Total Sn-2 Palmitic Triacylglycerols and the Ratio of OPL to OPO in Human Milk Fat Substitute Modulated Bile Acid Metabolism and Intestinal Microbiota Composition in Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals and Diets
2.3. Collection of Animal Samples
2.4. Hepatic and Ileal BA Contents
2.5. Protein Level Analysis
2.6. DNA Extraction from Intestinal Contents
2.7. PCR Amplification and Sequencing
2.8. Bioinformatics Analysis
2.9. Statistical Analyses
3. Results
3.1. Hepatic and Ileal BA Contents
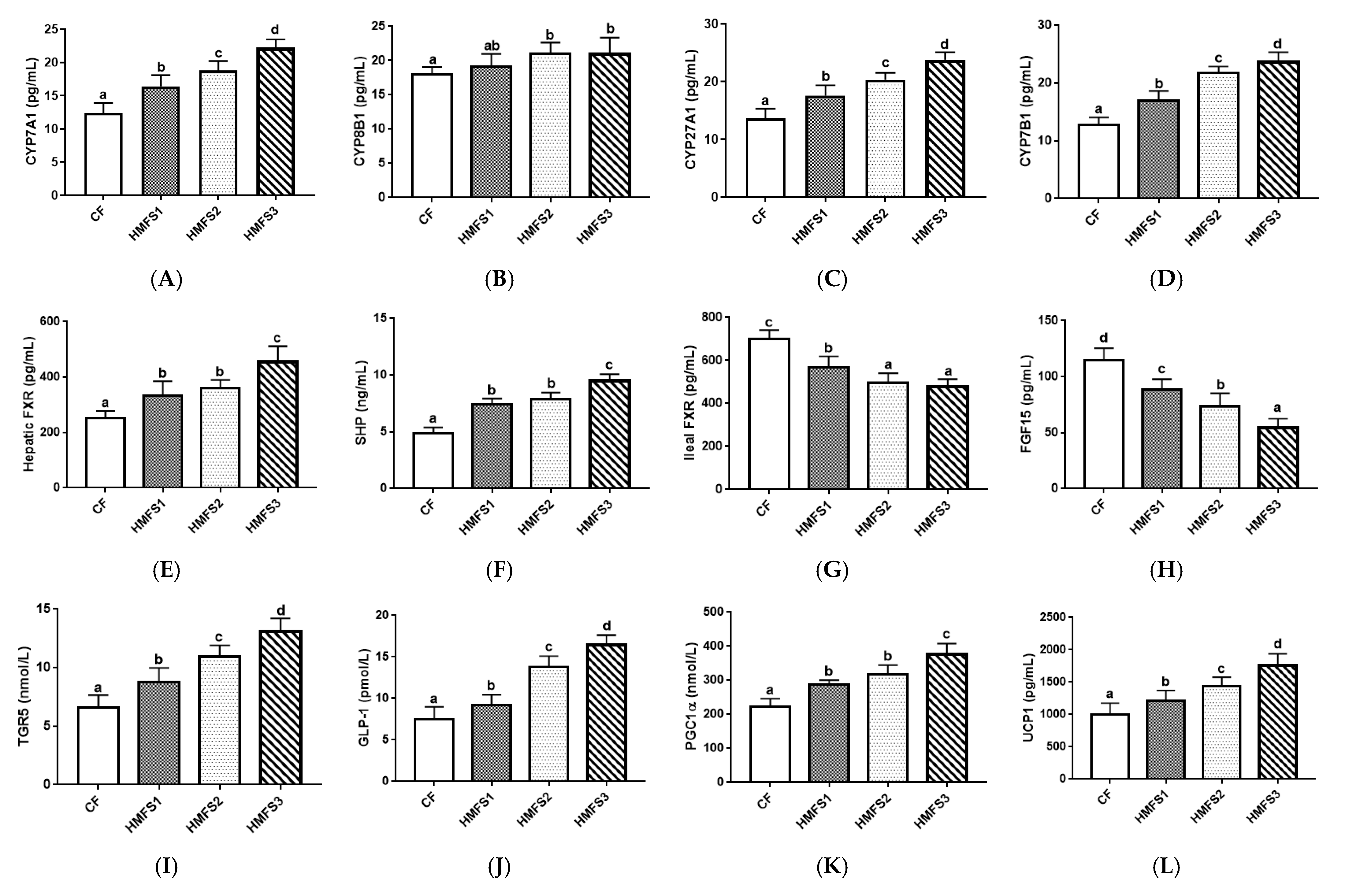
3.2. Protein Levels Related to BA Metabolism
3.3. Gut Microbiota Community
3.3.1. α-Diversity of Gut Microbiota
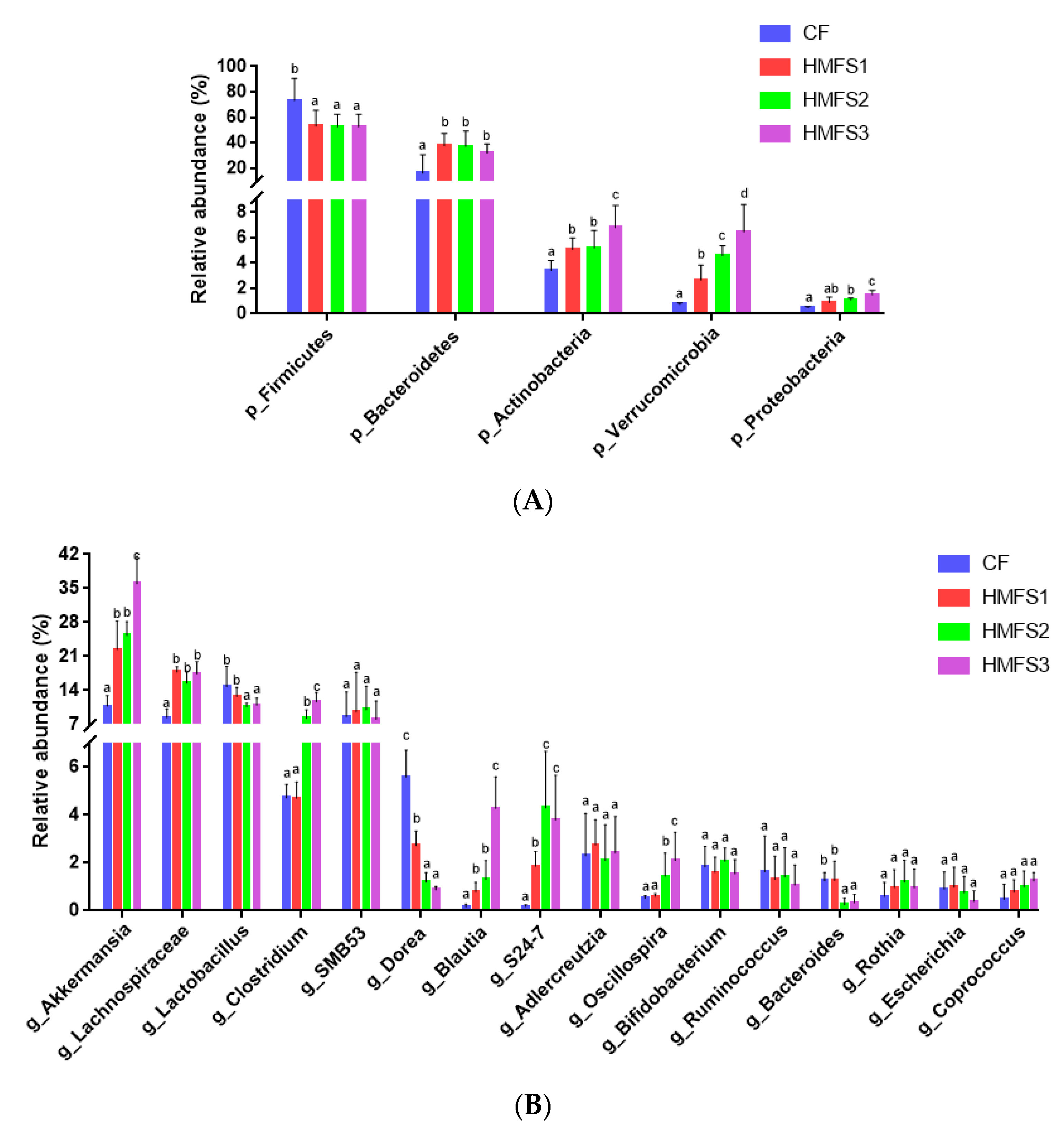
3.3.2. Relative Abundance of Gut Microbiota on Phylum Level
3.3.3. Relative Abundance of Gut Microbiota on Genus Level
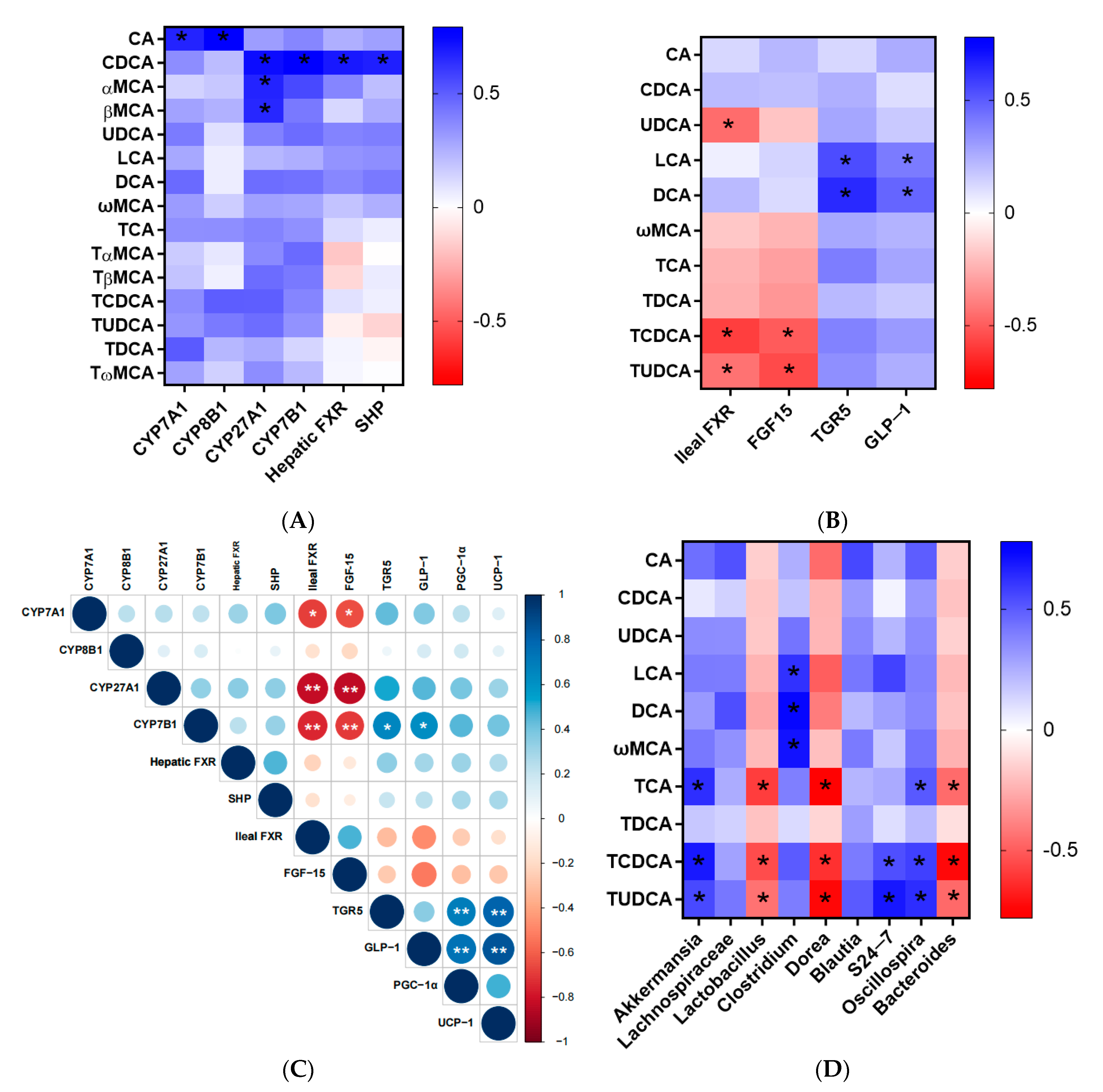
3.4. Correlation Analysis
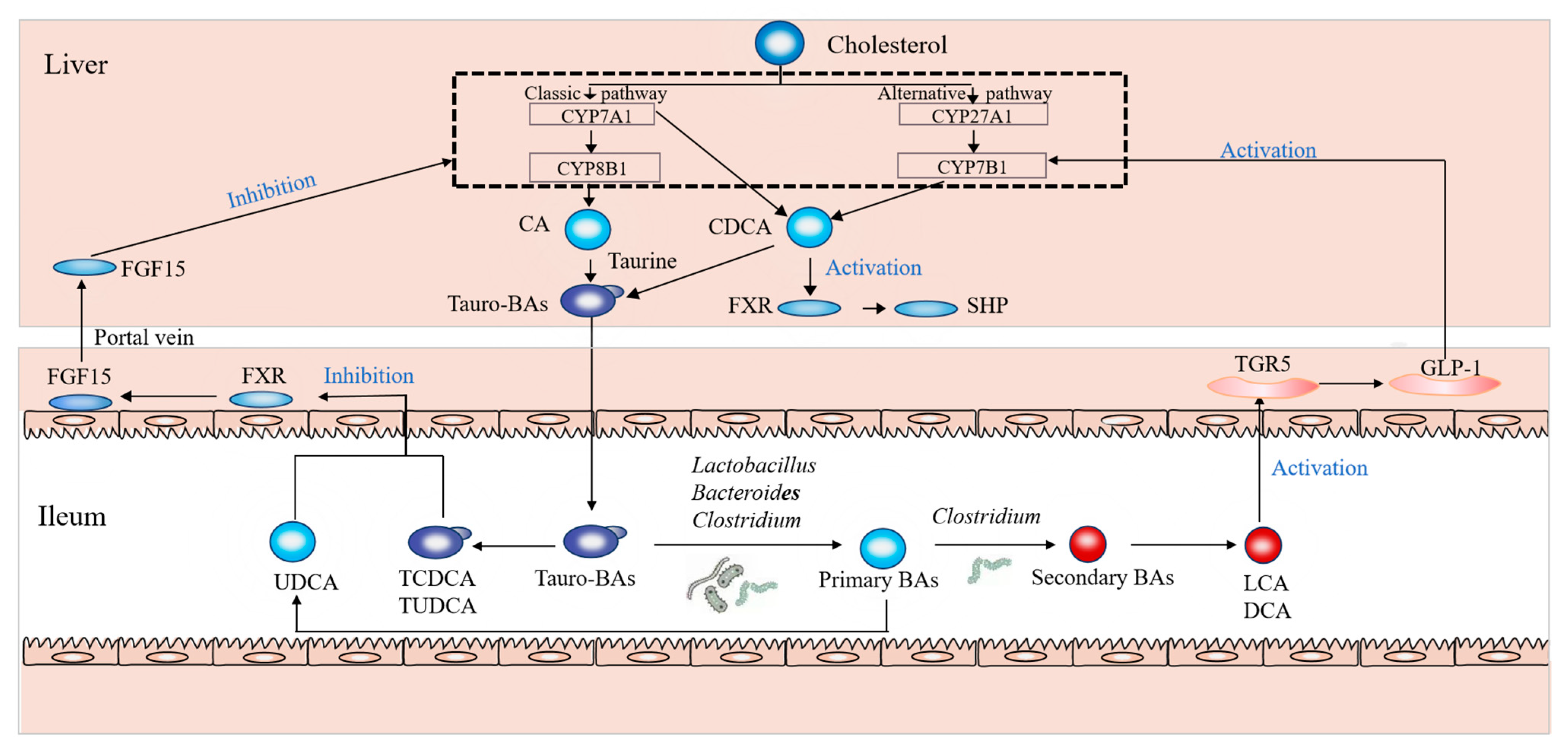
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BA | Bile acid |
| BAs | Bile acids |
| BLASTs | Basic local alignment search tools |
| BSH | Bile salt hydrolase |
| CA | Cholic acid |
| CDCA | Chendoxycholic acid |
| CF | Control fat |
| CYP7A1 | Cholesterol 7α-hydroxylase |
| CYP8B1 | Sterol 12α-hydroxylase |
| CYP27A1 | Sterol-27-hydroxylase |
| CYP7B1 | Oxysterol 7α-hydroxylase |
| DCA | Deoxycholic acid |
| EC | Ethic Committee |
| ELISA | Enzyme-linked Immunosorbent Assay |
| FGF15 | Fibroblast growth factor 15 |
| FXR | Farnesoid X receptor |
| GLP-1 | Glucagon-like peptide-1 |
| HMFS | Human milk fat substitute |
| LCA | Lithocholic acid |
| αMCA | α Muricholic acid |
| βMCA | β Muricholic acid |
| ωMCA | ω Muricholic acid |
