Evaluation of a Plant-Based Infant Formula Containing Almonds and Buckwheat on Gut Microbiota Composition, Intestine Morphology, Metabolic and Immune Markers in a Neonatal Piglet Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Animal Care
2.2. Sample Collection
2.3. Microbiome Library Preparation, Sequencing, and Analysis
2.4. Gastrointestinal Tract Histomorphometric Analyses
2.5. Plasma Cytokines Measurements
2.6. ELISA
2.7. Statistical Analysis
3. Results
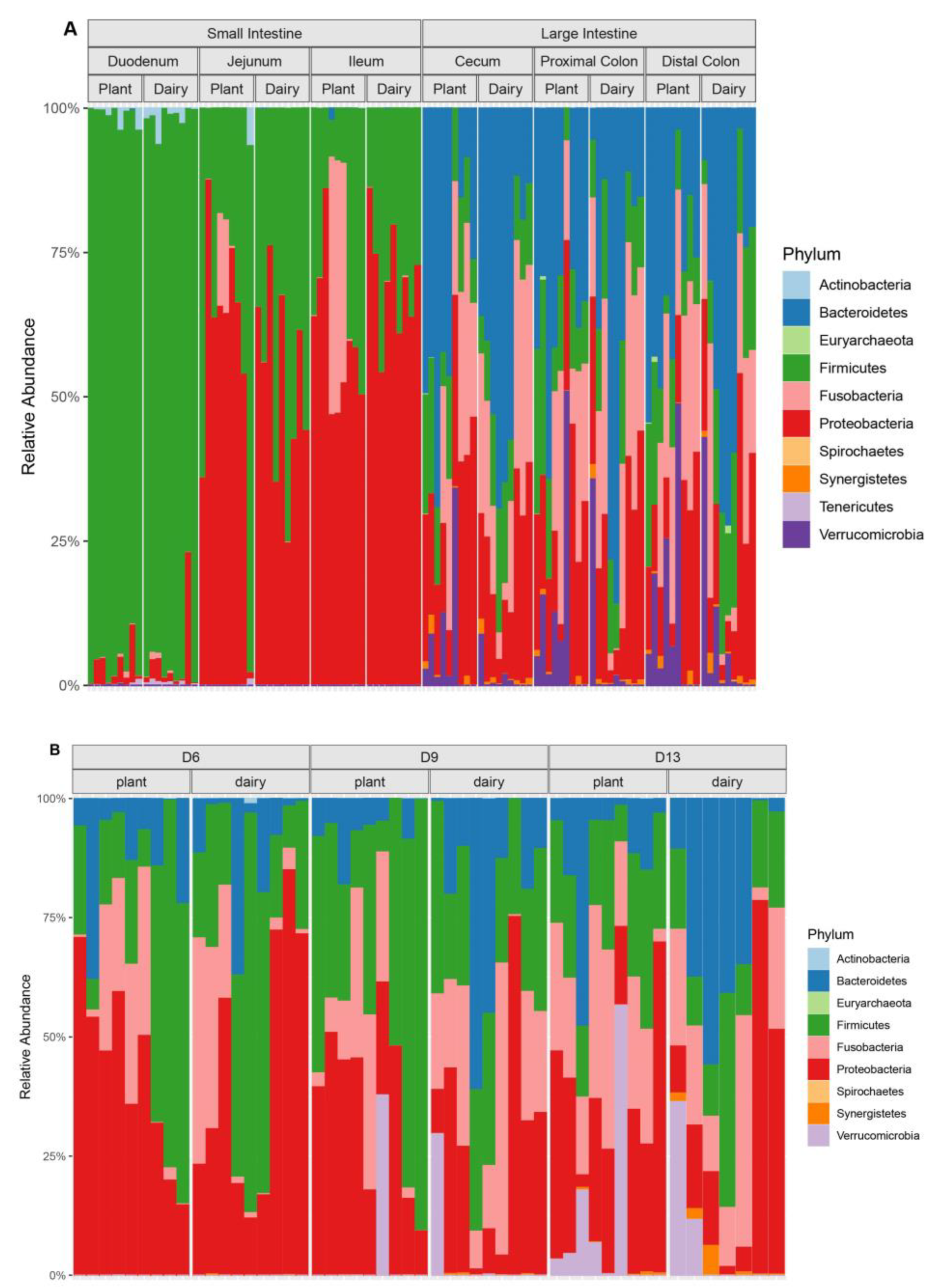
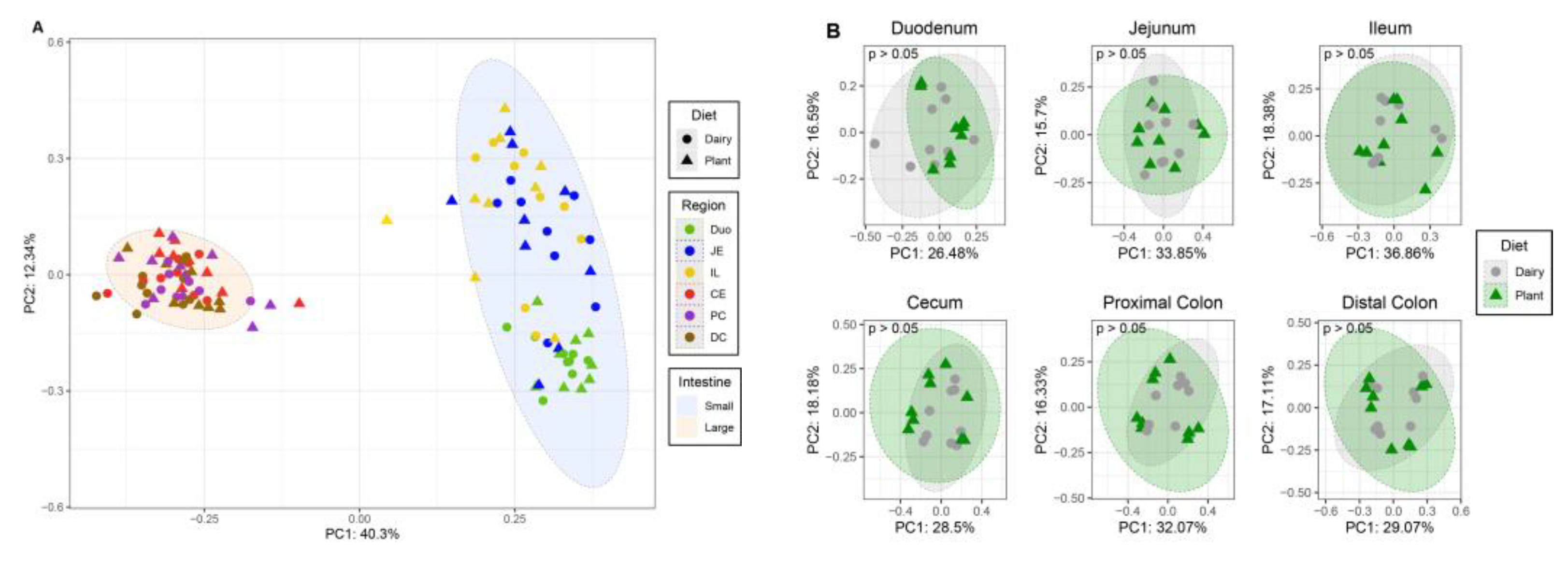
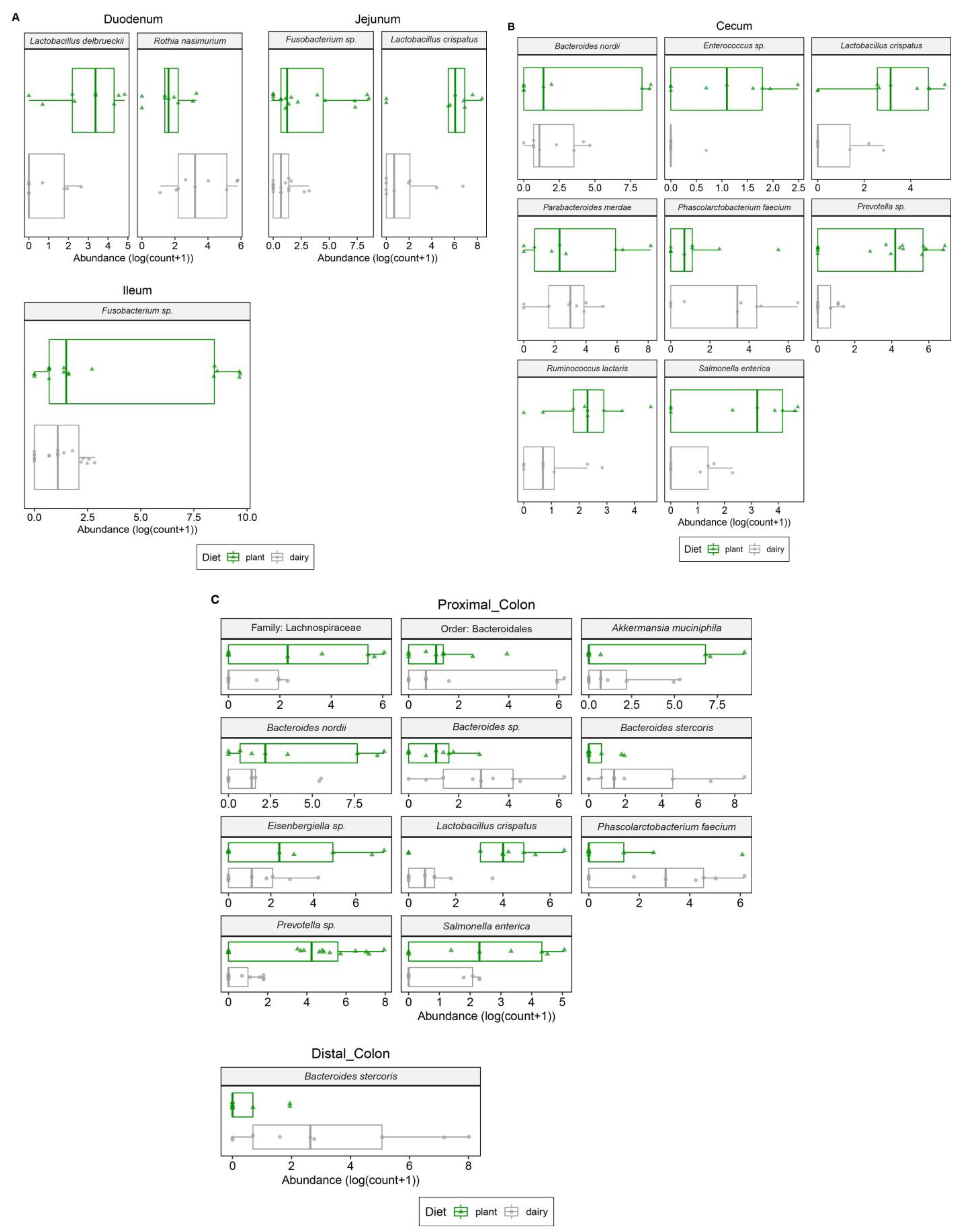
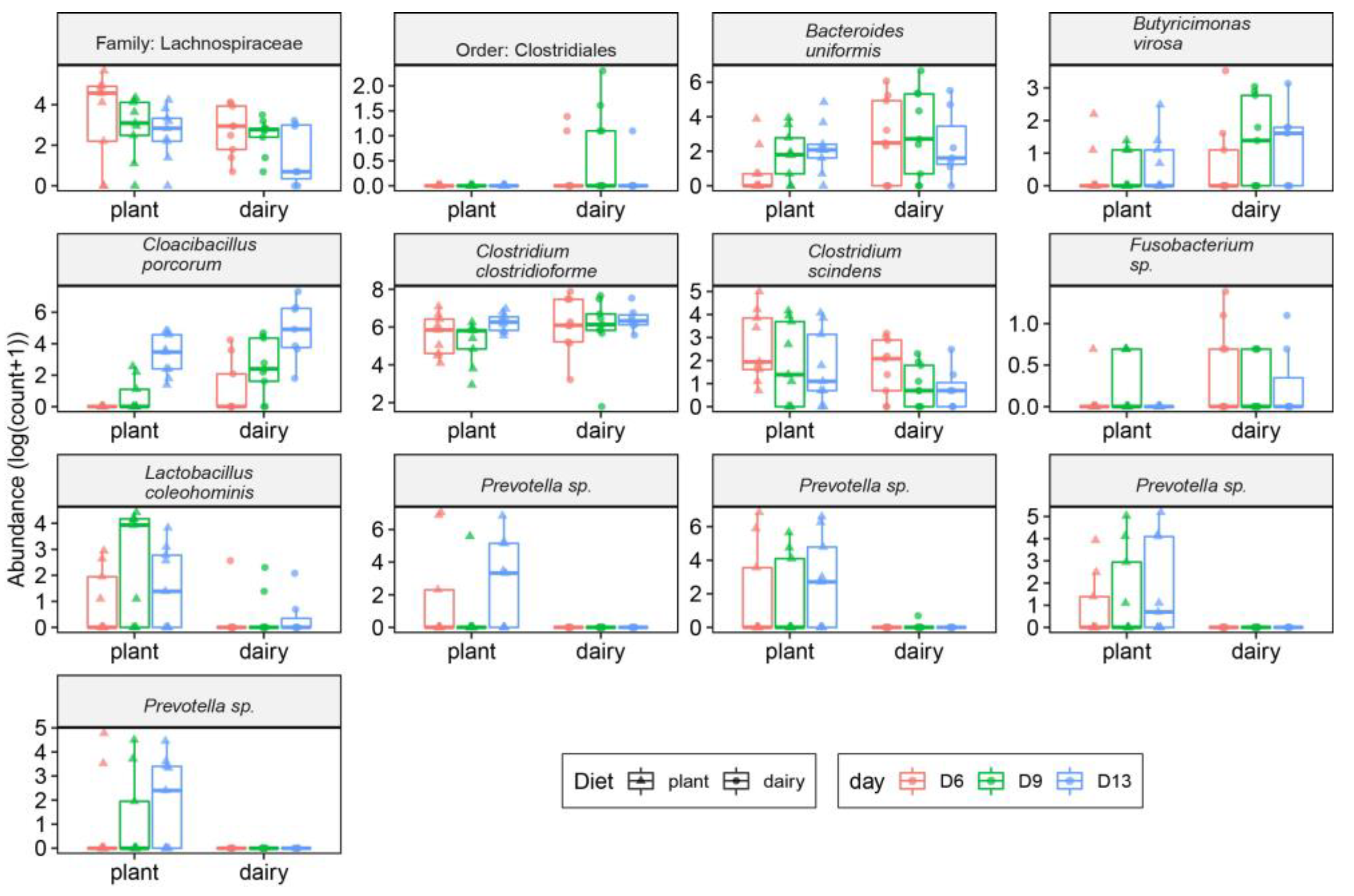
3.1. Plant-Based Formula Altered Microbiota Composition in the Gastrointestinal Tract
3.2. Microbiota Diversity
3.3. Gut and Fecal Microbiota Composition at Taxonomic Level
3.4. Plant-Based Infant Formula Impact on Gut Morphology
3.5. Plant-Based Formula and Its Effects on Circulatory Cytokines
3.6. Plant-Based Infant Formula Impact on Growth, Metabolism Related Parameters, and Immunoglobulin E
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albenberg, L.G.; Wu, G.D. Diet and the intestinal microbiome: Associations, functions, and implications for health and disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The Role of Microbiota in Infant Health: From Early Life to Adulthood. Front. Immunol. 2021, 12, 708472. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Pluske, J.R.; Turpin, D.L.; Kim, J.C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2018, 4, 187–196. [Google Scholar] [CrossRef]
