Wheat Grains as a Sustainable Source of Protein for Health
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Determination of Total Seed Protein
2.3. Statistical Analysis
2.4. Genotyping and Marker Quality Control
2.5. GWAS Analysis
2.6. Genes’ Identification, Annotation, and Expression Analysis
3. Results
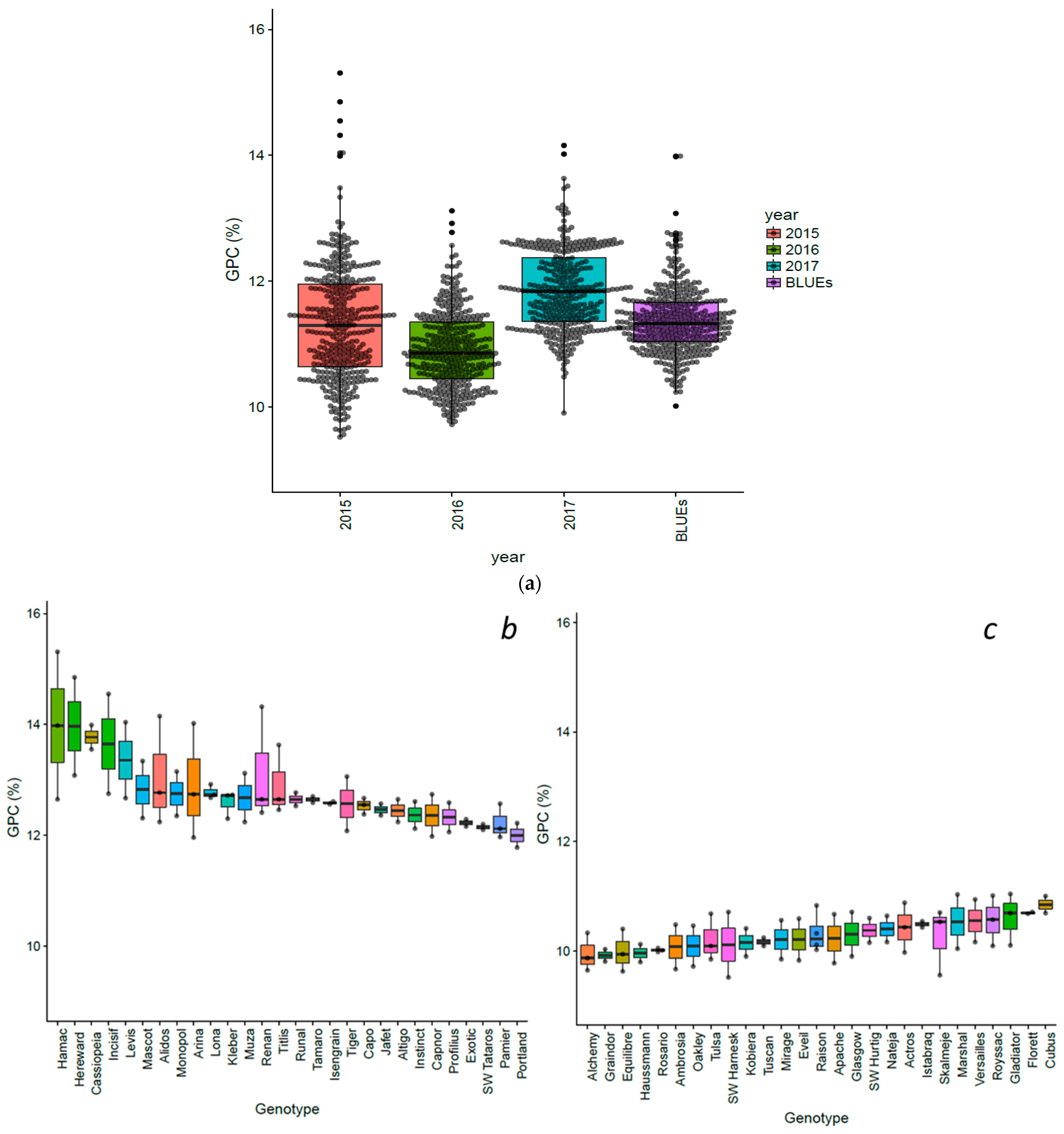
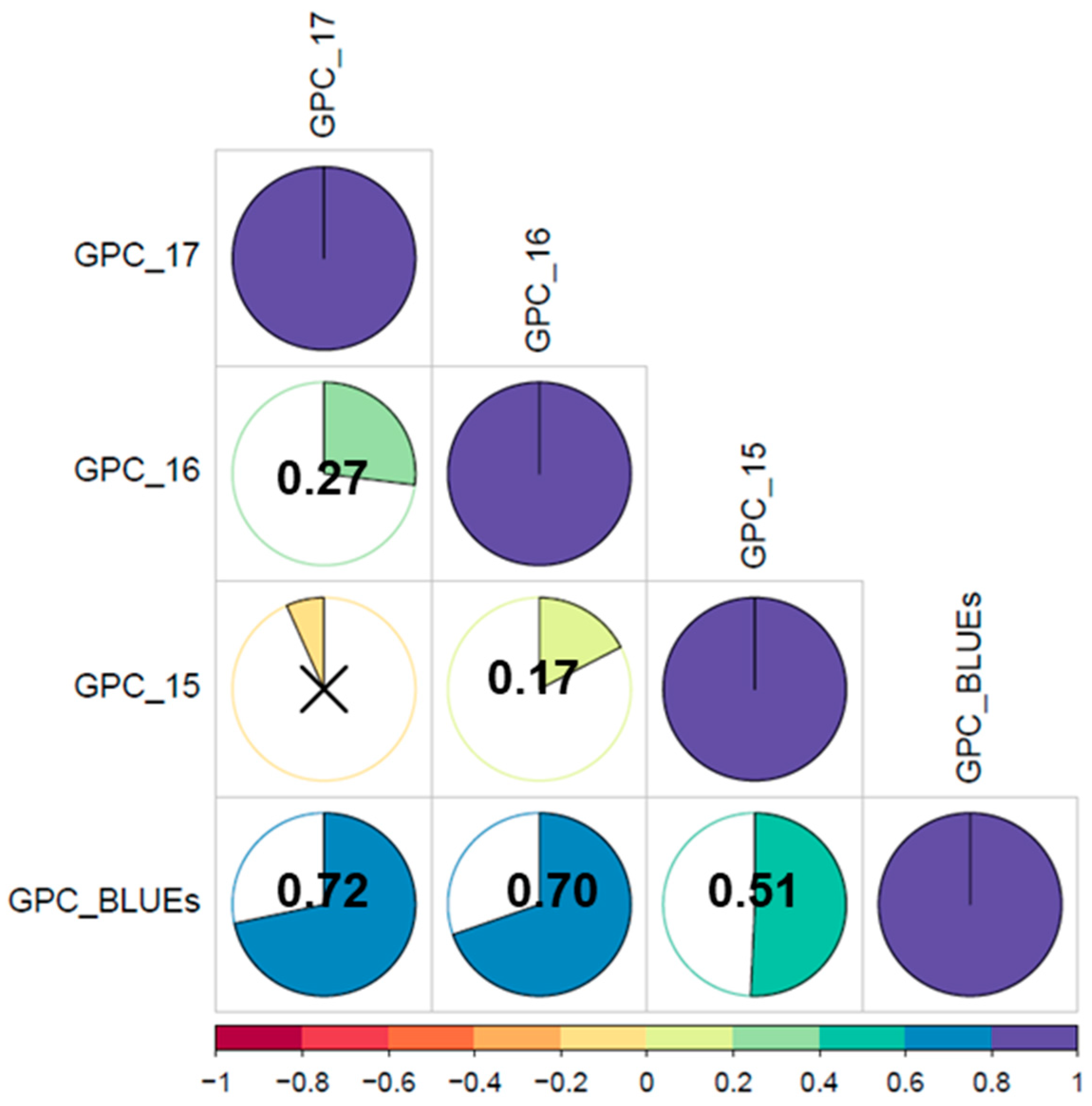
3.1. Variations of Grain Protein Content in a Worldwide Winter Wheat Panel
3.2. QTNs Underlying GPC Variations
3.3. High-Confidence Candidate Genes Related to GPC
4. Discussion
4.1. The Importance of GPC on Bread Wheat, the Main Source of Vegetable Protein Worldwide
4.2. Novel Candidates’ Genes with High Effect on GPC and High Expression during Grain Filling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Willett, W.; Rockström, J.; Loken, B.; Springmann, M.; Lang, T.; Vermeulen, S.; Murray, C.J. Food in the Anthropocene: The Eat–lancet Commission on Healthy Diets from Sustainable Food Systems. Lancet 2019, 393, 447–492. [Google Scholar] [CrossRef]
- Erenstein, O.; Jaleta, M.; Mottaleb, K.A.; Sonder, K.; Donovan, J.; Braun, H.J. Global Trends in Wheat Production, Consumption and Trade; Springer International Publishing: Cham, Switzerland, 2022; pp. 47–66. [Google Scholar]
- Shewry, P.R.; Hey, S.J. The Contribution of Wheat to Human Diet and Health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food Security and the Dynamics of Wheat and Maize Value Chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- Poutanen, K.S.; Kårlund, A.O.; Gómez-Gallego, C.; Johansson, D.P.; Scheers, N.M.; Marklinder, I.M.; Eriksen, A.K.; Silventoinen, P.C.; Nordlund, E.; Sozer, N.; et al. Grains—A major source of sustainable protein for health. Nutr. Rev. 2022, 80, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, A.; Sharma, N.K.; Kaur, N.; Chunduri, V.; Chawla, M.; Sharma, S.; Singh, K.; Garg, M. Soft and hard textured wheat differ in starch properties as indicated by trimodal distribution, morphology, thermal and crystalline properties. PLoS ONE 2016, 11, e0147622. [Google Scholar] [CrossRef] [PubMed]
- Schaible, U.E.; Kaufmann, S.H.E. Malnutrition and infection: Complex mechanisms and global impacts. PLoS Med. 2007, 4, e115. [Google Scholar] [CrossRef] [PubMed]
- Kar, B.R.; Rao, S.L.; Chandramouli, B.A. Cognitive Development in Children with Chronic Protein Energy Malnutrition. Behav. Brain Funct. 2008, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Müller, O.; Krawinkel, M. Malnutrition and health in developing countries. Can. Med. Assoc. J. 2005, 173, 279–286. [Google Scholar] [CrossRef]
- Simón, M.R.; Fleitas, M.C.; Castro, A.C.; Schierenbeck, M. How Foliar Fungal Diseases Affect Nitrogen Dynamics, Milling, and End-Use Quality of Wheat. Front. Plant Sci. 2020, 11, 569401. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Hameed, A.; Tahir, M.F. Wheat Quality: A Review on Chemical Composition, Nutritional Attributes, Grain Anatomy, Types, Classification, and Function of Seed Storage Proteins in Bread Making Quality. Front. Nutr. 2023, 10, 1053196. [Google Scholar] [CrossRef]