| OPL | 1-Oleoyl-2-palmitoyl-3-linoleoylglycerol |
| OPO | 1,3-Dioleoyl-2-palmitoylglycerol |
| OTUs | Operational taxonomic units |
| PA | Palmitic acid |
| PCA | Principal component analysis |
| PGC1α | Peroxisome proliferators-activated receptor γ coactivator l-alpha |
| SD rats | Sprague–Dawley rats |
| SCFA | Short-chain fatty acid |
| SHP | Small heterodimer partner |
| TAGs | Triacyclglycerols |
| TCA | Tauro cholic acid |
| TCDCA | Tauro chendoxycholic acid |
| TDCA | Tauro deoxycholic acid |
| TGR5 | G protein-coupled receptor 5 |
| TαMCA | Tauro α muricholic acid |
| TβMCA | Tauro β muricholic acid |
| TωMCA | Tauro ω muricholic acid |
| TUDCA | Tauro ursodeoxycholic acid |
| UCP1 | Uncoupling protein 1 |
| UDCA | Ursodeoxycholic acid |
| UHPLC/TQ-MS | Ultrahigh performance liquid chromatography/triple quadrupole mass spectrometer |
References
- Ye, Z.; Xu, Y.J.; Liu, Y. Influences of dietary oils and fats, and the accompanied minor content of components on the gut microbiota and gut inflammation: A review. Trends Food Sci. Tech. 2021, 113, 255–276. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Y.; Jacoby, J.J.; Jiang, Y.R.; Zhang, Y.Q.; Yu, L.L. Effects of medium-and long-chain triacylglycerols on lipid metabolism and gut microbiota composition in C57BL/6J mice. J. Agric. Food Chem. 2017, 65, 6599–6607. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Sun, C.; Wang, X.; Jin, Q.; Xu, X.; Akoh, C.C.; Wang, X. Lipase-catalyzed synthesis of sn-2 palmitate: A review. Engineering 2020, 6, 406–414. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Li, Y. Effects of Sn-2-palmitate-enriched formula feeding on infants’ growth, stool characteristics, stool fatty acid soap contents and bone mineral content: A systematic review and meta-analysis of randomized controlled trials. Crit. Rev. Food Sci. 2022, 63, 10256–10266. [Google Scholar] [CrossRef]
