Integration with Transcriptomic and Metabolomic Analyses Reveals the In Vitro Cytotoxic Mechanisms of Chinese Poplar Propolis by Triggering the Glucose Metabolism in Human Hepatocellular Carcinoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation of the Chinese Poplar Propolis Ethanol Extract
2.3. UHPLC/Q-TOF-MS
2.4. Cell Culture
2.5. Cell Viability Assay
2.6. Quantitative Real-Time Reverse-Transcriptase Polymerase Chain Reaction
2.7. RNA Sequencing
2.8. Measurement of Glucose Consumption and Lactate Production
2.9. Western Blotting
2.10. Cellular Metabolite Extraction
2.11. Molecular Docking Analysis
2.12. Statistical Analysis
3. Results
3.1. Nontargeted UHPLC/Q-TOF-MS CP Analysis
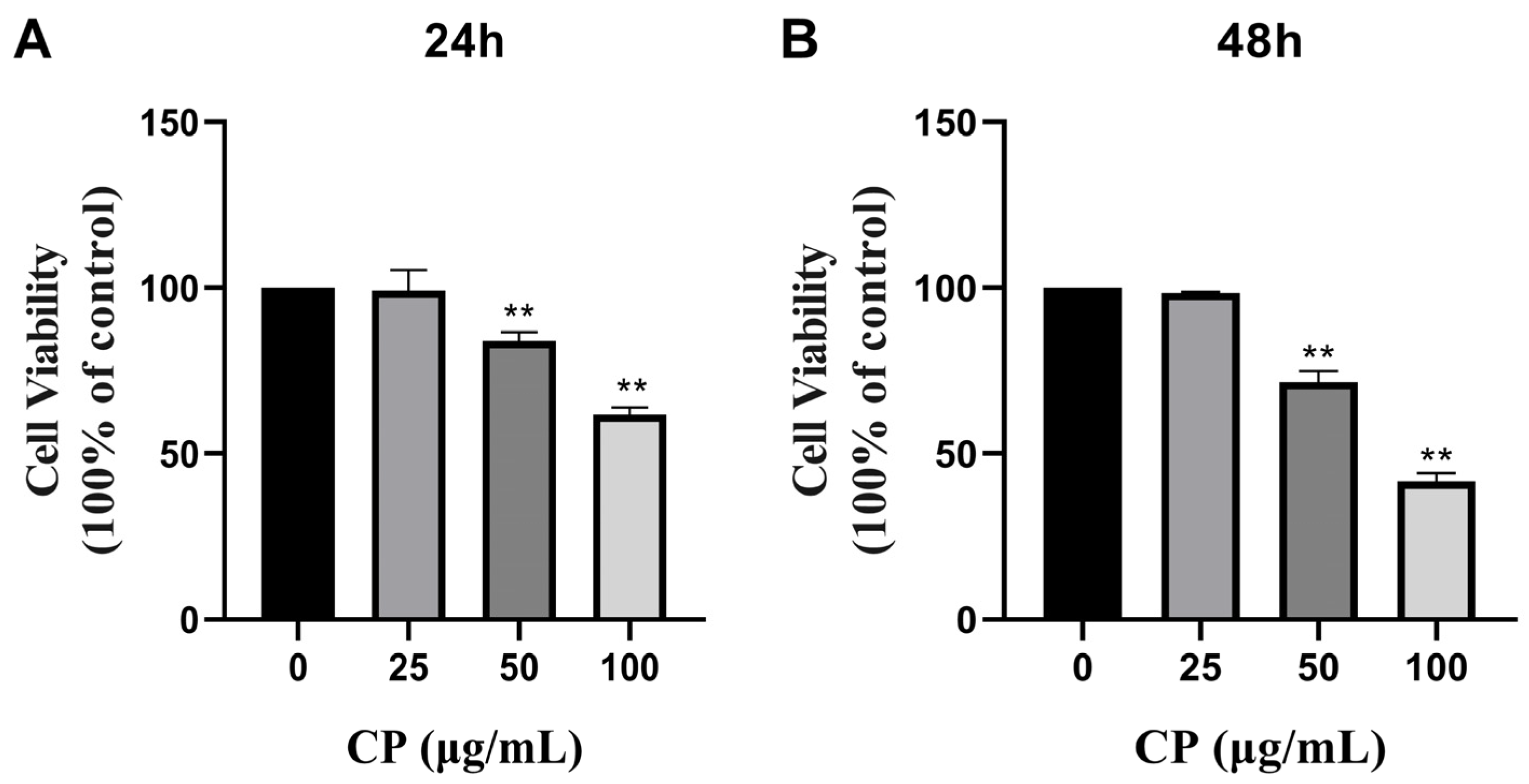
3.2. CP Inhibited the Cell Viability of HepG2 Cells
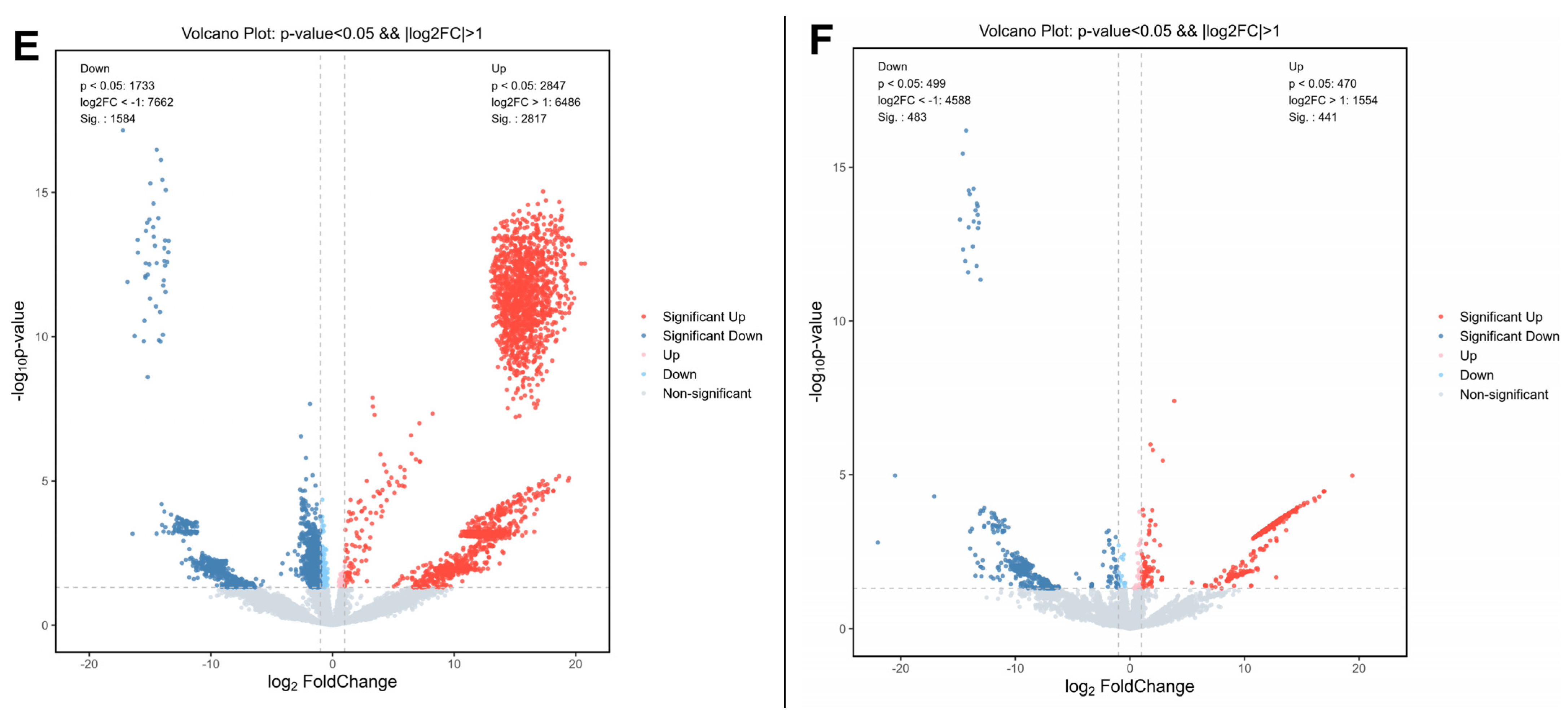
3.3. Propolis Affected Metabolism-Related Transcriptome Alterations in HepG2 Cells
3.4. CP Suppressed Glucose Consumption and Lactate Production
3.5. CP Inhibited the mRNA Expression of Glucose Transporter Proteins GLUT1 and GLUT3
3.6. CP Suppressed the Levels of Four Glycolytic Key Enzymes
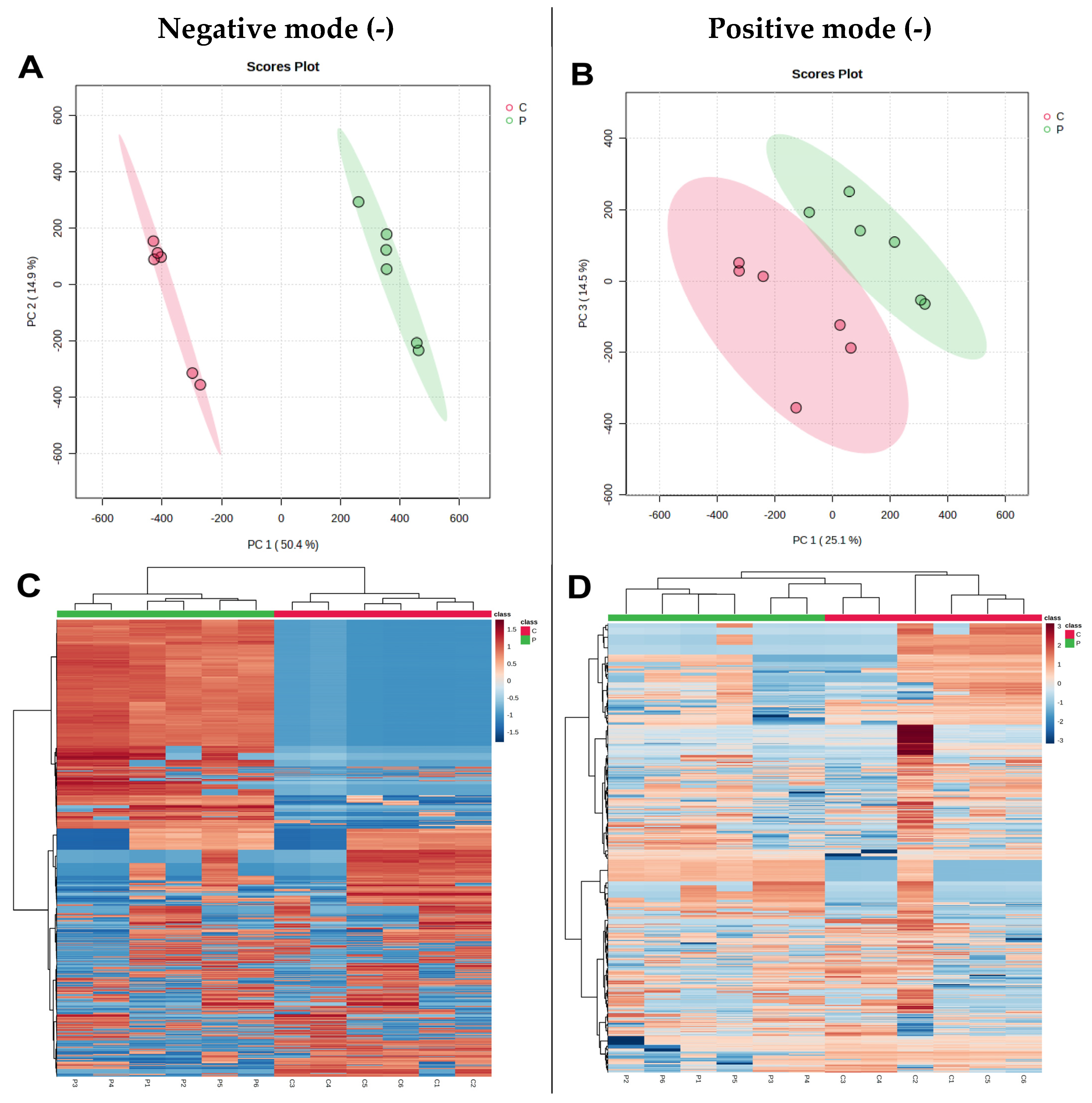
3.7. Differential Cellular Metabolite Analysis
3.8. Analysis of the Differential Metabolites Associated with Glycolysis
3.9. Molecular Docking Simulation of the Effects of Primary Active Components of Propolis on Glycolysis Key Enzymes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Xiang, D.; Cheng, Z.; Liu, H.; Wang, X.; Han, T.; Sun, W.; Li, X.; Yang, W.; Chen, C.; Xia, M.; et al. Shp2 promotes liver cancer stem cell expansion by augmenting β-catenin signaling and predicts chemotherapeutic response of patients. Hepatology 2017, 65, 1566–1580. [Google Scholar] [CrossRef]
- Bobiş, O. Plants: Sources of Diversity in Propolis Properties. Plants 2022, 11, 2298. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, C.-P.; Wang, K.; Li, G.Q.; Hu, F.-L. Recent Advances in the Chemical Composition of Propolis. Molecules 2014, 19, 19610–19632. [Google Scholar] [CrossRef]
- Sforcin, J.M.; Bankova, V. Propolis: Is there a potential for the development of new drugs? J. Ethnopharmacol. 2011, 133, 253–260. [Google Scholar] [CrossRef]
