Unraveling the Gut Microbiome–Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
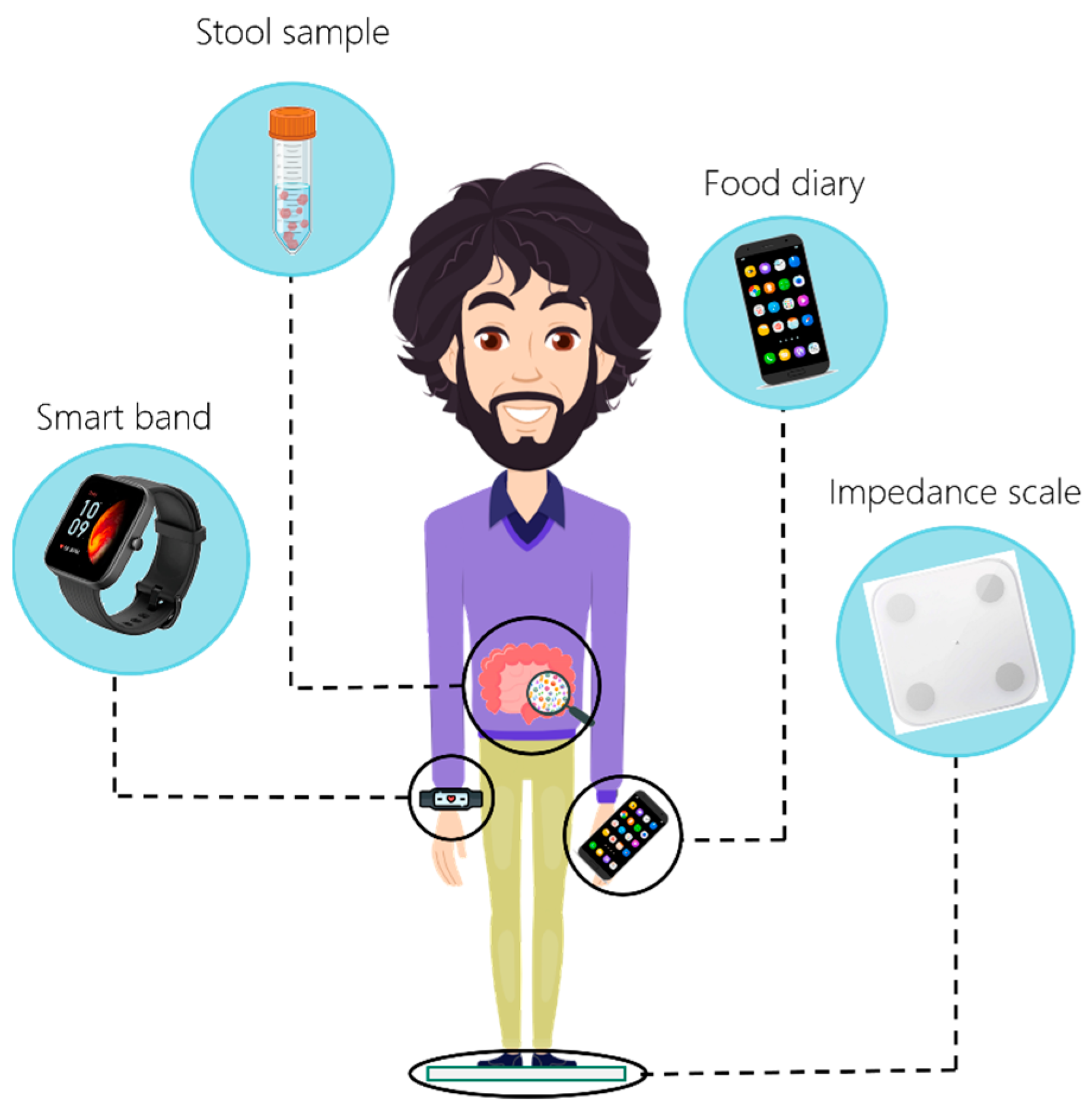
2.2. Timeline
2.3. Sample Collection Procedure for Nutrigenomics
2.4. Sample Collection Procedure and Analysis of Microbiome
2.5. Formulation of the Personalized Nutritional Plan
2.6. Data Organization and Analysis
2.7. Statistics
3. Results
3.1. Precision Nutrition
3.2. Changes in Food Intake
3.3. Changes in Nutritional Variables
3.4. Effects of the Nutritional Plan on Anthropometric and Physiological Parameters of the Participants
3.5. Effects of the Nutritional Plan on the GUT Microbiome
3.5.1. Evaluation of the Stability of the Microbial Composition
3.5.2. Evaluation of the Changes in Composition after the Nutritional Intervention
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional Interactions between the Gut Microbiota and Host Metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut Microbiota and Aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; Bäckhed, F. Diet-Microbiota Interactions as Moderators of Human Metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- O’Riordan, K.J.; Collins, M.K.; Moloney, G.M.; Knox, E.G.; Aburto, M.R.; Fülling, C.; Morley, S.J.; Clarke, G.; Schellekens, H.; Cryan, J.F. Short Chain Fatty Acids: Microbial Metabolites for Gut-Brain Axis Signalling. Mol. Cell. Endocrinol. 2022, 546, 111572. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.-H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-N.; Liu, X.-T.; Liang, Z.-H.; Wang, J.-H. Gut Microbiota in Obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The Gut Microbiota and Inflammatory Bowel Disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef]
- Altamirano-Barrera, A.; Uribe, M.; Chávez-Tapia, N.C.; Nuño-Lámbarri, N. The Role of the Gut Microbiota in the Pathology and Prevention of Liver Disease. J. Nutr. Biochem. 2018, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Slavin, J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef]
- Martín, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing Functional and Translational Microbiome Research Using Meta-Omics Approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B.-M.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.-H. Health Impact and Therapeutic Manipulation of the Gut Microbiome. High Throughput 2020, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.J.; Vangay, P.; Al-Ghalith, G.A.; Hillmann, B.M.; Ward, T.L.; Shields-Cutler, R.R.; Kim, A.D.; Shmagel, A.K.; Syed, A.N.; Personalized Microbiome Class Students; et al. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host. Microbe. 2019, 25, 789–802.e5. [Google Scholar] [CrossRef]
- Livingstone, K.M.; Ramos-Lopez, O.; Pérusse, L.; Kato, H.; Ordovas, J.M.; Martínez, J.A. Precision Nutrition: A Review of Current Approaches and Future Endeavors. Trends Food Sci. Technol. 2022, 128, 253–264. [Google Scholar] [CrossRef]