- Ghide, M.K.; Yan, Y. 1, 3-Dioleoyl-2-palmitoyl glycerol (OPO)—Enzymatic synthesis and use as an important supplement in infant formulas. J. Food Biochem. 2021, 45, e13799. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Fang, S.Z.; Liu, W.W.; Zhang, H.; Zhang, Y.Q.; Xie, Z.H.; Yang, P.Y.; Wan, J.C.; Gao, B.Y.; Yu, L.L. Triacylglycerol structure and composition of human milk fat substitute affect the absorption of fatty acids and calcium, lipid metabolism and bile acid metabolism in newly-weaned Sprague–Dawley rats. Food Funct. 2023, 14, 7574. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.Y.; Zhang, H.; Wan, J.C.; Hu, J.Y.; Liu, J.C.; Wang, J.; Zhang, Y.Q.; Yu, L.L. Dietary sn-2 palmitic triacylglycerols reduced faecal lipids, calcium contents and altered lipid metabolism in Sprague-Dawley rats. Int. J. Food Sci. Nutr. 2019, 70, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.R.; Haeusler, R.A. Bile acids in glucose metabolism and insulin signalling—Mechanisms and research needs. Nat. Rev. Endocrinol. 2019, 15, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Huang, F.; Zhao, A.; Lei, S.; Zhang, Y.; Xie, G.; Jia, W. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017, 15, 120. [Google Scholar] [CrossRef] [PubMed]
- Bao, Q.; Yu, L.; Chen, D.; Li, L. Variation in the gut microbial community is associated with the progression of liver regeneration. Hepatol. Res. 2020, 50, 121–136. [Google Scholar] [CrossRef]