- Perez-Cano, F.J.; Yaqoob, P.; Martin, R.; Castell Escuer, M.; Juarez-Rubio, C. Immunonutrition in early life: Diet and immune development. Clin. Dev. Immunol. 2012, 2012, 207509. [Google Scholar] [CrossRef]
- Halpern, S.R.; Sellars, W.A.; Johnson, R.B.; Anderson, D.W.; Saperstein, S.; Reisch, J.S. Development of childhood allergy in infants fed breast, soy, or cow milk. J. Allergy Clin. Immunol 1973, 51, 139–151. [Google Scholar] [CrossRef]
- Stintzing, G.; Zetterstrom, R. Cow’s milk allergy, incidence and pathogenetic role of early exposure to cow’s milk formula. Acta Paediatr Scand. 1979, 68, 383–387. [Google Scholar] [CrossRef]
- Schoemaker, A.A.; Sprikkelman, A.B.; Grimshaw, K.E.; Roberts, G.; Grabenhenrich, L.; Rosenfeld, L.; Siegert, S.; Dubakiene, R.; Rudzeviciene, O.; Reche, M.; et al. Incidence and natural history of challenge-proven cow’s milk allergy in European children--EuroPrevall birth cohort. Allergy 2015, 70, 963–972. [Google Scholar] [CrossRef]
- Flom, J.D.; Sicherer, S.H. Epidemiology of Cow’s Milk Allergy. Nutrients 2019, 11, 1051. [Google Scholar] [CrossRef]
- Mousan, G.; Kamat, D. Cow’s Milk Protein Allergy. Clin. Pediatr. 2016, 55, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Brozek, J.; Schunemann, H.; Bahna, S.L.; von Berg, A.; Beyer, K.; Bozzola, M.; Bradsher, J.; Compalati, E.; Ebisawa, M.; et al. World Allergy Organization (WAO) Diagnosis and Rationale for Action against Cow’s Milk Allergy (DRACMA) Guidelines. World Allergy Organ. J. 2010, 3, 57–161. [Google Scholar] [CrossRef] [PubMed]