- Fleitas, M.C.; Schierenbeck, M.; Gerard, G.S.; Dietz, J.I.; Golik, S.I.; Campos, P.E.; Simón, M.R. How Leaf Rust Disease and Its Control with Fungicides Affect Dough Properties, Gluten Quality and Loaf Volume under Different N Rates in Wheat. J. Cereal Sci. 2018, 80, 119–127. [Google Scholar] [CrossRef]
- Biesiekierski, J.R. What Is Gluten? J. Gastroenterol. Hepatol. 2017, 32 (Suppl. S1), 78–81. [Google Scholar] [CrossRef]
- Laidig, F.; Hüsken, A.; Rentel, D.; Piepho, H.-P. Protein Use Efficiency and Stability of Baking Quality in Winter Wheat Based on the Relation of Loaf Volume and Grain Protein Content. Züchter Genet. Breed. Res. 2022, 135, 1331–1343. [Google Scholar] [CrossRef]
- Ooms, N.; Delcour, J.A. How to Impact Gluten Protein Network Formation during Wheat Flour Dough Making. Curr. Opin. Food Sci. 2019, 25, 88–97. [Google Scholar] [CrossRef]
- Baardseth, P.; Kvaal, K.; Lea, P.; Ellekjær, M.R.; Færgestad, E.M. The Effects of Bread Making Process and Wheat Quality on French Baguettes. J. Cereal Sci. 2000, 32, 73–87. [Google Scholar] [CrossRef]
- Kartseva, T.; Alqudah, A.M.; Aleksandrov, V.; Alomari, D.Z.; Doneva, D.; Arif, M.A.R.; Börner, A.; Misheva, S. Nutritional Genomic Approach for Improving Grain Protein Content in Wheat. Foods 2023, 12, 1399. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Chhuneja, P.; Jaiswal, J.P.; Tamhankar, S.; Mishra, V.K.; Bains, N.S.; Chand, R.; Joshi, A.K.; Kaur, S.; et al. Pyramiding of Genes for Grain Protein Content, Grain Quality, and Rust Resistance in Eleven Indian Bread Wheat Cultivars: A Multi-Institutional Effort. Mol. Breed. 2022, 42, 21. [Google Scholar] [CrossRef] [PubMed]
- Tanin, M.J.; Sharma, A.; Saini, D.K.; Singh, S.; Kashyap, L.; Srivastava, P.; Mavi, G.S.; Kaur, S.; Kumar, V.; Kumar, V.; et al. Ascertaining Yield and Grain Protein Content Stability in Wheat Genotypes Having the Gpc-B1 Gene Using Univariate, Multivariate, and Correlation Analysis. Front. Genet. 2022, 13, 1001904. [Google Scholar] [CrossRef]
- Tshikunde, N.M.; Mashilo, J.; Shimelis, H.; Odindo, A. Agronomic and Physiological Traits, and Associated Quantitative Trait Loci (QTL) Affecting Yield Response in Wheat (Triticum aestivum L.): A Review. Front. Plant Sci. 2019, 10, 1428. [Google Scholar] [CrossRef]
- Schierenbeck, M.; Alqudah, A.M.; Lohwasser, U.; Tarawneh, R.A.; Simón, M.R.; Börner, A. Genetic dissection of grain architecture-related traits in a winter wheat population. BMC Plant Biol. 2021, 21, 417. [Google Scholar]
- Nelson, J.C.; Andreescu, C.; Breseghello, F.; Finney, P.L.; Gualberto, D.G.; Bergman, C.J.; Sorrells, M.E. Quantitative trait locus analysis of wheat quality traits. Euphytica 2006, 149, 145–159. [Google Scholar] [CrossRef]