- Walther, B.; Lett, A.M.; Bordoni, A.; Tomás-Cobos, L.; Nieto, J.A.; Dupont, D.; Danesi, F.; Shahar, D.R.; Echaniz, A.; Re, R.; et al. GutSelf: Interindividual Variability in the Processing of Dietary Compounds by the Human Gastrointestinal Tract. Mol. Nutr. Food Res. 2019, 63, 1900677. [Google Scholar] [CrossRef]
- Nogal, B.; Blumberg, J.B.; Blander, G.; Jorge, M. Gut Microbiota–Informed Precision Nutrition in the Generally Healthy Individual: Are We There Yet? Curr. Dev. Nutr. 2021, 5, nzab107. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.J.; Zheng, J.J.; Kang, J.W.; Saboe, A.; Knights, D.; Zivkovic, A.M. A Guide to Diet-Microbiome Study Design. Front. Nutr. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Kuhnle, G.G.C. Nutritional Biomarkers for Objective Dietary Assessment. J. Sci. Food Agric. 2012, 92, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.-S.; Oh, K.; Kim, H.C. Dietary Assessment Methods in Epidemiologic Studies. Epidemiol. Health 2014, 36, e2014009. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Menichetti, G.; Loscalzo, J. The Unmapped Chemical Complexity of Our Diet. Nat. Food 2020, 1, 33–37. [Google Scholar] [CrossRef]
- Bianchetti, G.; Taralli, S.; Vaccaro, M.; Indovina, L.; Mattoli, M.V.; Capotosti, A.; Scolozzi, V.; Calcagni, M.L.; Giordano, A.; De Spirito, M.; et al. Automated Detection and Classification of Tumor Histotypes on Dynamic PET Imaging Data through Machine-Learning Driven Voxel Classification. Comput. Biol. Med. 2022, 145, 105423. [Google Scholar] [CrossRef]
- Bianchetti, G.; Di Giacinto, F.; De Spirito, M.; Maulucci, G. Machine-Learning Assisted Confocal Imaging of Intracellular Sites of Triglycerides and Cholesteryl Esters Formation and Storage. Anal. Chim. Acta 2020, 1121, 57–66. [Google Scholar] [CrossRef]
- Mancin, L.; Rollo, I.; Mota, J.F.; Piccini, F.; Carletti, M.; Susto, G.A.; Valle, G.; Paoli, A. Optimizing Microbiota Profiles for Athletes. Exerc. Sport Sci. Rev. 2021, 49, 42. [Google Scholar] [CrossRef]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the Gut Microbiota in Nutrition and Health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed]
- Ramos Meyers, G.; Samouda, H.; Bohn, T. Short Chain Fatty Acid Metabolism in Relation to Gut Microbiota and Genetic Variability. Nutrients 2022, 14, 5361. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Reglero, G.; Ordovás, J.M.; Dávalos, A. NutriGenomeDB: A Nutrigenomics Exploratory and Analytical Platform. Database 2019, 2019, baz097. [Google Scholar] [CrossRef]
- Posteraro, P.; De Maio, F.; Menchinelli, G.; Palucci, I.; Errico, F.M.; Carbone, M.; Sanguinetti, M.; Gasbarrini, A.; Posteraro, B. First Bloodstream Infection Caused by Prevotella Copri in a Heart Failure Elderly Patient with Prevotella-Dominated Gut Microbiota: A Case Report. Gut. Pathog. 2019, 11, 44. [Google Scholar] [CrossRef]
