cantoral, A.; Pizano-Zárate, M.L.; Baccarelli, A.A.; Just, A.C.; Colicino, E.; Deierlein, A.L.; Wright, R.O.; Oken, E.; et al. Patterns of Weight Change One Year after Delivery Are Associated with Cardiometabolic Risk Factors at Six Years Postpartum in Mexican Women. Nutrients 2020, 12, 170. [Google Scholar] [CrossRef]
- de la Garza, A.L.; Romero-Delgado, B.; Martínez-Tamez, A.M.; Cárdenas-Tueme, M.; Camacho-Zamora, B.D.; Matta-Yee-Chig, D.; Sánchez-Tapia, M.; Torres, N.; Camacho-Morales, A. Maternal Sweeteners Intake Modulates Gut Microbiota and Exacerbates Learning and Memory Processes in Adult Male Offspring. Front. Pediatr. 2022, 9, 491. [Google Scholar] [CrossRef]
| Variable | Q1 | Q2 | Q3 | Q4 | Fa/Hb/χ2 | p |
|---|---|---|---|---|---|---|
| <4 t/wk | 4 to <8 t/wk | 8 to <16.5 t/wk | ≥16.5 t/wk | |||
| n = 16 | n = 25 | n = 20 | n = 21 | |||
| Age, years | 26[21;30] | 23[20;23] | 24[20;31] | 20[18;26] | 6.391 | 0.094 b,d |
| Children, n | 2[1;2] | 2[1;2] | 1[1;2] | 1[1;1] | 11.600 | 0.009 b,d,f |
| First child rate, n (%) | 4(25) | 11(44) | 11(55) | 17(81) | 12.442 | 0.006 c |
| Menarche age, years | 13[11;14] | 13[12.8;14.3] | 12.5[11;13] | 12[11;12] | 10.079 | 0.018 b |
| Gestational age (Capurro), weeks | 39.8[39;41.1] | 40[39.2;41.1] | 39.1[38.1;40.3] | 39.5[38;40] | 4.340 | 0.227 b |
| Gestational age (USG/LMP), weeks | 39.8[39.5;40.4] | 39[37.8;40.1] | 38.5[37.1;40.2] | 39.1[36;39.6] | 3.449 | 0.327 b |
| Systolic pressure, mmHg | 105[100;112.5] | 110[100;120] | 105[100;110] | 100[100;110] | 0.767 | 0.857 b |
| Diastolic pressure, mmHg | 70[63.8;80] | 70[63.8;80] | 70[60;70] | 67.5[60;75] | 3.334 | 0.343 b |
| Height, cm | 159.5[155.5;161] | 157[153.5;162.3] | 153[147.8;163.8] | 160[158;166] | 8.961 | 0.030 b,e,g |
| Weight, kg | 71.9 ± 21.5 | 67.2 ± 14.2 | 69.7 ± 13.1 | 70.7 ± 19.1 | 0.274 | 0.844 a |
| Corrected weight †, kg | 58[54.8;68.5] | 57[48.6;67.7] | 60.8[52.3;74.8] | 56.8[52;81] | 0.694 | 0.875 b |
| BMI (postpartum), kg/m2 | 26[24.1;32.4] | 25.7[23.1;30.9] | 29.4[25.7;34.3] | 26.3[23.4;34.3] | 2.635 | 0.451 b |
| Corrected BMI †, kg/cm2 | 26[24.1;32.4] | 25.6[23.1;30.9] | 29.4[25.7;34.3] | 26.3[23.4;34.3] | 2.748 | 0.432 b |
| Fat mass, % | 27.8[27.4;34.8] | 28.5[24.3;34.3] | 26.7[21.4;35.7] | 27.7[23;35.7] | 0.292 | 0.962 b |
| Lean mass, % | 72.2[65.2;73] | 71.5[65.7;75.8] | 73.4[64.4;78.6] | 72.3[64.3;77] | 1.274 | 0.735 b |
| Total body water, % | 49.6 ± 11.6 | 51.8 ± 7.9 | 51.4 ± 6.2 | 48.8 ± 8.8 | 0.480 | 0.697 a |
| Phase angle | 7.12 ± 2.77 | 5.90 ± 1.57 | 6.64 ± 2.88 | 7.40 ± 3.46 | 1.298 | 0.282 a |
| Newborn’s temperature, °C | 36.9[36.8;37] | 36.7[36.4;37] | 36.9[36.7;37] | 36.9[36.8;37.1] | 3.342 | 0.342 b |
| NB length, cm | 49.7 ± 2.0 | 49.7 ± 1.8 | 48.3 ± 3.5 | 48.9 ± 1.8 | 1.627 | 0.190 a |
| NB weight, kg | 3.30 ± 0.30 | 3.29 ± 0.49 | 3.04 ± 0.43 | 3.09 ± 0.38 | 2.058 | 0.113 a |