- Liu, J.; Huang, L.; Wang, C.; Liu, Y.; Yan, Z.; Wang, Z.; Xiang, L.; Zhong, X.; Gong, F.; Zheng, Y.; et al. Genome-wide association study reveals novel genomic regions associated with high grain protein content in wheat lines derived from wild emmer wheat. Front. Plant Sci. 2019, 10, 464. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and Predictive Breeding for European Winter Wheat’s Grain Protein Content, Grain Starch Content, and Grain Hardness. Sci. Rep. 2020, 10, 12541. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.; Suliman, S.; Hagras, A.; Thabet, S.; Al-Abdallat, A.; Abdelmula, A.A.; Tadesse, W. Multi-model genome-wide association and genomic prediction analysis of 16 agronomic, physiological and quality related traits in ICARDA spring wheat. Euphytica 2021, 217, 205. [Google Scholar] [CrossRef]
- Kumar, A.; Jain, S.; Elias, E.M.; Ibrahim, M.; Sharma, L.K. An Overview of QTL Identification and Marker-Assisted Selection for Grain Protein Content in Wheat. In Eco-Friendly Agro-Biological Techniques for Enhancing Crop Productivity; Sengar, R., Singh, A., Eds.; Springer: Singapore, 2018; pp. 245–274. [Google Scholar]
- Rathan, N.D.; Krishna, H.; Ellur, R.K.; Sehgal, D.; Govindan, V.; Ahlawat, A.K.; Krishnappa, G.; Jaiswal, J.P.; Singh, J.B.; Sv, S.; et al. Genome-Wide Association Study Identifies Loci and Candidate Genes for Grain Micronutrients and Quality Traits in Wheat (Triticum aestivum L.). Sci. Rep. 2022, 12, 7037. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Mohler, V.; Hartl, L. Genetics of the Inverse Relationship between Grain Yield and Grain Protein Content in Common Wheat. Plants 2022, 11, 2146. [Google Scholar] [CrossRef]
- Alomari, D.; Eggert, K.; von Wirén, N.; Polley, A.; Plieske, J.; Ganal, M.; Liu, F.; Pillen, K.; Röder, M. Whole-Genome Association Mapping and Genomic Prediction for Iron Concentration in Wheat Grains. Int. J. Mol. Sci. 2018, 20, 76. [Google Scholar] [CrossRef]
- Alomari, D.Z.; Alqudah, A.M.; Pillen, K.; von Wirén, N.; Röder, M.S. Toward Identification of a Putative Candidate Gene for Nutrient Mineral Accumulation in Wheat Grains for Human Nutrition Purposes. J. Exp. Bot. 2021, 72, 6305–6318. [Google Scholar] [CrossRef]
- Alomari, D.Z.; Eggert, K.; von Wirén, N.; Pillen, K.; Röder, M.S. Genome-Wide Association Study of Calcium Accumulation in Grains of European Wheat Cultivars. Front. Plant Sci. 2017, 8, 1797. [Google Scholar] [CrossRef]
- Delwiche, S.R.; Pierce, R.O.; Chung, O.K.; Seabourn, B.W. Protein Content of Wheat by Near-Infrared Spectroscopy of Whole Grain: Collaborative Study. J. AOAC Int. 1998, 81, 587–603. [Google Scholar] [CrossRef][Green Version]
- VSN International. Genstat for Windows, 18th ed.; VSN International Ltd.: Hemel Hempstead, UK, 2015. [Google Scholar]
- Falconer, D.S.; Mackay, T.F. Introduction to Quantitative Genetics, 4th ed.; Pearson Prentice Hall: Hoboken, NJ, USA, 2005. [Google Scholar]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E. Akhunov Characterization of Polyploid Wheat Genomic Diversity Using a High-Density 90 000 Single Nucleotide Polymorphism Array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Allen, A.M.; Winfield, M.O.; Burridge, A.J.; Downie, R.C.; Benbow, H.R.; Barker, G.L.A.