- De Maio, F.; Ianiro, G.; Coppola, G.; Santopaolo, F.; Abbate, V.; Bianco, D.M.; Del Zompo, F.; De Matteis, G.; Leo, M.; Nesci, A.; et al. Improved Gut Microbiota Features after the Resolution of SARS-CoV-2 Infection. Gut. Pathog. 2021, 13, 62. [Google Scholar] [CrossRef] [PubMed]
- De Maio, F.; Boru, C.E.; Avallone, M.; Velotti, N.; Bianco, D.M.; Capoccia, D.; Greco, F.; Guarisco, G.; Nogara, M.; Sanguinetti, M.; et al. Characterization of Gut Microbiota in Patients with Metabolic Syndrome Candidates for Bariatric/Metabolic Surgery: Preliminary Findings of a Multi-Center Prospective Study. Diabetes Res. Clin. Pract. 2021, 180, 109079. [Google Scholar] [CrossRef]
- Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. Available online: https://pubmed.ncbi.nlm.nih.gov/23630581/ (accessed on 15 June 2023).
- World Health Organization (Ed.) Promoting a Healthy Diet for the WHO Eastern Mediterranean Region: User-Friendly Guide; World Health Organization, Regional Office for the Eastern Mediterranean: Cairo, Egypt, 2012; ISBN 978-92-9021-834-0.
- EFSA Panel on Dietetic Products, Nutrition, and Allergies (NDA). Scientific Opinion on Establishing Food-Based Dietary Guidelines. EFS2 2010, 8, 1460. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martín, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the Fecal Microbiota: An Optimized Ion Torrent 16S RRNA Gene-Based Analysis Protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bianchetti, G.; Abeltino, A.; Serantoni, C.; Ardito, F.; Malta, D.; De Spirito, M.; Maulucci, G. Personalized Self-Monitoring of Energy Balance through Integration in a Web-Application of Dietary, Anthropometric, and Physical Activity Data. J. Pers. Med. 2022, 12, 568. [Google Scholar] [CrossRef] [PubMed]
- Abeltino, A.; Bianchetti, G.; Serantoni, C.; Ardito, C.F.; Malta, D.; De Spirito, M.; Maulucci, G. Personalized Metabolic Avatar: A Data Driven Model of Metabolism for Weight Variation Forecasting and Diet Plan Evaluation. Nutrients 2022, 14, 3520. [Google Scholar] [CrossRef]
- Abeltino, A.; Bianchetti, G.; Serantoni, C.; Riente, A.; De Spirito, M.; Maulucci, G. Putting the Personalized Metabolic Avatar into Production: A Comparison between Deep-Learning and Statistical Models for Weight Prediction. Nutrients 2023, 15, 1199. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAO/WHO Global Individual Food Consumption Data Tool (GIFT): Methodological Document; FAO: Rome, Italy, 2022. [Google Scholar]
- Pu, F.; Chen, N.; Xue, S. Calcium Intake, Calcium Homeostasis and Health. Food Sci. Hum. Wellness 2016, 5, 8–16. [Google Scholar] [CrossRef]
- Stone, M.S.; Martyn, L.; Weaver, C.M. Potassium Intake, Bioavailability, Hypertension, and Glucose Control. Nutrients 2016, 8, 444. [Google Scholar] [CrossRef] [PubMed]
