Cholecystectomy Significantly Alters Gut Microbiota Homeostasis and Metabolic Profiles: A Cross-Sectional Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.1.1. Open Database Search
2.1.2. CCE Volunteer Recruitment
2.1.3. Inclusion and Exclusion Criteria
2.2. Dietary Assessment and Questionnaire Processing
2.3. Untargeted Metabolomics
2.4. Determination of Fecal SCFA Contents
2.5. Determination of BA Levels in Fecal Samples
2.6. Determination of Calprotectin Levels in the Fecal Samples
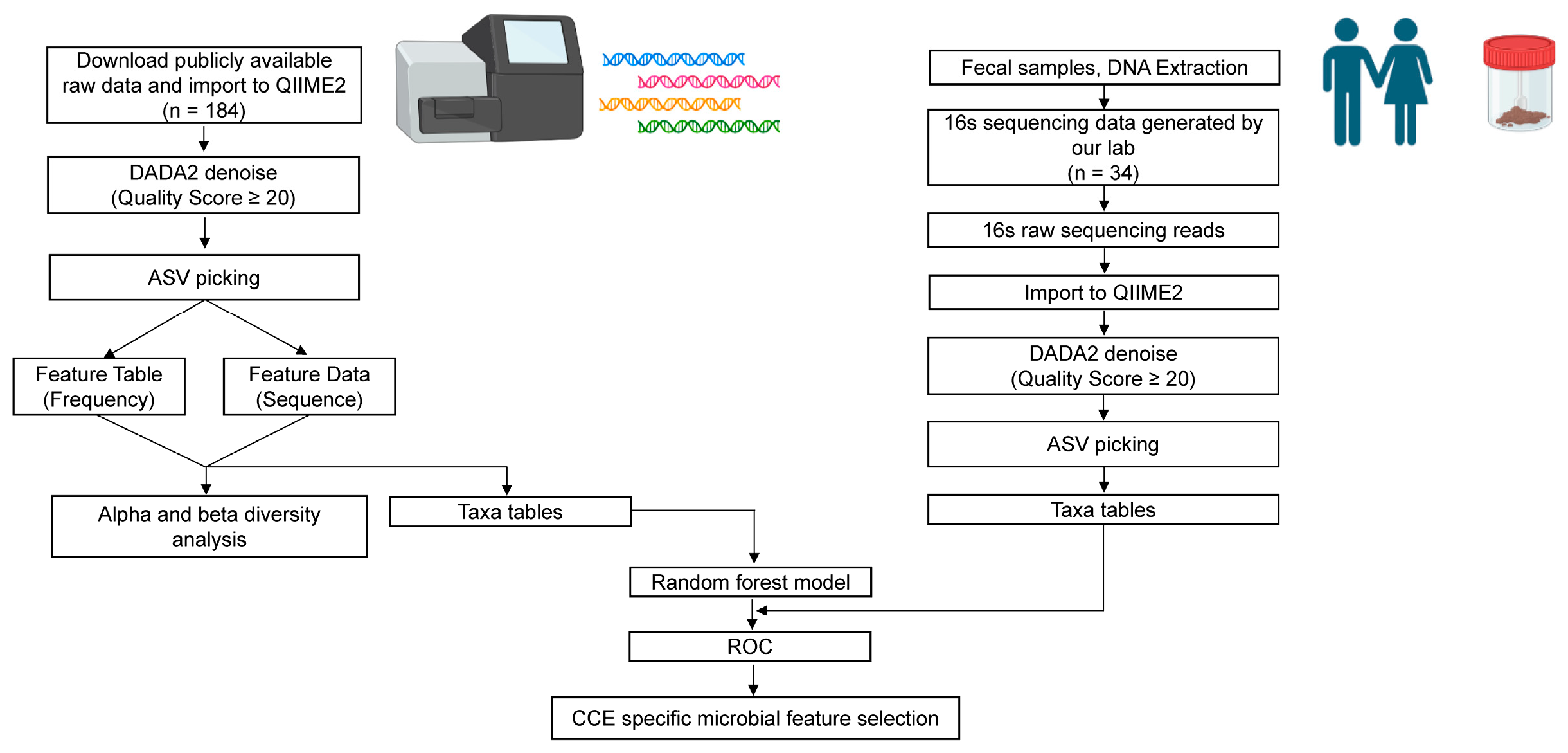
2.7. Data Sources
2.8. 16S rRNA Data Processing
2.9. Microbial Community Analysis
2.10. Statistical Analysis
3. Results
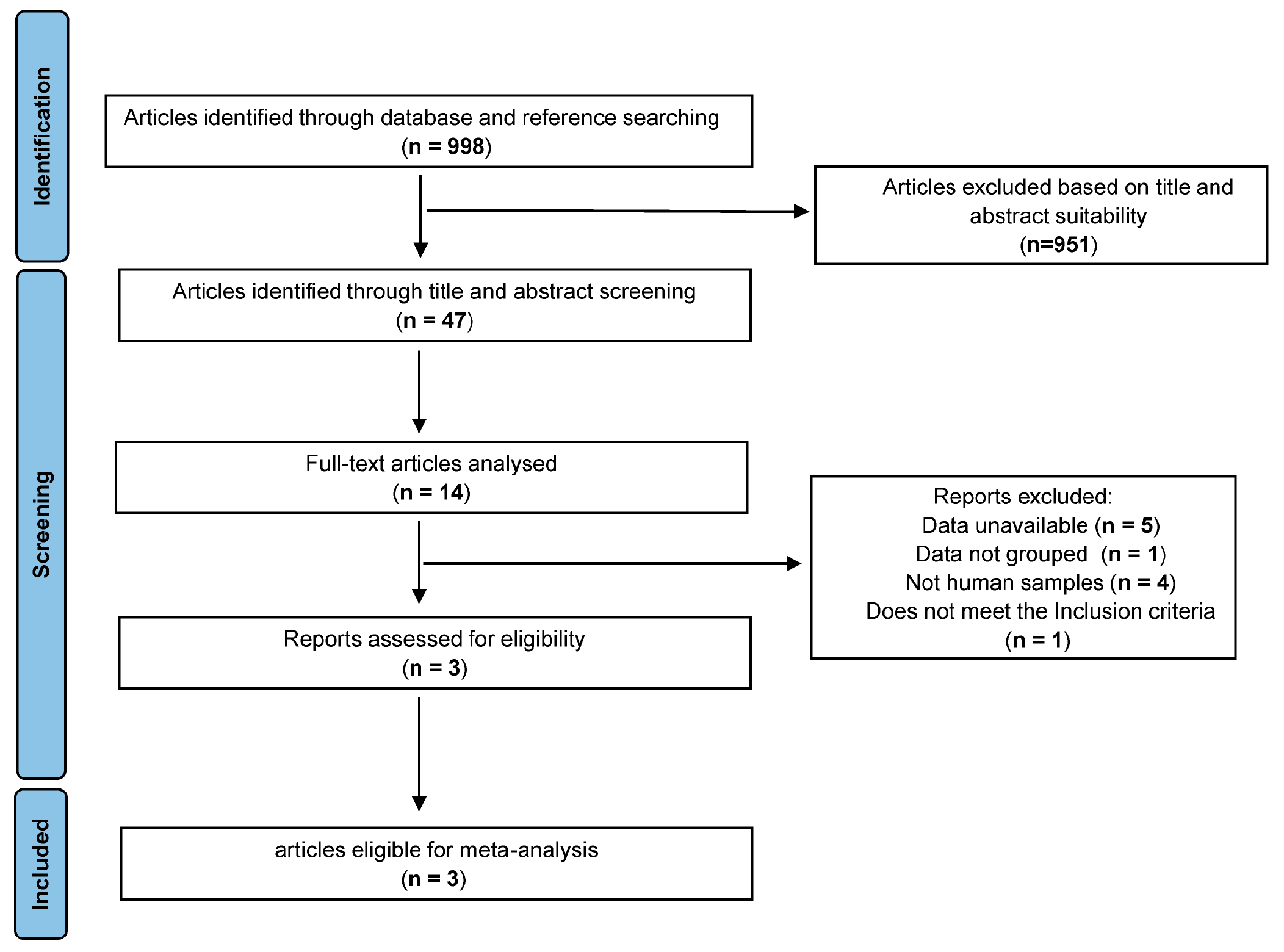
3.1. Study Selection
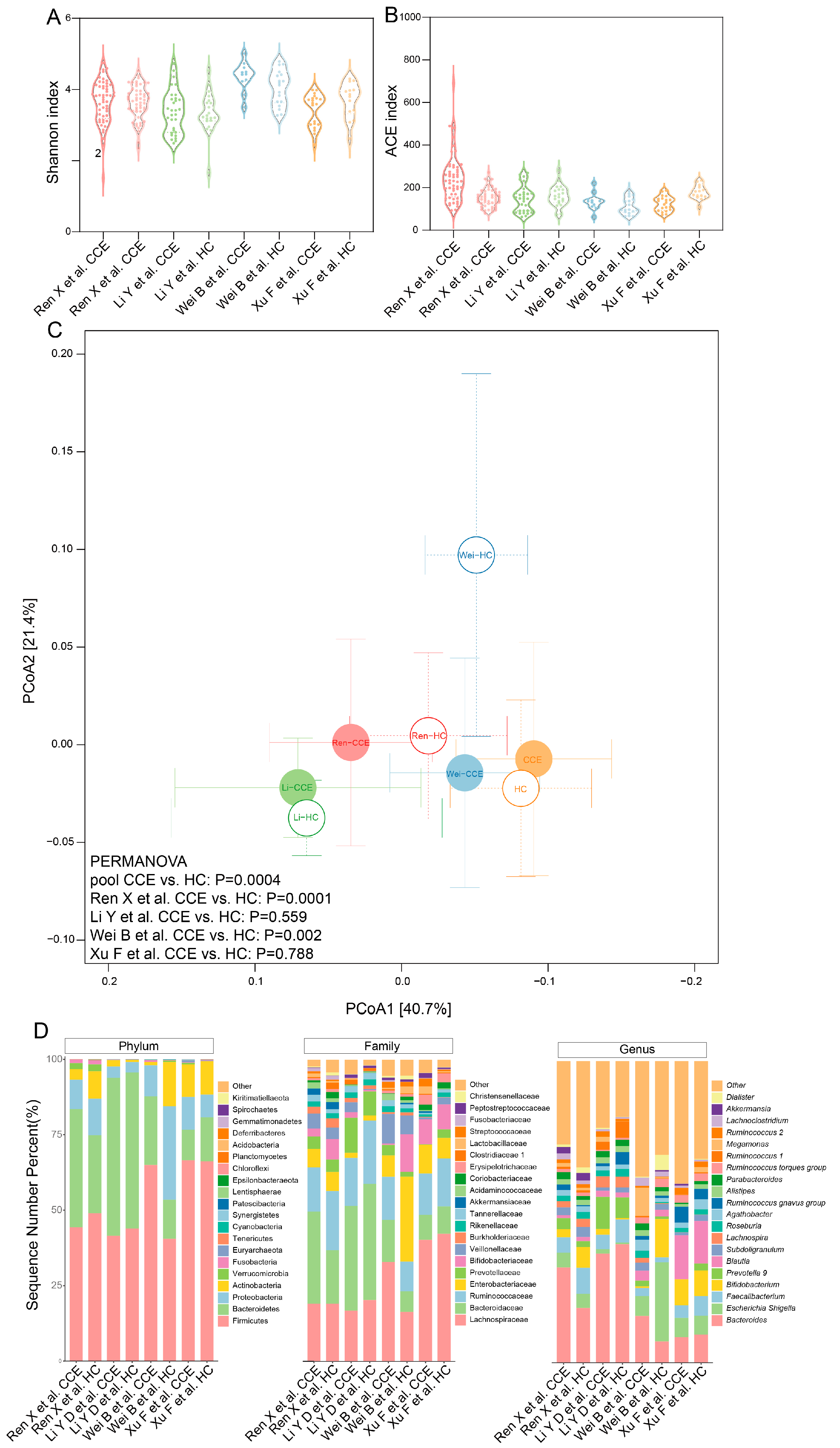
3.2. Microbial Diversity and Composition Comparison between CCE and HC Groups
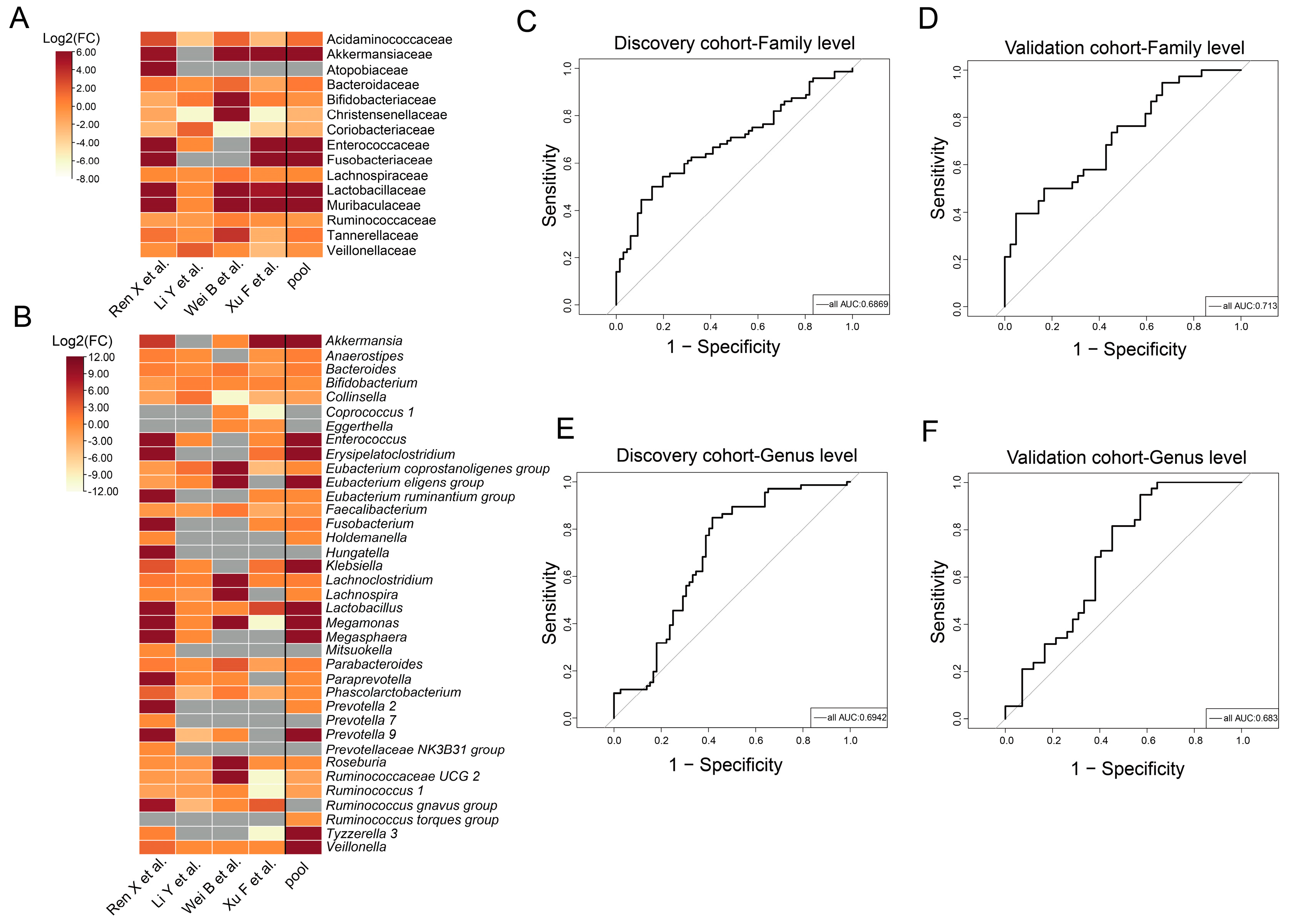
3.3. Taxa with Significantly Different Abundances between CCE and HC Groups
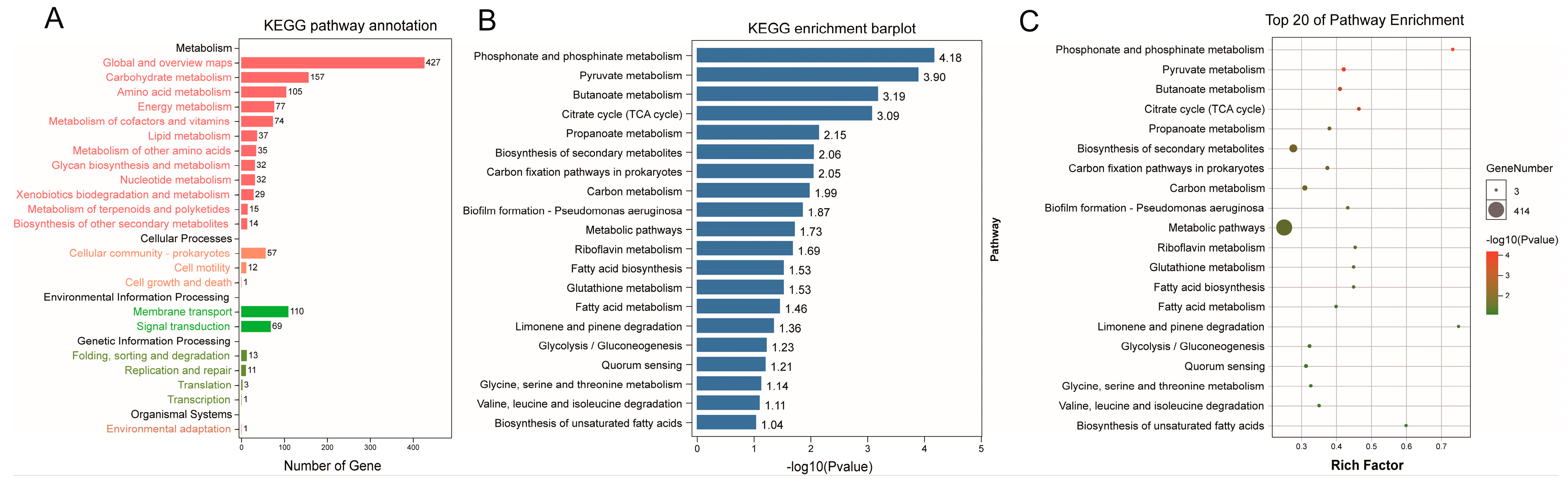
3.4. Functional Prediction of Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) Analysis between the CCE and HC Groups
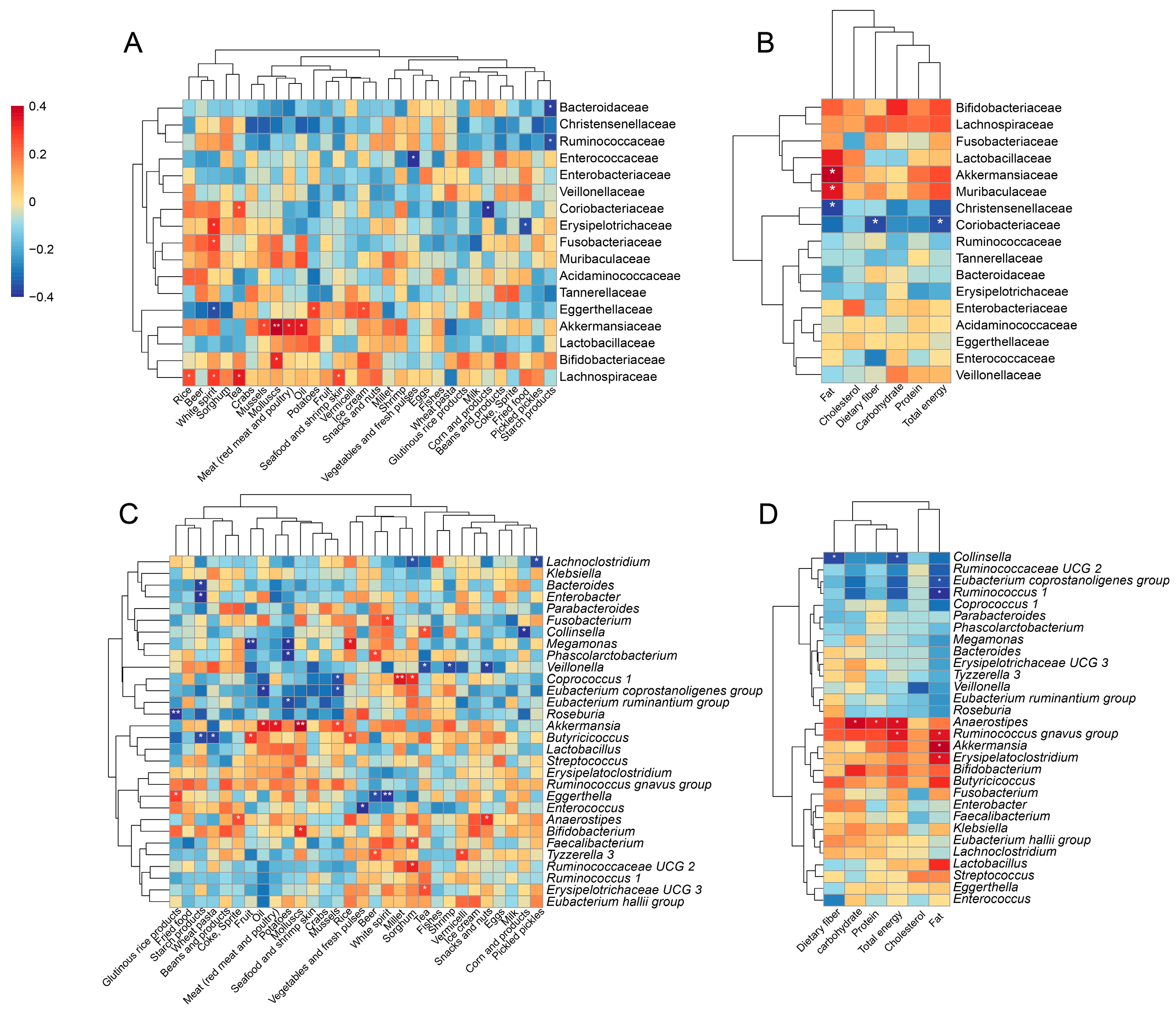
3.5. Alterations in Gut Microbiota Associated with Diets
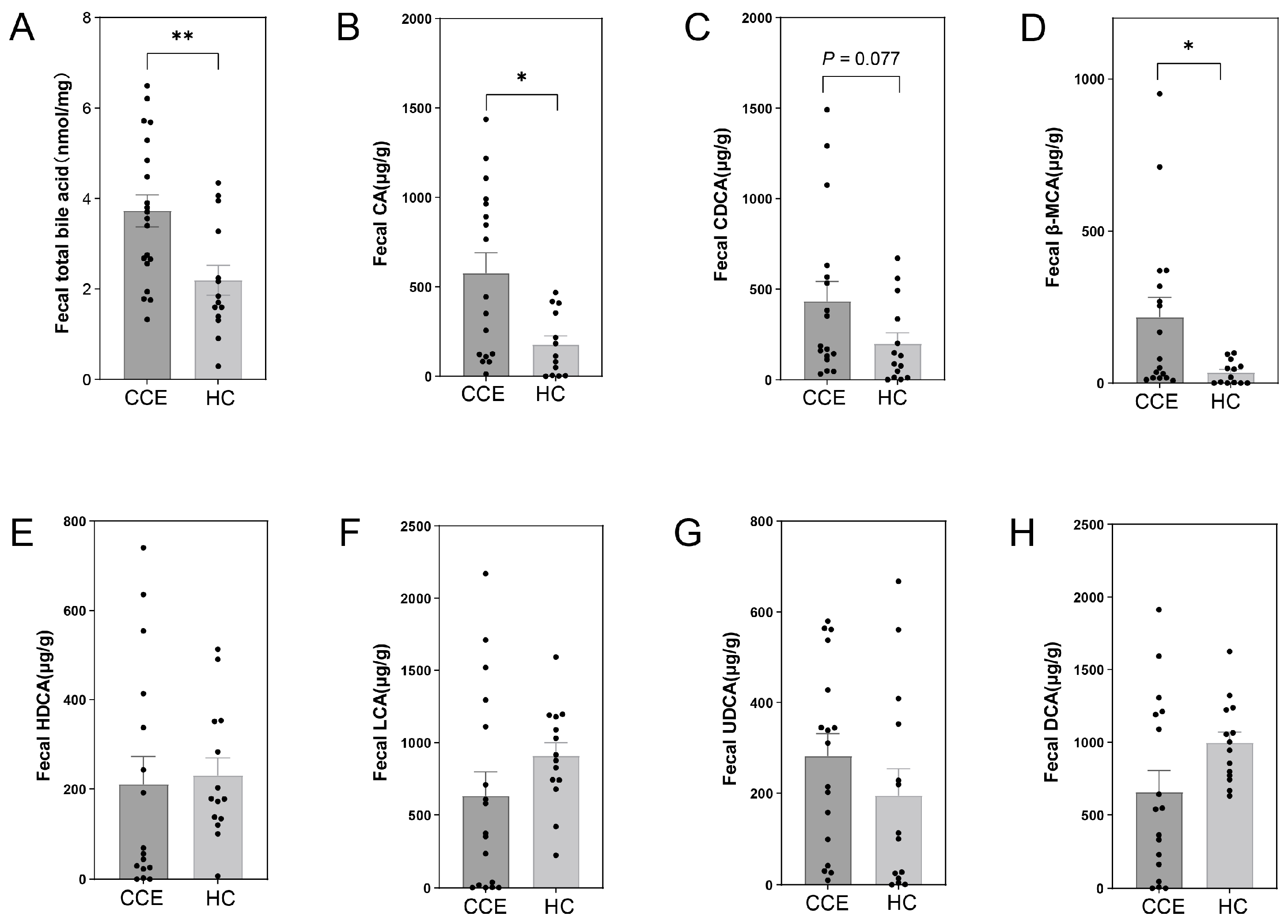
3.6. Alterations in Fecal BA between CCE and HC Groups
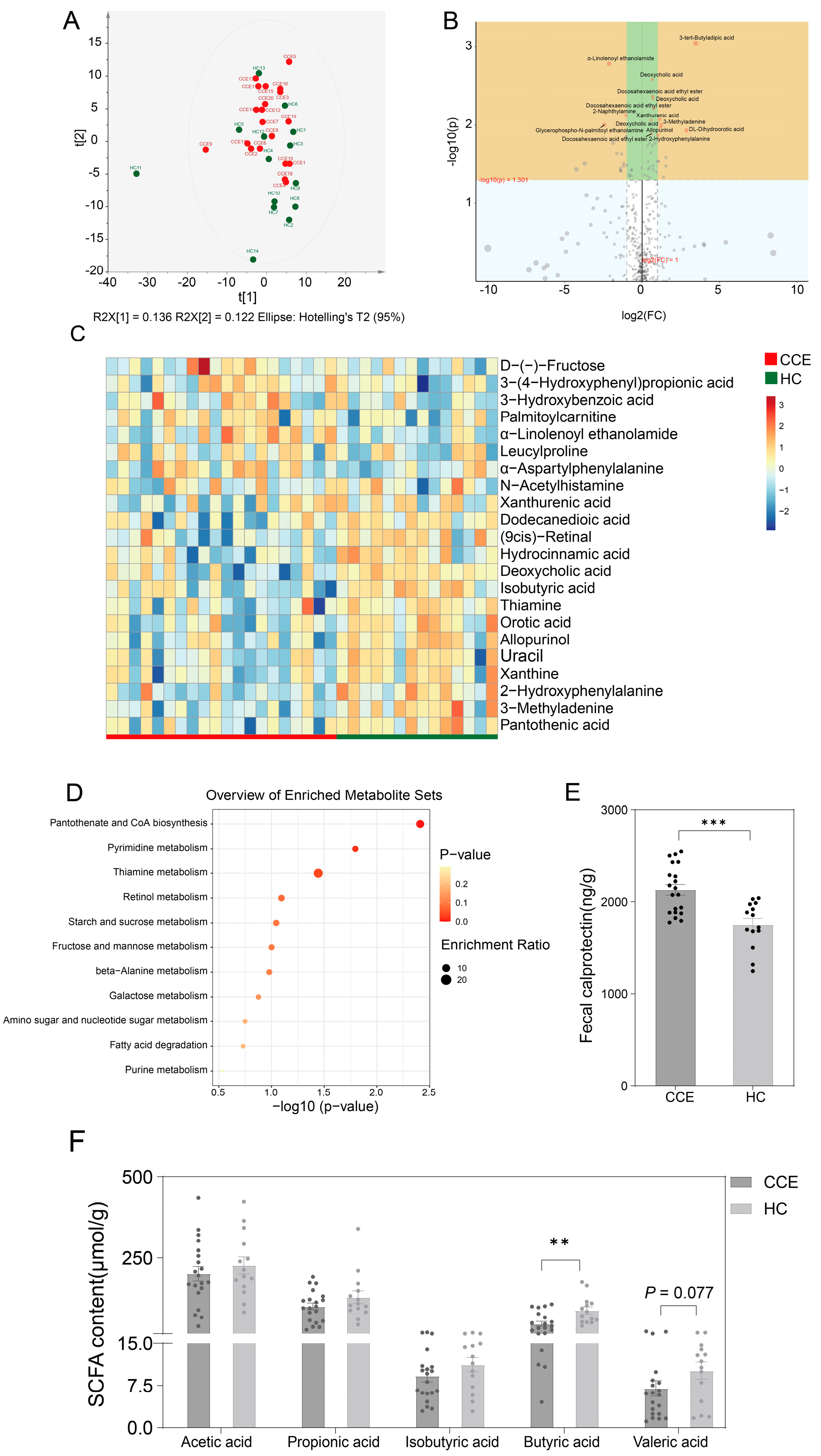
3.7. Alterations in Fecal Metabolome between CCE and HC Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Macierzanka, A.; Torcello-Gomez, A.; Jungnickel, C.; Maldonado-Valderrama, J. Bile salts in digestion and transport of lipids. Adv. Colloid Interface Sci. 2019, 274, 102045. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, R.; Hashem, J.; Walsh, T.N. A Review of Acute Cholecystitis. JAMA—J. Am. Med. Assoc. 2022, 328, 76–77. [Google Scholar] [CrossRef] [PubMed]
- Di Ciaula, A.; Garruti, G.; Wang, D.Q.H.; Portincasa, P. Cholecystectomy and risk of metabolic syndrome. Eur. J. Intern. Med. 2018, 53, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.S.; Kim, D.; Chung, G.E.; Kim, W.; Kim, Y.J.; Yoon, J.H. Cholecystectomy is independently associated with nonalcoholic fatty liver disease in an Asian population. World J. Gastroenterol. 2015, 21, 6287–6295. [Google Scholar] [CrossRef]