- Segueni, N.; Boutaghane, N.; Asma, S.T.; Tas, N.; Acaroz, U.; Arslan-Acaroz, D.; Shah, S.R.A.; Abdellatieff, H.A.; Akkal, S.; Peñalver, R.; et al. Review on Propolis Applications in Food Preservation and Active Packaging. Plants 2023, 12, 1654. [Google Scholar] [CrossRef]
- Asma, S.T.; Bobiş, O.; Bonta, V.; Acaroz, U.; Shah, S.R.A.; Istanbullugil, F.R.; Arslan-Acaroz, D. General Nutritional Profile of Bee Products and Their Potential Antiviral Properties against Mammalian Viruses. Nutrients 2022, 14, 3579. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Yuan, W.; Hao, S.; Wang, M.; Wang, F.; Xuan, H. Bioactive components and mechanisms of poplar propolis in inhibiting proliferation of human hepatocellular carcinoma HepG2 cells. Biomed. Pharmacother. 2021, 144, 112364. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Liu, X.; Hao, S.; Zhang, Z.; Xuan, H. Chinese Poplar Propolis Inhibits MDA-MB-231 Cell Proliferation in an Inflammatory Microenvironment by Targeting Enzymes of the Glycolytic Pathway. J. Immunol. Res. 2021, 2021, 6641341. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, Y.; Chen, X.; Jiang, X.; Wang, K.; Hu, F. Chinese Propolis Exerts Anti-Proliferation Effects in Human Melanoma Cells by Targeting NLRP1 Inflammatory Pathway, Inducing Apoptosis, Cell Cycle Arrest, and Autophagy. Nutrients 2018, 10, 1170. [Google Scholar] [CrossRef]
- Burns, J.S.; Manda, G. Metabolic Pathways of the Warburg Effect in Health and Disease: Perspectives of Choice, Chain or Chance. Int. J. Mol. Sci. 2017, 18, 2755. [Google Scholar] [CrossRef]
- Samec, M.; Liskova, A.; Koklesova, L.; Samuel, S.M.; Zhai, K.; Buhrmann, C.; Varghese, E.; Abotaleb, M.; Qaradakhi, T.; Zulli, A.; et al. Flavonoids against the Warburg phenotype—Concepts of predictive, preventive and personalised medicine to cut the Gordian knot of cancer cell metabolism. EPMA J. 2020, 11, 377–398. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, R.; Yu, W.; Min, Z.; Ye, M. The SIRT6-Autophagy-Warburg Effect Axis in Papillary Thyroid Cancer. Front. Oncol. 2020, 10, 1265. [Google Scholar] [CrossRef]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef]