- Li, W.; Zhang, K.; Yang, H. Pectin alleviates high fat (lard) diet-induced nonalcoholic fatty liver disease in mice: Possible role of short-chain fatty acids and gut microbiota regulated by pectin. J. Agric. Food Chem. 2018, 66, 8015–8025. [Google Scholar] [CrossRef]
- Wan, J.C.; Hu, S.Y.; Ni, K.F.; Chang, G.F.; Sun, X.J.; Yu, L.L. Characterisation of fecal soap fatty acids, calcium contents, bacterial community and short-chain fatty acids in sprague dawley rats fed with different sn-2 palmitic triacylglycerols diets. PLoS ONE 2016, 11, e0164894. [Google Scholar] [CrossRef]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Stephens, R.W.; Arhire, L.; Covasa, M. Gut microbiota: From microorganisms to metabolic organ influencing obesity. Obesity 2018, 26, 801–809. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Qi, L.; Sun, X.; Hu, D. Comparison between the fecal bacterial microbiota of healthy and diarrheic captive musk deer. Front. Microbiol. 2018, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, Y.; Xia, F.; Abudukerimu, B.; Zhang, W.; Guo, Y.; Lu, Y. A glucagon-like peptide-1 receptor agonist lowers weight by modulating the structure of gut microbiota. Front. Endocrinol. 2018, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Kliewer, S.A. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137. [Google Scholar] [CrossRef] [PubMed]
- Li-Hawkins, J.; Gafvels, M.; Olin, M.; Lund, E.G.; Andersson, U.; Schuster, G.; Eggertsen, G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J. Clin. Investig. 2002, 110, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.J.; Zheng, X.J.; Ma, X.H.; Jiang, R.Q.; Zhou, W.Y.; Zhou, S.P.; Zhang, Y.J.; Lei, S.; Wang, S.L.; Kuang, J.L.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef]