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; et al. Characterization of a Wheat Breeders’ Array Suitable for High-Throughput SNP Genotyping of Global Accessions of Hexaploid Bread Wheat (Triticum Aestivum). Plant Biotechnol. J. 2017, 15, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Sorrells, M.E.; Gustafson, J.P.; Somers, D.; Chao, S.; Benscher, D.; Guedira-Brown, G. Qualset Reconstruction of the Synthetic W7984× Opata M85 Wheat Reference Population. Genome 2011, 54, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dong, C.; Chen, Z.; Gui, L.; Chen, C.; Li, D.; Xie, Z.; Zhang, Q.; Zhang, X.; Xia, C.; et al. WheatGmap: A comprehensive platform for wheat gene mapping and genomic studies. Mol. Plant 2021, 14, 187–190. [Google Scholar] [CrossRef]
- Saint Pierre, C.; Peterson, C.J.; Ross, A.S.; Ohm, J.B.; Verhoeven, M.C.; Larson, M.; Hoefer, B. Winter Wheat Genotypes under Different Levels of Nitrogen and Water Stress: Changes in Grain Protein Composition. J. Cereal Sci. 2008, 47, 407–416. [Google Scholar] [CrossRef]
- Ross, A.S. Genetic and Other Factors Affecting Wheat Quality. In Achieving Sustainable Cultivation of Wheat; Burleigh Dodds Science Publishing: Cambridge, UK, 2017; Volume 1, pp. 233–268. [Google Scholar]
- Doroodian, P.; Hua, Z. The ubiquitin switch in plant stress response. Plants 2021, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Xie, M.; Hu, M.; Cui, X.; Wu, H.; Li, X.; Hu, P.; Tong, C.; Yu, X. Genome-Wide Characterization of Ubiquitin-Conjugating Enzyme Gene Family Explores Its Genetic Effects on the Oil Content and Yield of Brassica Napus. Front. Plant Sci. 2023, 14, 1118339. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y. Ubiquitin-mediated control of seed size in plants. Front. Plant Sci. 2014, 5, 332. [Google Scholar] [CrossRef]
- Brinton, J.; Simmonds, J.; Uauy, C. Ubiquitin-related genes are differentially expressed in isogenic lines contrasting for pericarp cell size and grain weight in hexaploid wheat. BMC Plant Biol. 2018, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Chen, N.; Dang, Y.; Wang, Y.; Wen, H.; Zheng, J.; Zheng, X.; Zhao, J.; Lu, J.; Qiao, L. Identification and Validation of Quantitative Trait Loci for Chlorophyll Content of Flag Leaf in Wheat under Different Phosphorus Supply. Front. Plant Sci. 2022, 13, 1019012. [Google Scholar] [CrossRef]
- Grasser, M.; Kane, C.M.; Merkle, T.; Melzer, M.; Emmersen, J.; Grasser, K.D. Transcript Elongation Factor TFIIS Is Involved in Arabidopsis Seed Dormancy. J. Mol. Biol. 2009, 386, 598–611. [Google Scholar] [CrossRef]
- Obermeyer, S.; Stöckl, R.; Schnekenburger, T.; Kapoor, H.; Stempfl, T.; Schwartz, U.; Grasser, K.D. TFIIS Is Crucial during Early Transcript Elongation for Transcriptional Reprogramming in Response to Heat Stress. J. Mol. Biol. 2023, 435, 167917. [Google Scholar] [CrossRef]
- Da Costa e Silva, O.; Lorbiecke, R.; Garg, P.; Müller, L.; Waßmann, M.; Lauert, P.; Scanlon, M.; Hsia, A.P.; Schnable, P.S.; Krupinska, K.; et al. The Etched1 Gene of Zea mays (L.) Encodes a Zinc Ribbon Protein That Belongs to the Transcriptionally Active Chromosome (TAC) of Plastids and Is Similar to the Transcription Factor TFIIS. Plant J. 2004, 38, 923–939. [Google Scholar] [CrossRef]