- Mirzaei, H.; Hamblin, M.R. Regulation of Glycolysis by Non-coding RNAs in Cancer: Switching on the Warburg Effect. Mol. Ther.-Oncolytics 2020, 19, 218–239. [Google Scholar] [CrossRef]
- Zambrano, A.; Molt, M.; Uribe, E.; Salas, M. Glut 1 in Cancer Cells and the Inhibitory Action of Resveratrol as A Potential Therapeutic Strategy. Int. J. Mol. Sci. 2019, 20, 3374. [Google Scholar] [CrossRef]
- Zheng, X.; Al Naggar, Y.; Wu, Y.; Liu, D.; Hu, Y.; Wang, K.; Jin, X.; Peng, W. Untargeted metabolomics description of propolis’s in vitro antibacterial mechanisms against Clostridium perfringens. Food Chem. 2023, 406, 135061. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion–switching, targeted mass spectrometry–based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Xuan, H.; Wang, Y.; Li, A.; Fu, C.; Wang, Y.; Peng, W. Bioactive Components of Chinese Propolis Water Extract on Antitumor Activity and Quality Control. Evid.-Based Complement. Altern. Med. 2016, 2016, 9641965. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Charoensiddhi, S.; Xue, X.; Sun, B.; Liu, Y.; El-Seedi, H.R.; Wang, K. A review on the gastrointestinal protective effects of tropical fruit polyphenols. Crit. Rev. Food Sci. Nutr. 2023, 63, 7197–7223. [Google Scholar] [CrossRef]
- Asma, S.T.; Acaroz, U.; Imre, K.; Morar, A.; Shah, S.R.A.; Hussain, S.Z.; Arslan-Acaroz, D.; Demirbas, H.; Hajrulai-Musliu, Z.; Istanbullugil, F.R.; et al. Natural Products/Bioactive Compounds as a Source of Anticancer Drugs. Cancers 2022, 14, 6203. [Google Scholar] [CrossRef]
- Wang, K.; Wan, Z.; Ou, A.; Liang, X.; Guo, X.; Zhang, Z.; Wu, L.; Xue, X. Monofloral honey from a medical plant, Prunella Vulgaris, protected against dextran sulfate sodium-induced ulcerative colitis via modulating gut microbial populations in rats. Food Funct. 2019, 10, 3828–3838. [Google Scholar] [CrossRef]
- Patel, S. Emerging Adjuvant Therapy for Cancer: Propolis and its Constituents. J. Diet. Suppl. 2016, 13, 245–268. [Google Scholar] [CrossRef]
- Xu, D.; Jin, J.; Yu, H.; Zhao, Z.; Ma, D.; Zhang, C.; Jiang, H. Chrysin inhibited tumor glycolysis and induced apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J. Exp. Clin. Cancer Res. 2017, 36, 44. [Google Scholar] [CrossRef]