- Chen, C.-H.; Lin, C.-L.; Kao, C.-H. The Effect of Cholecystectomy on the Risk of Colorectal Cancer in Patients with Gallbladder Stones. Cancers 2020, 12, 550. [Google Scholar] [CrossRef] [PubMed]
- Shabanzadeh, D.M.; Skaaby, T.; Sørensen, L.T.; Jørgensen, T. Screen-detected gallstone disease and cardiovascular disease. Eur. J. Epidemiol. 2017, 32, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Shi, N.; Li, N.; Duan, X.; Niu, H. Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 2017, 4, 14. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Ren, X.; Zhang, Y.; Ke, Z.; Zhou, J.; Wang, Y.; Zhang, Y.; Liu, Y. Cholecystectomy-induced secondary bile acids accumulation ameliorates colitis through inhibiting monocyte/macrophage recruitment. Gut Microbes 2022, 14, 2107387. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Wu, X.; Jing, H.; Zhang, S.; Hu, Z.; Rao, L.; Chang, Q.; Wang, L.; Zhang, Z. Gut microbiota alteration after cholecystectomy contributes to post-cholecystectomy diarrhea via bile acids stimulating colonic serotonin. Gut Microbes 2023, 15, 2168101. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Xu, J.; Zhang, Y.; Chen, G.; Zhang, Y.; Huang, Q.; Liu, Y. Bacterial Alterations in Post-Cholecystectomy Patients Are Associated With Colorectal Cancer. Front. Oncol. 2020, 10, 1418. [Google Scholar] [CrossRef]
- Xu, Y.; Jing, H.; Wang, J.; Zhang, S.; Chang, Q.; Li, Z.; Wu, X.; Zhang, Z. Disordered Gut Microbiota Correlates With Altered Fecal Bile Acid Metabolism and Post-cholecystectomy Diarrhea. Front. Microbiol. 2022, 13, 800604. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.D.; Liu, B.N.; Zhao, S.H.; Zhou, Y.L.; Bai, L.; Liu, E.Q. Changes in gut microbiota composition and diversity associated with post-cholecystectomy diarrhea. World J. Gastroenterol. 2021, 27, 391–403. [Google Scholar] [CrossRef]
- Frost, F.; Kacprowski, T.; Ruhlemann, M.; Weiss, S.; Bang, C.; Franke, A.; Pietzner, M.; Aghdassi, A.A.; Sendler, M.; Volker, U.; et al. Carrying asymptomatic gallstones is not associated with changes in intestinal microbiota composition and diversity but cholecystectomy with significant dysbiosis. Sci. Rep. 2021, 11, 6677. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, J.; Li, J.; Yan, P.; Jin, Y.; Zhang, R.; Yue, W.; Guo, Q.; Geng, J. Cholecystectomy Damages Aging-Associated Intestinal Microbiota Construction. Front. Microbiol. 2018, 9, 1402. [Google Scholar] [CrossRef]