- Dumont, M.; Lehner, A.; Bouton, S.; Kiefer-Meyer, M.C.; Voxeur, A.; Pelloux, J.; Lerouge, P.; Mollet, J.-C. The Cell Wall Pectic Polymer Rhamnogalacturonan-II Is Required for Proper Pollen Tube Elongation: Implications of a Putative Sialyltransferase-like Protein. Ann. Bot. 2014, 114, 1177–1188. [Google Scholar] [CrossRef]
- Cascallares, M.; Setzes, N.; Marchetti, F.; López, G.A.; Distéfano, A.M.; Cainzos, M.; Zabaleta, E.; Pagnussat, G.C. A Complex Journey: Cell Wall Remodeling, Interactions, and Integrity during Pollen Tube Growth. Front. Plant Sci. 2020, 11, 599247. [Google Scholar] [CrossRef]
- Rodríguez-Salazar, J.; Suárez, R.; Caballero-Mellado, J.; Iturriaga, G. Trehalose Accumulation in Azospirillum Brasilense Improves Drought Tolerance and Biomass in Maize Plants. FEMS Microbiol. Lett. 2009, 296, 52–59. [Google Scholar] [CrossRef]
- Griffiths, C.A.; Sagar, R.; Geng, Y.; Primavesi, L.F.; Patel, M.K.; Passarelli, M.K.; Gilmore, I.S.; Steven, R.T.; Bunch, J.; Paul, M.J.; et al. Chemical intervention in plant sugar signalling increases yield and resilience. Nature 2016, 540, 574–578. [Google Scholar] [CrossRef]
- Paul, M.J.; Watson, A.; Griffiths, C.A. Trehalose 6-Phosphate Signalling and Impact on Crop Yield. Biochem. Soc. Trans. 2020, 48, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, S.; Ding, L.; Cheng, X.; Kang, Z.; Mao, H. Genome-wide analysis of trehalose-6-phosphate phosphatases (TPP) gene family in wheat indicates their roles in plant development and stress response. BMC Plant Biol. 2022, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Louis, J.; Ayre, B.G.; Reese, J.C.; Pegadaraju, V.; Shah, J. TREHALOSE PHOSPHATE SYNTHASE11-Dependent Trehalose Metabolism Promotes Arabidopsis Thaliana Defense against the Phloem-Feeding Insect Myzus Persicae: Trehalose Signaling in Plant Defense. Plant J. 2011, 67, 94–104. [Google Scholar] [CrossRef]
- Huda, K.M.K.; Yadav, S.; Banu, M.S.A.; Trivedi, D.K.; Tuteja, N. Genome-Wide Analysis of Plant-Type II Ca2+ ATPases Gene Family from Rice and Arabidopsis: Potential Role in Abiotic Stresses. Plant Physiol. Biochem. 2013, 65, 32–47. [Google Scholar] [CrossRef]
- Hoffmann, R.D.; Portes, M.T.; Olsen, L.I.; Damineli, D.S.C.; Hayashi, M.; Nunes, C.O.; Pedersen, J.T.; Lima, P.T.; Campos, C.; Feijó, J.A.; et al. Plasma Membrane H+-ATPases Sustain Pollen Tube Growth and Fertilization. Nat. Commun. 2020, 11, 2395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wu, H.; Xu, W.; Zhang, W.; Chen, X.; Zhu, Y.; Chen, H.; Zeng, H. Genome-wide identification, characterization, and expression analyses of P-type ATPase superfamily genes in soybean. Agronomy 2020, 11, 71. [Google Scholar] [CrossRef]
- Taneja, M.; Upadhyay, S.K. Molecular Characterization and Differential Expression Suggested Diverse Functions of P-Type II Ca2+ ATPases in Triticum aestivum L. BMC Genom. 2018, 19, 389. [Google Scholar] [CrossRef]