- Sepúlveda, C.; Núñez, O.; Torres, A.; Guzmán, L.; Wehinger, S. Antitumor Activity of Propolis: Recent Advances in Cellular Perspectives, Animal Models and Possible Applications. Food Rev. Int. 2020, 36, 429–455. [Google Scholar] [CrossRef]
- Chang, H.; Wang, Y.; Yin, X.; Liu, X.; Xuan, H. Ethanol extract of propolis and its constituent caffeic acid phenethyl ester inhibit breast cancer cells proliferation in inflammatory microenvironment by inhibiting TLR4 signal pathway and inducing apoptosis and autophagy. BMC Complement. Altern. Med. 2017, 17, 471. [Google Scholar] [CrossRef] [PubMed]
- Forma, E.; Bryś, M. Anticancer Activity of Propolis and Its Compounds. Nutrients 2021, 13, 2594. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Chen, S.; Peng, H.; Xiao, L.; Du, E.; Liu, Y.; Lin, D.; Wang, Y.; Xu, Y.; et al. PKMYT1 is associated with prostate cancer malignancy and may serve as a therapeutic target. Gene 2020, 744, 144608. [Google Scholar] [CrossRef]
- Town, J.; Pais, H.; Harrison, S.; Stead, L.F.; Bataille, C.; Bunjobpol, W.; Zhang, J.; Rabbitts, T.H. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc. Natl. Acad. Sci. USA 2016, 113, 3603–3608. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.; Mueller-Klieser, W. Lactate: A metabolic key player in cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, X.; Wang, Y.; Sun, Q.; Chen, M.; Liu, S.; Zou, X. Licochalcone A suppresses hexokinase 2-mediated tumor glycolysis in gastric cancer via downregulation of the Akt signaling pathway. Oncol. Rep. 2018, 39, 1181–1190. [Google Scholar] [CrossRef]
- Ojelabi, O.A.; Lloyd, K.P.; De Zutter, J.K.; Carruthers, A. Red wine and green tea flavonoids are cis-allosteric activators and competitive inhibitors of glucose transporter 1 (GLUT1)-mediated sugar uptake. J. Biol. Chem. 2018, 293, 19823–19834. [Google Scholar] [CrossRef]