- Merritt, R.J.; Fleet, S.E.; Fifi, A.; Jump, C.; Schwartz, S.; Sentongo, T.; Duro, D.; Rudolph, J.; Turner, J.; NASPGHAN Committee on Nutrition. North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition Position Paper: Plant-based Milks. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 276–281. [Google Scholar] [CrossRef]
- Bhatia, J.; Greer, F.; Committee on Nutrition. Use of soy protein-based formulas in infant feeding. Pediatrics 2008, 121, 1062–1068. [Google Scholar] [CrossRef]
- Tuohy, P.G. Soy infant formula and phytoestrogens. J. Paediatr. Child Health 2003, 39, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Andres, A.; Casey, P.H.; Cleves, M.A.; Badger, T.M. Body fat and bone mineral content of infants fed breast milk, cow’s milk formula, or soy formula during the first year of life. J. Pediatr. 2013, 163, 49–54. [Google Scholar] [CrossRef]
- Rosa, F.; Yelvington, B.; Terry, N.; Tripp, P.; Pittman, H.E., 3rd; Fay, B.L.; Ross, T.J.; Sikes, J.D.; Flowers, J.B.; Bar-Yoseph, F.; et al. Evaluation of the Safety of a Plant-Based Infant Formula Containing Almonds and Buckwheat in a Neonatal Piglet Model. Nutrients 2022, 14, 1499. [Google Scholar] [CrossRef]
- Council, N.R. Nutrient Requirements of Swine: Eleventh Revised Edition; The National Academies Press: Washington, DC, USA, 2012; p. 420. [Google Scholar] [CrossRef]
- Yoav Benjamini, Y.H. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Section on Breastfeeding; Eidelman, A.I.; Schanler, R.J.; Johnston, M.; Landers, S.; Noble, L.; Szucs, K.; Viehmann, L. Breastfeeding and the use of human milk. Pediatrics 2012, 129, e827–e841. [Google Scholar] [CrossRef]