- Louriki, S.; Rehman, S.; El Hanafi, S.; Bouhouch, Y.; Al-Jaboobi, M.; Amri, A.; Douira, A.; Tadesse, W. Identification of Resistance Sources and Genome-Wide Association Mapping of Septoria Tritici Blotch Resistance in Spring Bread Wheat Germplasm of ICARDA. Front. Plant Sci. 2021, 12, 600176. [Google Scholar] [CrossRef]
- Liu, Y.; Imai, R. Function of Plant DExD/H-Box RNA Helicases Associated with Ribosomal RNA Biogenesis. Front. Plant Sci. 2018, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, H.; Zhang, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef]
- Tuteja, N.; Sahoo, R.K.; Garg, B.; Tuteja, R. O s SUV 3 Dual Helicase Functions in Salinity Stress Tolerance by Maintaining Photosynthesis and Antioxidant Machinery in Rice (O Ryza Sativa L. Cv. IR 64). Plant J. 2013, 76, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.; Sement, F.M.; Gagliardi, D. MTR4, a Putative RNA Helicase and Exosome Co-Factor, Is Required for Proper RRNA Biogenesis and Development in Arabidopsis Thaliana: Role of AtMTR4 in RRNA Biogenesis. Plant J. 2011, 68, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.; Zuber, H.; Sement, F.M.; Chicher, J.; Kuhn, L.; Hammann, P.; Brunaud, V.; Bérard, C.; Bouteiller, N.; Balzergue, S.; et al. The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in Arabidopsis Thaliana. PLoS Genet. 2014, 10, e1004564. [Google Scholar] [CrossRef]
- Guo, B.; Jin, X.; Chen, J.; Xu, H.; Zhang, M.; Lu, X.; Wu, R.; Zhao, Y.; Guo, Y.; An, Y.; et al. ATP-Dependent DNA Helicase (TaDHL), a Novel Reduced-Height (Rht) Gene in Wheat. Genes 2022, 13, 979. [Google Scholar] [CrossRef]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; et al. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef]



| Env | Chr | Marker | Effect (%GPC) | −log10 | Position (bp) | Candidate Gene and Genomic Location (bp) | Annotation (Superfamily and PANTHER) |
|---|---|---|---|---|---|---|---|
| 2017 | 1A | AX-94392216 | −0.344 | 4.59 | 22611655 | TraesCS1A02G041100 (22612227-2614080) RGA5 gene | P-loop containing nucleoside triphosphate hydrolase |
| 2017 | 1A | AX-158560740 | −0.266 | 4.17 | 27275836 | NA | |
| blue | 1A | AX-158556547 | 0.119 | 5.58 | 476981928 | TraesCS1A02G279600 (476972557-476981741) | Josephin domain |
| 2015 | 1A | RAC875_c46551_339 | 0.172 | 5.04 | 506283718 | TraesCS1A02G314400 (506281816-506286607) | Homeobox-like domain superfamily |
| 2015 | 1A | IAAV6234 | 0.154 | 4.11 | 513893374 | TraesCS1A02G323500 (513879955-513894399) | P-loop containing nucleoside triphosphate hydrolase; DNA/RNA polymerase superfamily |
| blue | 2B | AX-158536988 | −0.123 | 4.47 | 122713731 | TraesCS2B02G154500 (122710128-122714573) | Protein Rolling Stone-like |
| 2016 | 3A | AX-94451685 | 0.240 | 4.22 | 14045695 | TraesCS3A02G026800 (14045083-14049309) | DEK C-terminal domain |
| 2016 | 3A | AX-94486651 | 0.240 | 4.31 | 14045732 | ||
| 2016 | 3A | Excalibur_c10383_432 | 0.240 | 4.31 | 14047699 | ||