- Serna, J.; Bergwitz, C. Importance of Dietary Phosphorus for Bone Metabolism and Healthy Aging. Nutrients 2020, 12, 3001. [Google Scholar] [CrossRef]
- Farquhar, W.B.; Edwards, D.G.; Jurkovitz, C.T.; Weintraub, W.S. Dietary Sodium and Health: More Than Just Blood Pressure. J. Am. Coll. Cardiol. 2015, 65, 1042–1050. [Google Scholar] [CrossRef]
- Prasad, A.S. Zinc in Human Health: Effect of Zinc on Immune Cells. Mol. Med. 2008, 14, 353–357. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.H.; Wu, J.H.Y.; Appel, L.J.; Creager, M.A.; Kris-Etherton, P.M.; Miller, M.; Rimm, E.B.; Rudel, L.L.; Robinson, J.G.; et al. Dietary Fats and Cardiovascular Disease: A Presidential Advisory From the American Heart Association. Circulation 2017, 136, e1–e23. [Google Scholar] [CrossRef] [PubMed]
- Hever, J.; Cronise, R.J. Plant-Based Nutrition for Healthcare Professionals: Implementing Diet as a Primary Modality in the Prevention and Treatment of Chronic Disease. J. Geriatr. Cardiol. 2017, 14, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.J.; Racette, S.B. The Utility of Body Composition Assessment in Nutrition and Clinical Practice: An Overview of Current Methodology. Nutrients 2021, 13, 2493. [Google Scholar] [CrossRef]
- Saxena, A.; Minton, D.; Lee, D.; Sui, X.; Fayad, R.; Lavie, C.J.; Blair, S.N. Protective Role of Resting Heart Rate on All-Cause and Cardiovascular Disease Mortality. Mayo Clin. Proc. 2013, 88, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- Serantoni, C.; Zimatore, G.; Bianchetti, G.; Abeltino, A.; De Spirito, M.; Maulucci, G. Unsupervised Clustering of Heartbeat Dynamics Allows for Real Time and Personalized Improvement in Cardiovascular Fitness. Sensors 2022, 22, 3974. [Google Scholar] [CrossRef]
- Eugene, A.R.; Masiak, J. The Neuroprotective Aspects of Sleep. MEDtube Sci. 2015, 3, 35–40. [Google Scholar]
- Peterson, L.W.; Artis, D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host. Microbe. 2015, 17, 260–273. [Google Scholar] [CrossRef]
- Torii, A.; Torii, S.; Fujiwara, S.; Tanaka, H.; Inagaki, N.; Nagai, H. Lactobacillus Acidophilus Strain L-92 Regulates the Production of Th1 Cytokine as Well as Th2 Cytokines. Allergol. Int. 2007, 56, 293–301. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Mazmanian, S.K. Control of Brain Development, Function, and Behavior by the Microbiome. Cell. Host. Microbe. 2015, 17, 565–576. [Google Scholar] [CrossRef]
- Jiang, S.; Fan, Q.; Zhang, Z.; Deng, Y.; Wang, L.; Dai, Q.; Wang, J.; Lin, M.; Zhou, J.; Long, Z.; et al. Biodegradation of Oil by a Newly Isolated Strain Acinetobacter Junii WCO-9 and Its Comparative Pan-Genome Analysis. Microorganisms 2023, 11, 407. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Napoli, S.; Alessandri, G.; Mancabelli, L.; Anzalone, R.; Longhi, G.; Viappiani, A.; Mangifesta, M.; Lugli, G.A.; et al. Colonization of the Human Gut by Bovine Bacteria Present in Parmesan Cheese. Nat. Commun. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Haskey, N.; Estaki, M.; Ye, J.; Shim, R.K.; Singh, S.; Dieleman, L.A.; Jacobson, K.; Gibson, D.L. A Mediterranean Diet Pattern Improves Intestinal Inflammation Concomitant with Reshaping of the Bacteriome in Ulcerative Colitis: A Randomised Controlled Trial. J. Crohn’s Colitis 2023, jjad073. [Google Scholar] [CrossRef]
- Samanta, I.; Bandyopadhyay, S. Chapter 14—Klebsiella. In Antimicrobial Resistance in Agriculture; Samanta, I., Bandyopadhyay, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 153–169. ISBN 978-0-12-815770-1. [Google Scholar]
- Dong, N.; Yang, X.; Chan, E.W.-C.; Zhang, R.; Chen, S. Klebsiella Species: Taxonomy, Hypervirulence and Multidrug Resistance. eBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef]
- Noriega, B.S.; Sanchez-Gonzalez, M.A.; Salyakina, D.; Coffman, J. Understanding the Impact of Omega-3 Rich Diet on the Gut Microbiota. Case Rep. Med. 2016, 2016, e3089303. [Google Scholar] [CrossRef]
- Filippis, F.D.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; Storia, A.L.; Laghi, L.; Serrazanetti, D.I.; Cagno, R.D.; Ferrocino, I.; Lazzi, C.; et al. High-Level Adherence to a Mediterranean Diet Beneficially Impacts the Gut Microbiota and Associated Metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Zhao, Y.; Cui, Y.; Zhong, C.; Zha, Y.; Li, S.; Cao, G.; Li, M.; Zhang, L.; Ning, K.; et al. Stage-Specific Roles of Microbial Dysbiosis and Metabolic Disorders in Rheumatoid Arthritis. Ann. Rheum. Dis. 2022, 81, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced Dietary Intake of Carbohydrates by Obese Subjects Results in Decreased Concentrations of Butyrate and Butyrate-Producing Bacteria in Feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia Intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front. Cell. Infect. Microbiol. 2021, 11, 757718. [Google Scholar] [CrossRef]
- Duncan, S.H.; Aminov, R.I.; Scott, K.P.; Louis, P.; Stanton, T.B.; Flint, H.J. Proposal of Roseburia Faecis Sp. Nov., Roseburia Hominis Sp. Nov. and Roseburia Inulinivorans Sp. Nov., Based on Isolates from Human Faeces. Int. J. Syst. Evol. Microbiol. 2006, 56, 2437–2441. [Google Scholar] [CrossRef]
- Haro, C.; Montes-Borrego, M.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Tinahones, F.J.; Landa, B.B.; et al. Two Healthy Diets Modulate Gut Microbial Community Improving Insulin Sensitivity in a Human Obese Population. J. Clin. Endocrinol. Metab. 2016, 101, 233–242. [Google Scholar] [CrossRef]
- Moura, G.B.; Silva, M.G.; Marconi, C. Milk and Dairy Consumption and Its Relationship With Abundance of Lactobacillus Crispatus in the Vaginal Microbiota: Milk Intake and Vaginal Lactobacillus. J. Low. Genit. Tract. Dis. 2023, 27, 280–285. [Google Scholar] [CrossRef]