- Dawod, B.; Marshall, J.S.; Azad, M.B. Breastfeeding and the developmental origins of mucosal immunity: How human milk shapes the innate and adaptive mucosal immune systems. Curr. Opin. Gastroenterol. 2021, 37, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Praveen, P.; Jordan, F.; Priami, C.; Morine, M.J. The role of breast-feeding in infant immune system: A systems perspective on the intestinal microbiome. Microbiome 2015, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Ficara, M.; Pietrella, E.; Spada, C.; Della Casa Muttini, E.; Lucaccioni, L.; Iughetti, L.; Berardi, A. Changes of intestinal microbiota in early life. J. Matern.-Fetal Neonatal Med. 2020, 33, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Fabiano, V.; Indrio, F.; Verduci, E.; Calcaterra, V.; Pop, T.L.; Mari, A.; Zuccotti, G.V.; Cullu Cokugras, F.; Pettoello-Mantovani, M.; Goulet, O. Term Infant Formulas Influencing Gut Microbiota: An Overview. Nutrients 2021, 13, 4200. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016, 6, 27427. [Google Scholar] [CrossRef]
- Valeriano, V.D.; Balolong, M.P.; Kang, D.K. Probiotic roles of Lactobacillus sp. in swine: Insights from gut microbiota. J. Appl. Microbiol. 2017, 122, 554–567. [Google Scholar] [CrossRef]