| 2016 | 3A | Excalibur_c11505_155 | 0.240 | 4.31 | 14850594 | TraesCS3A02G027700 (14848772-14852646) | Tetratricopeptide-like helical domain superfamily |
| 2016 | 3A | RAC875_c20134_535 | 0.240 | 4.31 | 14851011 | ||
| 2016 | 3A | IAAV1155 | 0.240 | 4.31 | 14851251 | ||
| 2016 | 3A | Excalibur_c92401_157 | 0.240 | 4.31 | 15089050 | TraesCS3A02G028300 (15086445-15089436) | Alpha-ketoglutarate-dependent dioxygenase AlkB-like superfamily |
| 2016 | 3A | CAP11_c6193_232 | 0.239 | 4.20 | 15090085 | TraesCS3A02G028400 (15089868-15092852) | A0A077RAM9 (hypothetical protein wheat) |
| blue | 3A | BS00065734_51 | 0.228 | 4.90 | 711095135 | NA | |
| 2017 | 3A | BS00065734_51 | 0.485 | 4.03 | 711095135 | NA | |
| 2016 | 3B | AX-108848182 | −0.213 | 4.46 | 511035835 | TraesCS3B02G317300 (511034602-511051546) | Peptidase S8/S53 domain superfamily |
| 2016 | 3B | AX-158537019 | −0.220 | 4.69 | 511074018 | TraesCS3B02G317600 (511072080-511076195) | Galactose-binding-like domain superfamily |
| 2016 | 3B | AX-111060338 | −0.214 | 4.42 | 511507665 | NA | |
| 2016 | 3B | AX-158538466 | −0.214 | 4.35 | 519416654 | TraesCS3B02G320500 (519415064-519417176) | ATPase, nucleotide binding domain |
| 2016 | 3B | AX-158558088 | −0.204 | 4.04 | 522280255 | NA | |
| 2016 | 3B | AX-110467694 | −0.220 | 4.59 | 524450613 | TraesCS3B02G323900 (524449173-52445430) | UDP-Glycosyltransferase/glycogen phosphorylase HAD-like superfamily |
| 2017 | 4A | AX-108845109 | 0.642 | 5.18 | 712225082 | NA | |
| blue | 5A | AX-158542530 | 0.156 | 5.64 | 382113600 | NA | |
| 2017 | 5A | AX-94552678 | 0.501 | 4.51 | 613543528 | TraesCS5A02G429000 (613543346-613547572) UBC2 Gene | Ubiquitin-conjugating enzyme/RWD-like |
| 2017 | 5A | BobWhite_rep_c64315_180 | 0.501 | 4.51 | 613543528 | ||
| 2017 | 5A | AX-109292583 | 0.515 | 4.44 | 613544399 | ||
| blue | 5B | AX-158525605 | −0.156 | 5.88 | 488112608 | TraesCS5B02G303800 (488111479-488113567) | Polyketide synthase, enoylreductase domain |
| 2016 | 6A | AX-158552362 | 0.305 | 4.19 | 10493939 | TraesCS6A02G021300 (10491634-10496460) | Sam-dependent Methyltransferase |
| 2016 | 6A | AX-108894863 | 0.313 | 4.27 | 10494137 | ||
| 2016 | 6A | RAC875_c22627_315 | 0.295 | 5.15 | 10560290 | TraesCS6A02G021600 (10559982-10561177) | Uncharacterized protein |
| 2016 | 6A | AX-95007092 | 0.301 | 4.45 | 11965153 | TraesCS6A02G024000 (11963972-11966414) | C2H2 zinc finger transcription factor |
| 2016 | 6A | AX-111512288 | −0.277 | 4.70 | 12058265 | TraesCS6A02G024100 (12055217-12058516) | LRRNT_2 domain-containing protein |
| 2016 | 6A | AX-110469066 | −0.287 | 4.83 | 12078919 | TraesCS6A02G024200 (12077818-12079854) | OS10G0469600 PROTEIN |
| 2016 | 6A | AX-158588344 | −0.262 | 4.03 | 12312937 | TraesCS6A02G024800 (12313087-12314289) | F-box domain-containing protein |
| 2015 | 6A | BS00073124_51 | −0.220 | 4.19 | 57728595 | TraesCS6A02G089400 (57725544-57732746) | Calcium-dependent protein kinase 16 |
| 2016 | 6A | AX-158530854 | −0.456 | 4.42 | 571851398 | TraesCS6A02G338300 (571851256-571855462) | E3 ubiquitin ligase |
| 2016 | 6A | AX-110545207 | −0.456 | 4.42 | 571851707 | ||