- Tanaka, H.; Fujita, N.; Sugimoto, R.; Urawa, N.; Horiike, S.; Kobayashi, Y.; Iwasa, M.; Ma, N.; Kawanishi, S.; Watanabe, S.; et al. Hepatic oxidative DNA damage is associated with increased risk for hepatocellular carcinoma in chronic hepatitis C. Br. J. Cancer 2008, 98, 580–586. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Ghoshal, K.; Fernandez, S.; Li, L. Identification of a Subtype of Hepatocellular Carcinoma with Poor Prognosis Based on Expression of Genes within the Glucose Metabolic Pathway. Cancers 2019, 11, 2023. [Google Scholar] [CrossRef]
- Paul, S.; Ghosh, S.; Kumar, S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol. 2022, 86, 1216–1230. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Enríquez, S.; Carreño-Fuentes, L.; Gallardo-Pérez, J.C.; Saavedra, E.; Quezada, H.; Vega, A.; Marín-Hernández, A.; Olín-Sandoval, V.; Torres-Márquez, M.E.; Moreno-Sánchez, R. Oxidative phosphorylation is impaired by prolonged hypoxia in breast and possibly in cervix carcinoma. Int. J. Biochem. Cell Biol. 2010, 42, 1744–1751. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.C.; DeNicola, G.M.; Nixon, C.; Blyth, K.; Labuschagne, C.F.; Tuveson, D.A.; Vousden, K.H. Dynamic ROS Control by TIGAR Regulates the Initiation and Progression of Pancreatic Cancer. Cancer Cell 2020, 37, 168–182.e4. [Google Scholar] [CrossRef]
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Chandel, N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Glycolysis. Cold Spring Harb. Perspect. Biol. 2021, 13, a040535. [Google Scholar] [CrossRef]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 Is Required for Tumor Initiation and Maintenance and Its Systemic Deletion Is Therapeutic in Mouse Models of Cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef]
- DeWaal, D.; Nogueira, V.; Terry, A.R.; Patra, K.C.; Jeon, S.M.; Guzman, G.; Au, J.; Long, C.P.; Antoniewicz, M.R.; Hay, N. Author Correction: Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 2018, 9, 2539. [Google Scholar] [CrossRef]
- Lee, J.-H.; Liu, R.; Li, J.; Wang, Y.; Tan, L.; Li, X.-J.; Qian, X.; Zhang, C.; Xia, Y.; Xu, D.; et al. EGFR-Phosphorylated Platelet Isoform of Phosphofructokinase 1 Promotes PI3K Activation. Mol. Cell 2018, 70, 197–210.e7. [Google Scholar] [CrossRef]