- Urizar, N.L.; Liverman, A.B.; Dodds, D.N.T.; Silva, F.V.; Ordentlich, P.; Yan, Y.; Moore, D.D. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 2002, 296, 1703–1706. [Google Scholar] [CrossRef]
- Jia, W.; Wei, M.L.; Rajani, C.; Zheng, X.J. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell 2021, 12, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fujino, M. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, G.; Golovko, A.; Markowitz, L.; Abbondanzo, S.J.; Zeng, M.; Yang, S.; Gustafson, E.L. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem. J. 2006, 398, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lou, G.; Meng, Z.; Huang, W. TGR5: A novel target for weight maintenance and glucose metabolism. Exp. Diabetes Res. 2011, 2011, 853501. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Bioph. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Auwerx, J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W.P. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Santos-Marcos, J.A.; Perez-Jimenez, F.; Camargo, A. The role of diet and intestinal microbiota in the development of metabolic syndrome. J. Nutr. Biochem. 2019, 70, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chassaing, B.; Singh, V.; Pellizzon, M.; Ricci, M.; Fythe, M.D.; Gewirtz, A.T. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe 2018, 23, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Arif, M.; Li, X.; Altay, O.; Yang, H.; Shi, M.; Mardinoglu, A. Multiomics analysis reveals the impact of microbiota on host metabolism in hepatic steatosis. Adv. Sci. 2022, 9, 2104373. [Google Scholar] [CrossRef]