- Patrignani, F.; Siroli, L.; Parolin, C.; Serrazanetti, D.I.; Vitali, B.; Lanciotti, R. Use of Lactobacillus Crispatus to Produce a Probiotic Cheese as Potential Gender Food for Preventing Gynaecological Infections. PLoS ONE 2019, 14, e0208906. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between Diet, Gut Microbiota Composition and Gut Metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Forslund, S.K.; Gudmundsdottir, V.; Petersen, A.Ø.; Hildebrand, F.; Hyötyläinen, T.; Nielsen, T.; Hansen, T.; Bork, P.; Ehrlich, S.D.; et al. A Computational Framework to Integrate High-Throughput “-Omics” Datasets for the Identification of Potential Mechanistic Links. Nat. Protoc. 2018, 13, 2781–2800. [Google Scholar] [CrossRef] [PubMed]
- Plaza Oñate, F.; Le Chatelier, E.; Almeida, M.; Cervino, A.C.L.; Gauthier, F.; Magoulès, F.; Ehrlich, S.D.; Pichaud, M. MSPminer: Abundance-Based Reconstitution of Microbial Pan-Genomes from Shotgun Metagenomic Data. Bioinformatics 2019, 35, 1544–1552. [Google Scholar] [CrossRef] [PubMed]


| Super-Category | Category | Gene | rs_Number |
|---|---|---|---|
| Weight management | Carbohydrates | ADRB2 | rs1042713 |
| TCF7L2 | rs7903146 | ||
| Proteins | FTO | rs1558902 | |
| FTO | rs9930506 | ||
| FTO | rs9939609 | ||
| Fats | TCF7L2 | rs12255372 | |
| FTO | rs9930506 | ||
| PPM1K | rs1440581 | ||
| PPARG | rs1801282 | ||
| FTO | rs9939609 | ||
| Snacking between meals | MC4R | rs17782313 | |
| Sweet taste preference | SLC2A2 | rs5400 | |
| Biological clock | CLOCK | rs1801260 | |
| Salt sensitivity | ACE | rs4343 | |
| AGT | rs699 | ||
| ATP2B1 | rs2681472 | ||
| Saturated fats | APOE | rs7412 | |
| APO3 | rs429358 | ||
| APOA2 | rs5082 | ||
| ω6/ω3 fatty acids | FADS1 | rs174546 | |
| FADS2 | rs174570 | ||
| Trans fats | FADS1 | rs174546 | |
| LIPC | rs1800588 | ||
| APOC3 | rs5128 | ||
| Sensitivities | Caffeine | ADORA2A | rs2298383 |
| ADORA2A | rs5751876 | ||
| CYP1A2 | rs762551 | ||
| Alcohol | ADH1C | rs283411 | |
| GABRA2 | rs279858 | ||
| Lactose | MCM6 | rs4988235 | |
| Gluten | HLA DQ 2.2 | rs2395182 | |
| HLA DQ 2.2 | rs4713586 | ||
| HLA–DQA1 | rs2187668 | ||
| HLA–DQB1 | rs7775228 | ||
| HLA DQ | rs7454108 | ||
| Detoxification capacity and antioxidant needs | Detoxification capacity | CYP1A2 | rs762551 |
| GSTP1 | rs1695 | ||
| Antioxidant needs | SOD2 | rs4880 | |
| CAT | rs1001179 | ||
| Vitamins | Vitamin A | BCO1 | rs6564851 |
| BCMO1 | rs7501331 | ||
| Vitamin B6 | ALPL | rs4654748 | |
| Vitamin B9—Folic and Folic acid | MTHFR | rs1801133 | |
| Vitamin B12 | FUT2 | rs492602 | |
| TCN1 | rs526934 | ||
| Vitamin C | SLC23A1 | rs10063949 | |
| SLC23A2 | rs6053005 | ||
| Vitamin D | CYP2R1 | rs10741657 | |
| GC | rs2282679 | ||
| VDR | rs2228570 | ||
| Vitamin E | SCARB1 | rs11057830 | |
| TRIP6 | rs964184 | ||
| Minerals | Low calcium levels | CYP2R1 | rs2060793 |
| GC | rs7041 | ||