- Grigor’eva, I.; Romanova, T.; Naumova, N.; Alikina, T.; Kuznetsov, A.; Kabilov, M. Gut Microbiome in a Russian Cohort of Pre- and Post-Cholecystectomy Female Patients. J. Pers. Med. 2021, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, D.; Caraba, A.; Ionita, I.; Lascu, A.; Hut, E.F.; Dragan, S.; Ancusa, O.E.; Suceava, I.; Lighezan, D. Dyspepsia and Gut Microbiota in Female Patients with Postcholecystectomy Syndrome. Int. J. Women’s Health 2022, 14, 41–56. [Google Scholar] [CrossRef]
- Kang, Z.; Lu, M.; Jiang, M.; Zhou, D.; Huang, H. Proteobacteria Acts as a Pathogenic Risk-Factor for Chronic Abdominal Pain and Diarrhea in Post-Cholecystectomy Syndrome Patients: A Gut Microbiome Metabolomics Study. Med. Sci. Monit. 2019, 25, 7312–7320. [Google Scholar] [CrossRef]
- Wei, B.; Wang, Y.; Xiang, S.; Jiang, Y.; Chen, R.; Hu, N. Alterations of gut microbiome in patients with type 2 diabetes mellitus who had undergone cholecystectomy. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E113–E121. [Google Scholar] [CrossRef] [PubMed]
- Yueh, T.P.; Chen, F.Y.; Lin, T.E.; Chuang, M.T. Diarrhea after laparoscopic cholecystectomy: Associated factors and predictors. Asian J. Surg. 2014, 37, 171–177. [Google Scholar] [CrossRef]
- Shin, Y.; Choi, D.; Lee, K.G.; Choi, H.S.; Park, Y. Association between dietary intake and postlaparoscopic cholecystectomic symptoms in patients with gallbladder disease. Korean J. Intern. Med. 2018, 33, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.L.; Norhaizan, M.E. Effect of High-Fat Diets on Oxidative Stress, Cellular Inflammatory Response and Cognitive Function. Nutrients 2019, 11, 2579. [Google Scholar] [CrossRef]
- Suriano, F.; Nystrom, E.E.L.; Sergi, D.; Gustafsson, J.K. Diet, microbiota, and the mucus layer: The guardians of our health. Front. Immunol. 2022, 13, 953196. [Google Scholar] [CrossRef]
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287–1298. [Google Scholar] [CrossRef]
- Fan, Y.; Støving, R.K.; Berreira Ibraim, S.; Hyötyläinen, T.; Thirion, F.; Arora, T.; Lyu, L.; Stankevic, E.; Hansen, T.H.; Déchelotte, P.; et al. The gut microbiota contributes to the pathogenesis of anorexia nervosa in humans and mice. Nat. Microbiol. 2023, 8, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hasegawa, K.; Chen, J. The use of food-frequency questionnaires for various purposes in China. Public Health Nutr. 2006, 5, 829–833. [Google Scholar] [CrossRef]