- Anhe, F.F.; Roy, D.; Pilon, G.; Dudonne, S.; Matamoros, S.; Varin, T.V.; Marette, A. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015, 64, 872–883. [Google Scholar] [CrossRef]
- Rao, Y.; Kuang, Z.; Li, C.; Guo, S.; Xu, Y.; Zhao, D.; Lu, Y. Gut Akkermansia muciniphila ameliorates metabolic dysfunction-associated fatty liver disease by regulating the metabolism of L-aspartate via gut-liver axis. Gut Microbes 2021, 13, 1927633. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Rodriguez, J.; Hiel, S.; Neyrinck, A.M.; Le Roy, T.; Potgens, S.A.; Leyrolle, Q.; Delzenne, N.M. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut 2020, 69, 1975–1987. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, Y.; Zhang, R.; Pan, J.; Qi, D.; Wang, J.; Yang, X. The protective effects of walnut green husk polysaccharide on liver injury, vascular endothelial dysfunction and disorder of gut microbiota in high fructose-induced mice. Int. J. Biol. Macromol. 2020, 162, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Yang, B.; Chen, K.; Kong, Y.; Fang, N.; Liu, J. Effect of Clostridium butyricum against microglia-mediated neuroinflammation in Alzheimer’s disease via regulating gut microbiota and metabolites butyrate. Mol. Nutr. Food Res. 2020, 64, 1900636. [Google Scholar] [CrossRef] [PubMed]

| CF | HMFS1 | HMFS2 | HMFS3 | |||
|---|---|---|---|---|---|---|
| Primary BAs | Free BAs | CA | 5.65 a ± 0.73 | 6.81 ab ± 1.09 | 7.29 b ± 1.02 | 7.64 b ± 0.84 |
| CDCA | 2.62 a ± 0.51 | 5.36 b ± 0.88 | 7.08 c ± 1.13 | 10.96 d ± 1.64 | ||
| UDCA | 5.62 a ± 0.90 | 5.89 a ± 1.13 | 6.21 a ± 1.18 | 6.67 a ± 0.84 | ||