- Yang, H.; Shu, Z.; Jiang, Y.; Mao, W.; Pang, L.; Redwood, A.; Jeter-Jones, S.L.; Jennings, N.B.; Ornelas, A.; Zhou, J.; et al. 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase-2 Regulates TP53-Dependent Paclitaxel Sensitivity in Ovarian and Breast Cancers. Clin. Cancer Res. 2019, 25, 5702–5716. [Google Scholar] [CrossRef]
- Umar, S.M.; Kashyap, A.; Kahol, S.; Mathur, S.R.; Gogia, A.; Deo, S.V.S.; Prasad, C.P. Prognostic and therapeutic relevance of phosphofructokinase platelet-type (PFKP) in breast cancer. Exp. Cell Res. 2020, 396, 112282. [Google Scholar] [CrossRef]
- Luo, W.; Semenza, G.L. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget 2011, 2, 551–556. [Google Scholar] [CrossRef]
- Ma, C.; Zu, X.; Liu, K.; Bode, A.M.; Dong, Z.; Liu, Z.; Kim, D.J. Knockdown of Pyruvate Kinase M Inhibits Cell Growth and Migration by Reducing NF-kB Activity in Triple-Negative Breast Cancer Cells. Mol. Cells 2019, 42, 628–636. [Google Scholar]
- Grover-McKay, M.; Walsh, S.A.; Seftor, E.A.; Thomas, P.A.; Hendrix, M.J.C. Role for glucose transporter 1 protein in human breast cancer. Pathol. Oncol. Res. 1998, 4, 115–120. [Google Scholar] [CrossRef]
- Brown, R.S.; Wahl, R.L. Overexpression of Glut-1 glucose transporter in human breast cancer. An immunohistochemical study. Cancer 1993, 72, 2979–2985. [Google Scholar] [CrossRef]
- Read, J.A.; Winter, V.J.; Eszes, C.M.; Sessions, R.B.; Brady, R.L. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins 2001, 43, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Zhao, J.K.; Ren, S.; Sun, W.W.; Zhang, W.J.; Zhang, J.N. Wogonin affects proliferation and the energy metabolism of SGC-7901 and A549 cells. Exp. Ther. Med. 2019, 17, 911–918. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef]
- Korga, A.; Ostrowska, M.; Jozefczyk, A.; Iwan, M.; Wojcik, R.; Zgorka, G.; Herbet, M.; Vilarrubla, G.G.; Dudka, J. Apigenin and hesperidin augment the toxic effect of doxorubicin against HepG2 cells. BMC Pharmacol. Toxicol. 2019, 20, 22. [Google Scholar] [CrossRef]
- Pralea, I.-E.; Petrache, A.-M.; Tigu, A.B.; Gulei, D.; Moldovan, R.-C.; Ilieș, M.; Nicoară, R.; Hegheș, S.-C.; Uifălean, A.; Iuga, C.-A. Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies. Pharmaceuticals 2022, 15, 808. [Google Scholar] [CrossRef]