| VDR | rs2228570 | ||
| Increased calcium concentration | CYP24A1 | rs1570669 | |
| Low iron levels | TMPRSS6 | rs4820268 | |
| TF | rs1799852 | ||
| TFR2 | rs7385804 | ||
| Iron overload | HFE | rs1799945 | |
| Magnesium | MUC1 | rs4072037 | |
| Sports profile | Endurance | ACE | rs4343 |
| PPARA | rs4253778 | ||
| HFE | rs1799945 | ||
| NFIA–AS2 | rs1572312 | ||
| ADRB3 | rs4994 | ||
| HIF1A | rs11549465 | ||
| PPARD | rs2016520 | ||
| NRF2 | rs7181866 | ||
| Strength | MSTN | rs1805086 | |
| PPARA | rs4253778 | ||
| ACTN3 | rs1815739 | ||
| AGT | rs699 | ||
| Power | ACTN3 | rs1815739 | |
| NOS3 | rs2070744 | ||
| ACE | rs4343 | ||
| AGT | rs699 | ||
| ADRB2 | rs1042713 | ||
| Aerobic capacity (VO2 max) | ADRB2 | rs1042713 | |
| CRP | rs1205 | ||
| GSTP1 | rs1695 | ||
| ACE | rs4343 | ||
| Muscle mass hypertrophy | LEPR | rs1137101 | |
| Motivation to exercise | BDNF | rs6265 | |
| COMT | rs4680 | ||
| Injury predisposition | Pain tolerance | COMT | rs4680 |
| Jumper’s knee and tennis elbow injuries | COL5A1 | rs12722 | |
| COL1A1 | rs1800012 | ||
| COL3A1 | rs1800255 | ||
| Achille’s tendon injury | COL5A1 | rs12722 | |
| Musculoskeletal health | BTNL2 | rs10947262 | |
| SPTBN1 | rs11898505 | ||
| Exercise rehabilitation | CRP | rs1205 | |
| SOD2 | rs4880 | ||
| ACTN3 | rs1815739 |
| Subject | Age | % Macronutrient Intake | Diet Type | % Macronutrients Diet-Provided |
|---|---|---|---|---|
| WL010114 | 26 | CHO: 62.8%—PRO: 18.3%—LIP: 18.9% | Mediterranean | CHO: 47.9%—PRO: 21.9%—LIP: 30.2% |
| WL010112 | 28 | CHO: 61.9%—PRO: 18.0%—LIP: 20.1% | Mediterranean (with high fish intake) | CHO: 47.1%—PRO: 23.3%—LIP: 29.5% |
| WL010111 | 44 | CHO: 64.2%—PRO: 19.0%—LIP: 16.8% | Mediterranean (with high fish intake) | CHO: 45.2%—PRO: 24.4%—LIP: 30.4% |
| WL010107 | 46 | CHO: 52.6%—PRO: 27.5%—LIP: 19.8% | Mediterranean | CHO: 47.5%—PRO: 23.0%—LIP: 29.5% |
| WL010106 | 52 | CHO: 67.5%—PRO: 20.0%—LIP: 12.5% | Mediterranean | CHO: 44.0%—PRO: 21.7%—LIP: 34.3% |
| WL010105 | 50 | CHO: 62.2%—PRO: 22.1%—LIP: 15.7% | Mediterranean (with high fish intake) | CHO: 42.2%—PRO: 22.8%—LIP: 34.9% |
| WL010108 | 40 | CHO: 61.2%—PRO: 20.5%—LIP: 18.3% | Ketogenic/Low Carb | CHO: 36.9%—PRO: 33.3%—LIP: 29.8% |
| Food Item | TCTRL Mean ± SD | TDIET Mean ± SD | t-Statistics | Trend | p-Value |
|---|---|---|---|---|---|
| Cereal Bars (g) | 13.1 ± 13.0 | 0.0 ± 0.0 | 2.673 | Decrease | 0.05 (*) |
| Chocolate (g) | 18.7 ± 18.8 | 0.6 ± 1.7 | 2.466 | Decrease | 0.05 (*) |
| Ice Cream (g) | 16.8 ± 23.1 | 60.7 ± 34.4 | −2.796 | Increase | 0.05 (*) |
| Parmesan Cheese (g) | 17.7 ± 4.4 | 24.2 ± 6.1 | −3.113 | Increase | 0.05 (*) |
| Oily Fish (g) | 109.4 ± 93.5 | 196.5 ± 131.5 | −3.672 | Increase | 0.05 (*) |
| Variable | TCTRL Mean ± SD | TDIET Mean ± SD | t-Statistics | Trend | p-Value |
|---|---|---|---|---|---|
| Intake (Kcal) | 1457 ± 554 | 1488 ± 549 | −0.580 | Increase | 0.583 |