| αMCA | 1.37 a ± 0.19 | 3.42 b ± 0.44 | 3.81 b ± 0.69 | 6.70 c ± 1.07 | ||
| βMCA | 2.53 a ± 0.15 | 6.69 b ± 1.31 | 7.23 b ± 0.94 | 10.47 c ± 1.57 | ||
| Tauro-conjugated BAs | TCA | 11.65 a ± 1.51 | 11.98 ab ± 1.82 | 12.29 b ± 1.29 | 13.89 c ± 2.03 | |
| TCDCA | 8.54 a ± 1.11 | 10.76 b ± 2.04 | 11.23 c ± 0.89 | 14.35 d ± 2.44 | ||
| TUDCA | 8.89 a ± 1.42 | 8.96 a ± 1.85 | 9.11 a ± 1.60 | 9.42 a ± 2.08 | ||
| TαMCA | 7.95 a ± 1.03 | 10.11 b ± 1.52 | 16.29 c ± 2.77 | 18.04 d ± 3.60 | ||
| TβMCA | 4.59 a ± 0.67 | 7.34 b ± 1.17 | 11.61 c ± 2.21 | 16.62 d ± 2.33 | ||
| Secondary BAs | Free BAs | DCA | 0.51 a ± 0.05 | 0.77 a ± 0.13 | 1.31 a ± 0.25 | 1.69 a ± 0.29 |
| LCA | 0.85 a ± 0.17 | 1.61 a ± 0.27 | 1.77 a ± 0.35 | 2.08 a ± 0.29 | ||
| ωMCA | 1.49 a ± 0.24 | 1.62 a ± 0.21 | 1.79 a ± 0.30 | 1.91 a ± 0.36 | ||
| Tauro-conjugated BAs | TDCA | 6.35 a ± 0.69 | 6.72 a ± 0.66 | 7.11 a ± 0.72 | 7.29 a ± 0.93 | |
| TωMCA | 5.61 a ± 0.73 | 6.18 ab ± 1.17 | 6.39 b ± 1.06 | 6.94 b ± 0.83 |
| CF | HMFS1 | HMFS2 | HMFS3 | |||
|---|---|---|---|---|---|---|
| Primary BAs | Free BAs | CA | 13.13 a ± 1.46 | 14.92 a ± 3.11 | 16.76 a ± 2.21 | 16.92 a ± 3.29 |
| CDCA | 8.39 a ± 1.05 | 8.87 a ± 1.66 | 10.73 a ± 1.69 | 11.92 a ± 2.01 | ||
| UDCA | 3.74 a ± 0.51 | 9.20 b ± 1.76 | 12.47 bc ± 2.62 | 14.82 c ± 1.98 | ||
| Tauro-conjugated BAs | TCA | 8.56 a ± 1.20 | 16.87 b ± 1.19 | 30.96 c ± 4.49 | 32.58 c ± 2.87 | |
| TCDCA | 6.65 a ± 0.99 | 9.26 b ± 1.16 | 11.44 b ± 2.10 | 25.70 c ± 2.49 | ||
| TUDCA | 3.94 a ± 0.55 | 9.15 b ± 0.62 | 23.03 c ± 2.27 | 34.41 d ± 5.84 | ||
| Secondary BAs | Free BAs | DCA | 6.65 a ± 0.99 | 9.26 a ± 1.16 | 16.44 b ± 3.10 | 19.72 c ± 4.49 |