| NB phase angle | 5.2[4.15;9.00] | 4.40[3.38;4.94] | 4.80[3.76;6.22] | 4.40[3.64;5.96] | 1.010 | 0.799 b |
| Mode of birth, n (%) | 3.008 | 0.390 c | ||||
| Vaginal | 9 (64.3) | 16 (66.7) | 7 (41.2) | 11(61.1) | ||
| C-section | 5 (35.7) | 8 (33.3) | 10 (58.8) | 7 (38.9) | ||
| NB sex, n (%) | 3.939 | 0.268 c | ||||
| Male | 8 (53.3) | 15 (65.2) | 10 (52.6) | 7 (35.0) | ||
| Female | 7 (46.7) | 8 (34.8) | 9 (47.4) | 13 (65.0) | ||
| Antibiotics *, n (%) | 4.291 | 0.232 c | ||||
| Yes | 11 (68.8) | 12 (48.0) | 8 (44.4) | 10 (47.6) | ||
| No | 5 (31.3) | 16 (64.0) | 10 (55.6) | 11 (52.4) |
| Variable | Q1 <4 t/wk n = 16 | Q2 4 to <8 t/wk n = 25 | Q3 8 to <16.5 t/wk) n = 20 | Q4 ≥16.5 t/wk n = 21 | H/χ2 | p |
|---|---|---|---|---|---|---|
| Energy, kcal | 1192[580;1475] | 1256[941;1559] | 1193[957;1673] | 944.8[685;1640] | 3.268 a | 0.352 a |
| Lipids, g | 38.7[14.3;46.3] | 53.1[31.1;72.6] | 42.9[36.1;68.9] | 33.2[15.2;71.1] | 4.911 a | 0.178 a |
| Proteins, g | 67.8[29.3;79.1] | 59.2[42.5;79.7] | 65[51.4;89.3] | 51[29.4;65.1] | 4.782 a | 0.188 a,f |
| Protein Def, n (%) | 9(56.3) | 17(68.0) | 14(70.0) | 19(90.5) | 5.757 b | 0.006 b |
| HCO, g | 151.7[82.4;196.5] | 147.5[101.7;173.6] | 130.1[92.9;190.7] | 112.2[92.1;208.6] | 1.691 a | 0.639 a |
| HCO Def, n (%) | 11(68.8) | 14(56.0) | 12(60.0) | 14(66.7) | 0.902 b | 0.821 b |
| Fiber, g | 10.7[4.1;21] | 7.6[4;15.8] | 6.2[3;14.7] | 10.1[7.4;13.6] | 4.480 a | 0.214 a |
| Iron, mg | 6.8[4.8;10.1] | 7.4[4.3;10] | 7.9[5.9;15.2] | 5.3[4.7;11.8] | 3.603 a | 0.308 a |
| Iron Def, n (%) | 16(100) | 25(100) | 20(100) | 21(100) | --- | --- |
| Sodium, mg | 788.3[183.2;1424.2] | 1083.4[694.6;1804.4] | 771.1[579.8;1952.7] | 506.9[138;1320] | 2.763 a | 0.430 a |
| Na Def, n (%) | 13(81.3) | 16(64.0) | 15(75.0) | 18(85.7) | 3.270 b | 0.352 b |
| Potassium, mg | 609.3[277.4;1130.2] | 286.3[55.8;1082] | 294.5[67.4;458.4] | 701.5[363.9;961.7] | 8.303 a | 0.040 a,f,f |
| K Def, n (%) | 15(93.8) | 24(96.0) | 20(100) | 21(100) | 2.258 b | 0.521 b |
| Calcium, mg | 493.8[18.1;637.5] | 689.1[356.6;945.6] | 567.7[460.1;1007.1] | 373.5[120.1;571.1] | 7.391 a | 0.060 a,e,f |
| Ca Def, n (%) | 15(93.8) | 21(84.0) | 16(80.0) | 21(100) | 5.268 b | 0.153 b |
| Phosphorus, mg | 0[0;0] | 0[0;116.5] | 0[0;0] | 0[0;93.2] | 6.630 a | 0.085 a,f |
| P Def, n (%) | 16(100) | 25(100) | 20(100) | 21(100) | --- | --- |
| Sugar, g | 32.1[17.3;62.3] | 17.2[7.5;39.1] | 7.9[0;18.5] | 17.6[8;63.9] | 11.234 a | 0.011 a,d,f |
| Vitamin A, µg | 232.3[105;553.2] | 290.9[153.7;433.8] | 251.3[166.6;432.2] | 144.8[44.4;412] | 3.070 a | 0.381 a |
| Vit A Def, n (%) | 14(87.5) | 24(96.0) | 17(85.0) | 20(95.2) | 2.434 b | 0.487 b |
| Vitamin B9, µg | 111.1[23.1;214.4] | 74.7[17.2;153.7] | 72.2[29.6;186.2] | 58.8[23.1;157.3] | 2.996 a | 0.392 a |
| Vit B9 Def, n (%) | 15(93.8) | 24(96.0) | 18(90.0) | 21(100) | 2.314 b | 0.510 b |