- Yang, Y.G.W.; Pan, X. China Food Composition; Peking University Medical Press: Beijing, China, 2009. [Google Scholar]
- Zhu, G.; Guo, M.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Bifidobacterium breve intervention combined with environmental enrichment alleviates cognitive impairment by regulating the gut microbiota and microbial metabolites in Alzheimer’s disease mice. Front. Immunol. 2022, 13, 1013664. [Google Scholar] [CrossRef]
- Qu, D.; Sun, F.; Feng, S.; Yu, L.; Tian, F.; Zhang, H.; Chen, W.; Zhai, Q. Protective effects of Bacteroides fragilis against lipopolysaccharide-induced systemic inflammation and their potential functional genes. Food Funct. 2022, 13, 1015–1025. [Google Scholar] [CrossRef]
- John, C.; Werner, P.; Worthmann, A.; Wegner, K.; Todter, K.; Scheja, L.; Rohn, S.; Heeren, J.; Fischer, M. A liquid chromatography-tandem mass spectrometry-based method for the simultaneous determination of hydroxy sterols and bile acids. J. Chromatogr. A 2014, 1371, 184–195. [Google Scholar] [CrossRef]
- Zhai, Q.; Liu, Y.; Wang, C.; Qu, D.; Zhao, J.; Zhang, H.; Tian, F.; Chen, W. Lactobacillus plantarum CCFM8661 modulates bile acid enterohepatic circulation and increases lead excretion in mice. Food Funct. 2019, 10, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Zhang, C.; Du, T.; Yi, S.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. Meta-analysis reveals different functional characteristics of human gut Bifidobacteria associated with habitual diet. Food Res. Int. 2023, 170, 112981. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Lagergren, J.; Ye, W.; Ekbom, A. Intestinal cancer after cholecystectomy: Is bile involved in carcinogenesis? Gastroenterology 2001, 121, 542–547. [Google Scholar] [CrossRef]
- Ruhl, C.E.; Everhart, J.E. Relationship of non-alcoholic fatty liver disease with cholecystectomy in the US population. Am. J. Gastroenterol. 2013, 108, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yu, Z.; Liu, Y.; Du, T.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. A High-Fat, High-Cholesterol Diet Promotes Intestinal Inflammation by Exacerbating Gut Microbiome Dysbiosis and Bile Acid Disorders in Cholecystectomy. Nutrients 2023, 15, 3829. [Google Scholar] [CrossRef] [PubMed]