| 2016 | 6A | AX-94973054 | −0.502 | 4.68 | 571852988 | ||
| 2016 | 6A | Tdurum_contig46828_730 | −0.502 | 4.68 | 571929129 | TraesCS6A02G338600 (571928076-571931078) | Aminotran_1_2 domain-containing protein |
| 2017 | 6B | RAC875_c18659_651 | 0.281 | 4.63 | 48348762 | TraesCS6B02G071500 (48347795-48354269) | Transcription elongation factor S-II, central domain superfamily |
| 2017 | 6B | wsnp_Ku_c8343_14190318 | 0.281 | 4.63 | 48348762 | ||
| 2017 | 6B | wsnp_Ex_c8011_13584847 | 0.286 | 4.73 | 48349076 | ||
| 2017 | 6B | wsnp_Ex_c13352_21044607 | 0.286 | 4.73 | 48352481 | ||
| 2017 | 6B | AX-95155979 | 0.286 | 4.73 | 48414777 | TraesCS6B02G071700 (48411470-48415030) | Sialyltransferase-like protein 2 |
| blue | 6B | CAP7_rep_c6771_332 | 0.146 | 6.41 | 49984647 | TraesCS6B02G073600 (49984316-49986442) | Protein lurp-one-related 1-related |
| 2017 | 6B | AX-94727470 | 0.472 | 4.34 | 659234876 | TraesCS6B02G384500 (659232852-659237118) TPS11 gene | Trehalose 6-phosphate phosphatase |
| 2017 | 6B | AX-158588655 | 0.539 | 4.16 | 665512073 | TraesCS6B02G391200 (665512071-665516262) | ATP-dependent RNA Helicase DDX51 |
| blue | 6D | RAC875_c64099_90 | −0.105 | 4.94 | 460570647 | TraesCS6D02G377900 (460567567-460573565) | Protein lutein deficient 5, chloroplastic |
| 2017 | 7A | AX-95203767 | 0.348 | 4.54 | 1281744 | TraesCS7A02G001700 (1280349-1295461) | Terpene cyclase/mutase family member |
| 2015 | 7A | wsnp_Ra_c4418_8012732 | 0.293 | 4.51 | 118156309 | TraesCS7A02G161500 (118145757-118159311) | PPR_long domain-containing protein |
| 2017 | 7A | BS00022169_51 | 0.283 | 4.04 | 691259601 | TraesCS7A02G501500 (691258415-691260048) | OS10G0469600 Protein |
| 2017 | 7A | AX-158567041 | 0.276 | 4.01 | 691474553 | TraesCS7A02G502400 (691472525-691473807) | Peptidase A1 domain-containing protein |
| 2016 | 7B | AX-94830265 | 0.525 | 4.23 | 47000619 | TraesCS7B02G047600 (47000524-47004008) | Plasma membrane ATPase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alomari, D.Z.; Schierenbeck, M.; Alqudah, A.M.; Alqahtani, M.D.; Wagner, S.; Rolletschek, H.; Borisjuk, L.; Röder, M.S. Wheat Grains as a Sustainable Source of Protein for Health. Nutrients 2023, 15, 4398. https://doi.org/10.3390/nu15204398
Alomari DZ, Schierenbeck M, Alqudah AM, Alqahtani MD, Wagner S, Rolletschek H, Borisjuk L, Röder MS. Wheat Grains as a Sustainable Source of Protein for Health. Nutrients. 2023; 15(20):4398. https://doi.org/10.3390/nu15204398
Chicago/Turabian StyleAlomari, Dalia Z., Matías Schierenbeck, Ahmad M. Alqudah, Mashael Daghash Alqahtani, Steffen Wagner, Hardy Rolletschek, Ljudmilla Borisjuk, and Marion S. Röder. 2023. "Wheat Grains as a Sustainable Source of Protein for Health" Nutrients 15, no. 20: 4398. https://doi.org/10.3390/nu15204398
APA StyleAlomari, D. Z., Schierenbeck, M., Alqudah, A. M., Alqahtani, M. D., Wagner, S., Rolletschek, H., Borisjuk, L., & Röder, M. S. (2023). Wheat Grains as a Sustainable Source of Protein for Health. Nutrients, 15(20), 4398. https://doi.org/10.3390/nu15204398