- Arora, S.; Joshi, G.; Chaturvedi, A.; Heuser, M.; Patil, S.; Kumar, R. A Perspective on Medicinal Chemistry Approaches for Targeting Pyruvate Kinase M2. J. Med. Chem. 2022, 65, 1171–1205. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Cui, R.; Ye, L.; Wang, Y.; Wang, X.; Zhang, Q.; Wang, K.; Dong, C.; Le, W.; Chen, B. LINC00941 promotes glycolysis in pancreatic cancer by modulating the Hippo pathway. Mol. Ther.-Nucleic Acids 2021, 26, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-S.; Lee, H.-T.; Lee, M.-H.; Pan, S.-C.; Ke, C.-Y.; Chiu, A.W.; Wei, Y.-H. Role of Mitochondrial DNA Copy Number Alteration in Human Renal Cell Carcinoma. Int. J. Mol. Sci. 2016, 17, 814. [Google Scholar] [CrossRef]
- Hung, B.P.; Hutton, D.L.; Kozielski, K.L.; Bishop, C.J.; Naved, B.; Green, J.J.; Caplan, A.I.; Gimble, J.M.; Dorafshar, A.H.; Grayson, W.L. Platelet-Derived Growth Factor BB Enhances Osteogenesis of Adipose-Derived but Not Bone Marrow-Derived Mesenchymal Stromal/Stem Cells. Stem Cells 2015, 33, 2773–2784. [Google Scholar] [CrossRef] [PubMed]





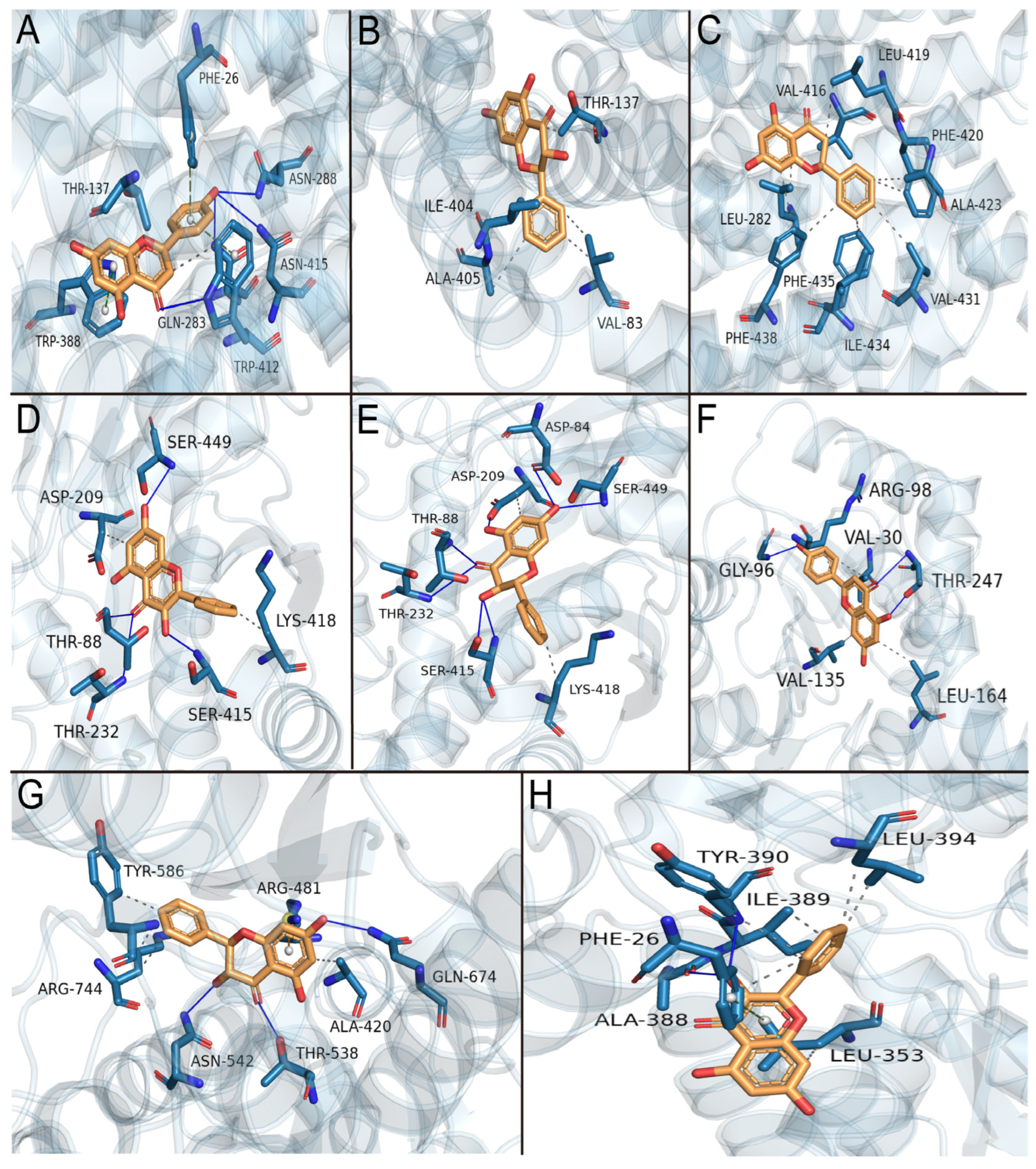
| Protein Targets | PDB ID | Affinity/(kcal·mol−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| Apigenin | Chrysin | Galangin | Caffeic Acid Phenethyl Ester | Caffeic Acid Benzyl Ester | Pinocembrin | Pinobanksin | ||
| GLUT1 | 6THA | −8.7 | −8.6 | −8.4 | −8.5 | −8.6 | −8.6 | −8.7 |
| GLUT3 | 5C65 | −6.2 | −6.4 | −6.4 | −5.7 | −6.0 | −6.5 | −6.4 |
| HK2 | 2NZT | −7.4 | −7.7 | −7.8 | −7.5 | −7.5 | −7.6 | −7.8 |
| LDHA | 4JNK | −8.0 | −7.6 | −7.6 | −7.0 | −7.3 | −7.6 | −7.5 |
| PFK | 4XZ2 | −7.4 | −7.4 | −7.6 | −6.6 | −6.9 | −7.4 | −7.7 |
| PKM2 | 3GQY | −7.1 | −7.3 | −7.4 | −7.3 | −7.2 | −7.2 | −7.2 |
| Apigenin–GLUT1 | Pinobanksin–GLUT1 | Pinocembrin–GLUT3 | Galangin–HK2 | Pinobanksin–HK2 | Apigenin–LDHA | Pinobanksin–PFK | Galangin–PKM2 | |
|---|---|---|---|---|---|---|---|---|
| Affinity (kcal/mol) | −8.7 | −8.7 | −6.5 | −7.8 | −7.8 | −8.0 | −7.7 | −7.4 |
| Number of hydrogen interactions | 4 | - | - | 4 | 7 | 3 | 3 | 2 |