- You, X.D.U.; Lenburg, M.E.; Kacena, M.A.; Charles, J.F. Murine Gut Microbiome Meta-analysis Reveals Alterations in Carbohydrate Metabolism in Response to Aging. mSystems 2022, 7, e0124821. [Google Scholar] [CrossRef]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A.; de Vos, W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 625–637. [Google Scholar] [CrossRef]
- Lievin-Le Moal, V.; Servin, A.L. Anti-infective activities of lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 2014, 27, 167–199. [Google Scholar] [CrossRef]
- Lei, W.; Cheng, Y.; Gao, J.; Liu, X.; Shao, L.; Kong, Q.; Zheng, N.; Ling, Z.; Hu, W. Akkermansia muciniphila in neuropsychiatric disorders: Friend or foe? Front. Cell. Infect. Microbiol. 2023, 13, 1224155. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Zheng, Y.; Huang, Y.; Feng, Y.; Xu, K.; Zhang, W.; Wang, Y.; Nie, K.; Qin, M. Excessive consumption of mucin by over-colonized Akkermansia muciniphila promotes intestinal barrier damage during malignant intestinal environment. Front. Microbiol. 2023, 14, 1111911. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.F.S.; Yokota, A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J. Lipid Res. 2017, 58, 1143–1152. [Google Scholar] [CrossRef]
- Ovadia, C.; Perdones-Montero, A.; Spagou, K.; Smith, A.; Sarafian, M.H.; Gomez-Romero, M.; Bellafante, E.; Clarke, L.C.D.; Sadiq, F.; Nikolova, V.; et al. Enhanced Microbial Bile Acid Deconjugation and Impaired Ileal Uptake in Pregnancy Repress Intestinal Regulation of Bile Acid Synthesis. Hepatology 2019, 70, 276–293. [Google Scholar] [CrossRef]
- Jones, H.; Alpini, G.; Francis, H. Bile acid signaling and biliary functions. Acta Pharm. Sin. B 2015, 5, 123–128. [Google Scholar] [CrossRef]
- Dong, S.; Zhu, M.; Wang, K.; Zhao, X.; Hu, L.; Jing, W.; Lu, H.; Wang, S. Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Res. 2021, 171, 105767. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Li, H.; Zhang, X.; Yang, T.; Yue, M.; Zhang, Y.; Chen, S.; Cui, N.; Yuan, C.; Li, J.; et al. Akkermansia muciniphila protects mice against an emerging tick-borne viral pathogen. Nat. Microbiol. 2023, 8, 91–106. [Google Scholar] [CrossRef]