- Chen, F.; Wang, H.; Chen, J.; Liu, Y.; Wen, W.; Li, Y.; Huang, X. Lactobacillus delbrueckii Ameliorates Intestinal Integrity and Antioxidant Ability in Weaned Piglets after a Lipopolysaccharide Challenge. Oxidative Med. Cell. Longev. 2020, 2020, 6028606. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Greer, R.L.; Dong, X.; Moraes, A.C.; Zielke, R.A.; Fernandes, G.R.; Peremyslova, E.; Vasquez-Perez, S.; Schoenborn, A.A.; Gomes, E.P.; Pereira, A.C.; et al. Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat. Commun. 2016, 7, 13329. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms 2020, 8, 1584. [Google Scholar] [CrossRef]
- Guevarra, R.B.; Hong, S.H.; Cho, J.H.; Kim, B.-R.; Shin, J.; Lee, J.H.; Kang, B.N.; Kim, Y.H.; Wattanaphansak, S.; Isaacson, R.E.; et al. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J. Anim. Sci. Biotechnol. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Correa, F.; Luise, D.; Bosi, P.; Trevisi, P. Weaning differentially affects the maturation of piglet peripheral blood and jejunal Peyer’s patches. Sci. Rep. 2022, 12, 1604. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, T.; Inoue, R.; Nakatani, M.; Fukuta, K.; Kishino, E.; Ito, T.; Ushida, K. Influence of weaning age on the villous height and disaccharidase activities in the porcine small intestine. Anim. Sci. J. 2016, 87, 67–75. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Schloss, P.D. Amplicon Sequence Variants Artificially Split Bacterial Genomes into Separate Clusters. mSphere 2021, 6, e0019121. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurung, M.; Rosa, F.; Yelvington, B.; Terry, N.; Read, Q.D.; Piccolo, B.D.; Moody, B.; Tripp, P.; Pittman, H.E., III; Fay, B.L.; et al. Evaluation of a Plant-Based Infant Formula Containing Almonds and Buckwheat on Gut Microbiota Composition, Intestine Morphology, Metabolic and Immune Markers in a Neonatal Piglet Model. Nutrients 2023, 15, 383. https://doi.org/10.3390/nu15020383
Gurung M, Rosa F, Yelvington B, Terry N, Read QD, Piccolo BD, Moody B, Tripp P, Pittman HE III, Fay BL, et al. Evaluation of a Plant-Based Infant Formula Containing Almonds and Buckwheat on Gut Microbiota Composition, Intestine Morphology, Metabolic and Immune Markers in a Neonatal Piglet Model. Nutrients. 2023; 15(2):383. https://doi.org/10.3390/nu15020383
Chicago/Turabian StyleGurung, Manoj, Fernanda Rosa, Brooke Yelvington, Nathan Terry, Quentin D. Read, Brian D. Piccolo, Becky Moody, Patricia Tripp, Hoy E. Pittman, III, Bobby L. Fay, and et al. 2023. "Evaluation of a Plant-Based Infant Formula Containing Almonds and Buckwheat on Gut Microbiota Composition, Intestine Morphology, Metabolic and Immune Markers in a Neonatal Piglet Model" Nutrients 15, no. 2: 383. https://doi.org/10.3390/nu15020383
APA StyleGurung, M., Rosa, F., Yelvington, B., Terry, N., Read, Q. D., Piccolo, B. D., Moody, B., Tripp, P., Pittman, H. E., III, Fay, B. L., Ross, T. J., Sikes, J. D., Flowers, J. B., Fox, R., LeRoith, T., Talatala, R., Bar-Yoseph, F., & Yeruva, L. (2023). Evaluation of a Plant-Based Infant Formula Containing Almonds and Buckwheat on Gut Microbiota Composition, Intestine Morphology, Metabolic and Immune Markers in a Neonatal Piglet Model. Nutrients, 15(2), 383. https://doi.org/10.3390/nu15020383