| Amino acid residues involved in hydrogen bonds | Gln283, Asn288, Trp412, Asn415 | - | - | Thr88, Thr232, Ser415, Ser449 | Asp84, Thr88, Asp209, Thr232, Ser415, Ser449 | Gly96, Thr247 | Thr538, Asn542, Gln674 | Ala388, Tyr390 |
| Number of hydrophobic interactions | 2 | 5 | 10 | 2 | 2 | 4 | 4 | 7 |
| Amino acid residues involved in hydrophobic interactions | Thr137, Trp412 | Val83, Thr137, Ile404, Ala405 | Leu282, Val416, Leu419, Phe420, Ala423, Val431, Ile434, Phe435, Phe438 | Asp209, Lys418 | Asp209, Lys418 | Val30, Arg98, Val13, Leu164 | Ala420, Tyr586, Arg744 | Phe26, Leu353, Ile389, Tyr390, Leu394 |
| Number of Π stacking | 3 | - | - | - | - | - | - | 1 |
| Amino acid residues involved in Π stacking | Phe26 (T), Trp412 (T), Trp388 (P) | - | - | - | - | - | - | Phe26 (T) |
| Amino acid residues involved in Π–cation interaction | - | - | - | - | - | - | Arg481 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Liu, Z.; Wu, Q.; Li, Z.; Yang, J.; Xuan, H. Integration with Transcriptomic and Metabolomic Analyses Reveals the In Vitro Cytotoxic Mechanisms of Chinese Poplar Propolis by Triggering the Glucose Metabolism in Human Hepatocellular Carcinoma Cells. Nutrients 2023, 15, 4329. https://doi.org/10.3390/nu15204329
Guo Y, Liu Z, Wu Q, Li Z, Yang J, Xuan H. Integration with Transcriptomic and Metabolomic Analyses Reveals the In Vitro Cytotoxic Mechanisms of Chinese Poplar Propolis by Triggering the Glucose Metabolism in Human Hepatocellular Carcinoma Cells. Nutrients. 2023; 15(20):4329. https://doi.org/10.3390/nu15204329
Chicago/Turabian StyleGuo, Yuyang, Zhengxin Liu, Qian Wu, Zongze Li, Jialin Yang, and Hongzhuan Xuan. 2023. "Integration with Transcriptomic and Metabolomic Analyses Reveals the In Vitro Cytotoxic Mechanisms of Chinese Poplar Propolis by Triggering the Glucose Metabolism in Human Hepatocellular Carcinoma Cells" Nutrients 15, no. 20: 4329. https://doi.org/10.3390/nu15204329
APA StyleGuo, Y., Liu, Z., Wu, Q., Li, Z., Yang, J., & Xuan, H. (2023). Integration with Transcriptomic and Metabolomic Analyses Reveals the In Vitro Cytotoxic Mechanisms of Chinese Poplar Propolis by Triggering the Glucose Metabolism in Human Hepatocellular Carcinoma Cells. Nutrients, 15(20), 4329. https://doi.org/10.3390/nu15204329