- He, N.; Wang, S.; Lv, Z.; Zhao, W.; Li, S. Low molecular weight chitosan oligosaccharides (LMW-COSs) prevent obesity-related metabolic abnormalities in association with the modification of gut microbiota in high-fat diet (HFD)-fed mice. Food Funct. 2020, 11, 9947–9959. [Google Scholar] [CrossRef]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef]
- He, Z.; Ma, Y.; Yang, S.; Zhang, S.; Liu, S.; Xiao, J.; Wang, Y.; Wang, W.; Yang, H.; Li, S.; et al. Gut microbiota-derived ursodeoxycholic acid from neonatal dairy calves improves intestinal homeostasis and colitis to attenuate extended-spectrum beta-lactamase-producing enteroaggregative Escherichia coli infection. Microbiome 2022, 10, 79. [Google Scholar] [CrossRef]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, X.; Liu, J.; Zhong, F.; Zhang, C.; Chen, Y.; Sun, T.; Ji, C.; Ma, D. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 2022, 13, 2522. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Shen, Y.; Zhang, H.; Cao, M.; Guo, M.; He, J.; Zhang, B.; Xiao, C. Gut Microbiota Characteristics of People with Obesity by Meta-Analysis of Existing Datasets. Nutrients 2022, 14, 2993. [Google Scholar] [CrossRef]
- Chu, Y.; Sun, S.; Huang, Y.; Gao, Q.; Xie, X.; Wang, P.; Li, J.; Liang, L.; He, X.; Jiang, Y.; et al. Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes 2021, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Schepper, J.D.; Collins, F.; Rios-Arce, N.D.; Kang, H.J.; Schaefer, L.; Gardinier, J.D.; Raghuvanshi, R.; Quinn, R.A.; Britton, R.; Parameswaran, N.; et al. Involvement of the Gut Microbiota and Barrier Function in Glucocorticoid-Induced Osteoporosis. J. Bone Miner. Res. 2020, 35, 801–820. [Google Scholar] [CrossRef]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; et al. Human Gut Microbiota and Its Metabolites Impact Immune Responses in COVID-19 and Its Complications. Gastroenterology 2023, 164, 272–288. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef]
- Ocvirk, S.; O’Keefe, S.J.D. Dietary fat, bile acid metabolism and colorectal cancer. Semin. Cancer Biol. 2021, 73, 347–355. [Google Scholar] [CrossRef]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.J.; Hsiao, E.Y. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018, 173, 1728–1741 e1713. [Google Scholar] [CrossRef]