| Vitamin C, mg | 25.5[14.4;143.4] | 14.3[1.1;149.7] | 20.7[3;65.4] | 24.9[9.6;75] | 3.081 a | 0.379 a |
| Vit CDef, n (%) | 10(62.5) | 18(72.0) | 16(80.0) | 15(71.4) | 1.353 b | 0.717 b |
| Selenium, µg | 32[24.4;55.6] | 24.6[11.8;33.3] | 27.1[19.6;57.1] | 23.1[1.5;46.6] | 4.057 a | 0.255 a |
| Sel Def, n (%) | 16(100) | 25(100) | 20(100) | 21(100) | --- | --- |
| Cholesterol, mg | 120.5[28.8;281.4] | 142.8[53.6;339.4] | 192.5[142.4;309.3] | 87.6[42.1;154.3] | 6.277 a | 0.099 a,f |
| Excess Chol, n (%) | 3(18.8) | 5(20.0) | 4(20.0) | 3(14.3) | 0.316 | 0.957 b |
| SFA, g | 0.7[0;0.8] | 2.3[0;6.3] | 1[0;1.5] | 0[0;5.1] | 7.438 a | 0.059 a,c,e |
| MFA, g | 1.1[0;5.8] | 5.8[0;12] | 4.4[0;5.8] | 0[0;9.3] | 3.752 a | 0.289 a |
| PFA, g | 0.4[0;3] | 3[0;4.6] | 1.4[0;2.8] | 0[0;2.6] | 3.808 a | 0.283 a |
| GI | 437.1[213;538.8] | 378.5[133;420.8] | 253.8[118.6;375.7] | 284[246;747.5] | 3.653 a | 0.302 a |
| GL | 87[40.5;184.2] | 90.9[32.9;94] | 69.9[29.8;93.3] | 80.4[66.7;148] | 2.563 a | 0.463 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tapia-González, A.; Vélez-Ixta, J.M.; Bueno-Hernández, N.; Piña-Escobedo, A.; Briones-Garduño, J.C.; de la Rosa-Ruiz, L.; Aguayo-Guerrero, J.; Mendoza-Martínez, V.M.; Snowball-del-Pilar, L.; Escobedo, G.; et al. Maternal Consumption of Non-Nutritive Sweeteners during Pregnancy Is Associated with Alterations in the Colostrum Microbiota. Nutrients 2023, 15, 4928. https://doi.org/10.3390/nu15234928
Tapia-González A, Vélez-Ixta JM, Bueno-Hernández N, Piña-Escobedo A, Briones-Garduño JC, de la Rosa-Ruiz L, Aguayo-Guerrero J, Mendoza-Martínez VM, Snowball-del-Pilar L, Escobedo G, et al. Maternal Consumption of Non-Nutritive Sweeteners during Pregnancy Is Associated with Alterations in the Colostrum Microbiota. Nutrients. 2023; 15(23):4928. https://doi.org/10.3390/nu15234928
Chicago/Turabian StyleTapia-González, Alejandro, Juan Manuel Vélez-Ixta, Nallely Bueno-Hernández, Alberto Piña-Escobedo, Jesús Carlos Briones-Garduño, Leticia de la Rosa-Ruiz, José Aguayo-Guerrero, Viridiana M. Mendoza-Martínez, Lenin Snowball-del-Pilar, Galileo Escobedo, and et al. 2023. "Maternal Consumption of Non-Nutritive Sweeteners during Pregnancy Is Associated with Alterations in the Colostrum Microbiota" Nutrients 15, no. 23: 4928. https://doi.org/10.3390/nu15234928
APA StyleTapia-González, A., Vélez-Ixta, J. M., Bueno-Hernández, N., Piña-Escobedo, A., Briones-Garduño, J. C., de la Rosa-Ruiz, L., Aguayo-Guerrero, J., Mendoza-Martínez, V. M., Snowball-del-Pilar, L., Escobedo, G., Meléndez-Mier, G., Méndez-García, L. A., García-Mena, J., & Esquivel-Velázquez, M. (2023). Maternal Consumption of Non-Nutritive Sweeteners during Pregnancy Is Associated with Alterations in the Colostrum Microbiota. Nutrients, 15(23), 4928. https://doi.org/10.3390/nu15234928