| LCA | 3.94 a ± 0.55 | 7.95 b ± 1.32 | 11.03 c ± 1.27 | 15.41 d ± 2.84 | ||
| ωMCA | 11.08 a ± 1.72 | 14.78 a ± 3.17 | 16.14 b ± 2.34 | 19.03 b ± 3.24 | ||
| Tauro-conjugated BAs | TDCA | 2.45 a ± 0.32 | 8.64 ab ± 0.39 | 10.81 b ± 2.09 | 11.65 b ± 2.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Fang, S.; Zhang, H.; Sun, X.; Yang, P.; Wan, J.; Zhang, Y.; Lu, W.; Yu, L. Total Sn-2 Palmitic Triacylglycerols and the Ratio of OPL to OPO in Human Milk Fat Substitute Modulated Bile Acid Metabolism and Intestinal Microbiota Composition in Rats. Nutrients 2023, 15, 4929. https://doi.org/10.3390/nu15234929
Zhu L, Fang S, Zhang H, Sun X, Yang P, Wan J, Zhang Y, Lu W, Yu L. Total Sn-2 Palmitic Triacylglycerols and the Ratio of OPL to OPO in Human Milk Fat Substitute Modulated Bile Acid Metabolism and Intestinal Microbiota Composition in Rats. Nutrients. 2023; 15(23):4929. https://doi.org/10.3390/nu15234929
Chicago/Turabian StyleZhu, Lin, Shuaizhen Fang, Hong Zhang, Xiangjun Sun, Puyu Yang, Jianchun Wan, Yaqiong Zhang, Weiying Lu, and Liangli Yu. 2023. "Total Sn-2 Palmitic Triacylglycerols and the Ratio of OPL to OPO in Human Milk Fat Substitute Modulated Bile Acid Metabolism and Intestinal Microbiota Composition in Rats" Nutrients 15, no. 23: 4929. https://doi.org/10.3390/nu15234929
APA StyleZhu, L., Fang, S., Zhang, H., Sun, X., Yang, P., Wan, J., Zhang, Y., Lu, W., & Yu, L. (2023). Total Sn-2 Palmitic Triacylglycerols and the Ratio of OPL to OPO in Human Milk Fat Substitute Modulated Bile Acid Metabolism and Intestinal Microbiota Composition in Rats. Nutrients, 15(23), 4929. https://doi.org/10.3390/nu15234929