- Ye, J.; Zhao, Y.; Chen, X.; Zhou, H.; Yang, Y.; Zhang, X.; Huang, Y.; Zhang, N.; Lui, E.M.K.; Xiao, M. Pu-erh tea ameliorates obesity and modulates gut microbiota in high fat diet fed mice. Food Res. Int. 2021, 144, 110360. [Google Scholar] [CrossRef]
- Pisanu, S.; Palmas, V.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Boi, F.; Loviselli, A.; et al. Impact of a Moderately Hypocaloric Mediterranean Diet on the Gut Microbiota Composition of Italian Obese Patients. Nutrients 2020, 12, 2707. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Wang, F.; Yuan, J.; Li, J.; Jiang, D.; Zhang, J.; Li, H.; Wang, R.; Tang, J.; Huang, T.; et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: A 6-month randomised controlled-feeding trial. Gut 2019, 68, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Li, P.; Hu, J.; Zhao, H.; Feng, J.; Chai, B. Multi-Omics Reveals Inhibitory Effect of Baicalein on Non-Alcoholic Fatty Liver Disease in Mice. Front. Pharmacol. 2022, 13, 925349. [Google Scholar] [CrossRef]
- Le, T.T.; Ziemba, A.; Urasaki, Y.; Hayes, E.; Brotman, S.; Pizzorno, G. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J. Lipid Res. 2013, 54, 1044–1057. [Google Scholar] [CrossRef]
- Peng, S.L.; Wang, R.; Zhou, Y.L.; Wei, W.; Zhong, G.H.; Huang, X.T.; Yang, S.; Liu, Q.D.; Liu, Z.G. Insight of a Metabolic Prognostic Model to Identify Tumor Environment and Drug Vulnerability for Lung Adenocarcinoma. Front. Immunol. 2022, 13, 872910. [Google Scholar] [CrossRef]
- Fang, H.; Fu, L.; Li, X.; Lu, C.; Su, Y.; Xiong, K.; Zhang, L. Long-term efficacy and safety of monotherapy with a single fresh fecal microbiota transplant for recurrent active ulcerative colitis: A prospective randomized pilot study. Microb. Cell Factories 2021, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Guo, M.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Integrative Metabolomic Characterization Reveals the Mediating Effect of Bifidobacterium breve on Amino Acid Metabolism in a Mouse Model of Alzheimer’s Disease. Nutrients 2022, 14, 735. [Google Scholar] [CrossRef]
| Characteristic | Ren X. et al. (n = 104) | Li Y. et al. (n = 51) | Wei B. et al. (n = 35) |
|---|---|---|---|
| Mean (SD) | Mean | Mean (SD) | |
| Sex (F/M) | 34/18 | 19/12 | 11/3 |
| 34/18 | 12/8 | 15/6 | |
| Age (years) | 60.02 (11.53) | 46.6 | 60.6 (10.1) |
| 59.71 (11.95) | 47.3 | 58.9 (9.9) | |
| BMI | 25.71 (3.47) | 22.15 | 25.2 (2.3) |
| 24.38 (3.63) | 22.1 | 24.8 (3.7) | |
| Design | Case–control | Case–control | Case–control |
| Country | China | China | China |
| DNA extr. | PSP-Spin Stool Kit | QIAamp DNA Stool Minikit | TIANamp Stool DNA kit |
| 16S region | V3-V4 | V3-V4 | V3-V4 |
| Seq. Tech. | MiSeq | HiSeq | MiSeq |
| PE vs. SE | PE | PE | PE |
| Accession number | PRJNA541484 | SRP247004 | RJCA002279 |
| Characteristic | Xu F. et al. CCE (n = 20) | Xu F. et al. HC (n = 14) |
|---|---|---|
| Sex (F/M) | 14/6 | 9/5 |
| Age (years) | 44.45 (10.52) | 42.71 (15.09) |
| BMI | 25.18 (2.73) * | 22.58 (2.88) * |
| Smokers (%) | 10 | 0 |
| BSS | 4.3 (1.1) | 4.07 (0.88) |
| Macronutrients g/day | ||
| Protein | 73.04 (26.07) | 63.04 (27.07) |
| Fat | 75.55 (29.44) * | 51.92 (20.58) * |
| Carbohydrates | 174.69 (54.94) | 149.74 (53.95) |
| Fiber | 9.08 (4.33) | 7.82 (5.31) |
| Cholesterol mg/day | 463.39 (196.31) | 413.69 (157.49) |
| Total energy | 1695.08 (537.54) * | 1326.91 (473.90) * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, F.; Chen, R.; Zhang, C.; Wang, H.; Ding, Z.; Yu, L.; Tian, F.; Chen, W.; Zhou, Y.; Zhai, Q. Cholecystectomy Significantly Alters Gut Microbiota Homeostasis and Metabolic Profiles: A Cross-Sectional Study. Nutrients 2023, 15, 4399. https://doi.org/10.3390/nu15204399
Xu F, Chen R, Zhang C, Wang H, Ding Z, Yu L, Tian F, Chen W, Zhou Y, Zhai Q. Cholecystectomy Significantly Alters Gut Microbiota Homeostasis and Metabolic Profiles: A Cross-Sectional Study. Nutrients. 2023; 15(20):4399. https://doi.org/10.3390/nu15204399
Chicago/Turabian StyleXu, Fusheng, Ruimin Chen, Chengcheng Zhang, Hao Wang, Zhijie Ding, Leilei Yu, Fengwei Tian, Wei Chen, Yongping Zhou, and Qixiao Zhai. 2023. "Cholecystectomy Significantly Alters Gut Microbiota Homeostasis and Metabolic Profiles: A Cross-Sectional Study" Nutrients 15, no. 20: 4399. https://doi.org/10.3390/nu15204399
APA StyleXu, F., Chen, R., Zhang, C., Wang, H., Ding, Z., Yu, L., Tian, F., Chen, W., Zhou, Y., & Zhai, Q. (2023). Cholecystectomy Significantly Alters Gut Microbiota Homeostasis and Metabolic Profiles: A Cross-Sectional Study. Nutrients, 15(20), 4399. https://doi.org/10.3390/nu15204399