| Carbohydrates (%) | 57.1 ± 5.1 | 51.7 ± 11.1 | 1.445 | Decrease | 0.198 |
| Proteins (%) | 20.8 ± 3.3 | 25.5 ± 8.5 | −1.380 | Increase | 0.217 |
| Fibers (%) | 4.7 ± 1.5 | 4.9 ± 1.5 | −0.633 | Increase | 0.550 |
| Calcium (mg) | 413 ± 121 | 601 ± 251 | −2.732 | Increase | 0.05 (*) |
| Potassium (mg) | 1476 ± 712 | 1815 ± 615 | −2.693 | Increase | 0.05 (*) |
| Phosphorus (mg) | 572 ± 241 | 709 ± 292 | −2.556 | Increase | 0.05 (*) |
| Sodium (mg) | 1200 ± 717 | 1388 ± 760 | −3.027 | Increase | 0.05 (*) |
| Zinc (mg) | 4 ± 2 | 5 ± 2 | −2.565 | Increase | 0.05 (*) |
| Group | Variable | TCTRL Mean ± SD | TDIET Mean ± SD | t-Statistics | Trend | p-Value |
|---|---|---|---|---|---|---|
| Anthropometric | Weight (kg) | 69.1 ± 12.6 | 67.2 ± 11.5 | 2.116 | Decrease | 0.08 |
| BMI (kg/m2) | 23.1 ± 2.8 | 22.4 ± 2.2 | 2.343 | Decrease | 0.05 (*) | |
| Basal Metabolism (kcal) | 1327 ± 279 | 1317 ± 249 | 0.486 | Decrease | 0.64 | |
| Physical Activity (kcal) | 402 ± 254 | 441 ± 280 | −0.804 | Increase | 0.45 | |
| Body Fat (%) | 27.1 ± 7.2 | 26.2 ± 6.3 | 1.604 | Decrease | 0.16 | |
| Muscle (kg) | 47.1 ± 10.8 | 47.1 ± 10.0 | −0.001 | Increase | 0.10 | |
| Bone Mass (kg) | 2.7 ± 0.4 | 2.7 ± 0.4 | 0.427 | Decrease | 0.68 | |
| Water (%) | 50.0 ± 3.9 | 51.4 ± 3.5 | −2.300 | Increase | 0.06 | |
| Physiological | Resting Heart Rate (bpm) | 60.9 ± 7.1 | 57.9 ± 7.3 | 2.571 | Decrease | 0.05 (*) |
| Average Heart Rate (bpm) | 73.8 ± 4.5 | 73.1 ± 5.0 | 1.033 | Decrease | 0.34 | |
| Deep Sleep (min) | 85.3 ± 12.3 | 88.0 ± 8.2 | −0.685 | Increase | 0.52 | |
| Shallow Sleep (min) | 281.5 ± 20.8 | 261.2 ± 14.4 | 3.682 | Decrease | 0.05 (*) | |
| REM (min) | 59.5 ± 15.2 | 58.7 ± 17.1 | 0.252 | Increase | 0.81 |
| Group | Variable | TCTRL Mean ± SD | TDIET Mean ± SD | t-Statistics | Trend | p-Value |
|---|---|---|---|---|---|---|
| Diversity | Richness | 409.7 ± 44.8 | 528.1 ± 110.9 | −4.355 | Increase | 0.05 (*) |
| Pielou’s evenness | 0.627 ± 0.026 | 0.634 ± 0.029 | −0.626 | Increase | 0.55 | |
| Shannon diversity | 3.76 ± 0.12 | 3.96 ± 0.11 | −3.396 | Increase | 0.05 (*) | |
| Phyla | Firmicutes | 0.54 ± 0.14 | 0.60 ± 0.15 | −1.895 | Increase | 0.11 |
| Bacteroidetes | 0.27 ± 0.20 | 0.21 ± 0.19 | 0.837 | Decrease | 0.43 | |
| Proteobacteria | 0.08 ± 0.14 | 0.06 ± 0.05 | 0.682 | Decrease | 0.52 | |
| Actinobacteria | 0.07 ± 0.10 | 0.08 ± 0.07 | −0.283 | Increase | 0.79 | |
| Verrucomicrobia | 0.02 ± 0.04 | 0.04 ± 0.05 | −0.896 | Increase | 0.40 | |
| Species | Acinetobacter junii | (1.2 ± 1.6) × 10−5 | (2.8 ± 2.0) × 10−5 | −2.525 | Increase | 0.05 (*) |
| Alistipes finegoldii | (2.9 ± 3.1) × 10−4 | (1.3 ± 2.0) × 10−4 | 2.662 | Decrease | 0.05 (*) | |
| Alistipes finegoldii DSM 17242 | (2.4 ± 4.5) × 10−6 | (6.9 ± 6.8) × 10−6 | −2.602 | Increase | 0.05 (*) | |
| Bacteroides plebeius | (5.6 ± 5.5) × 10−5 | (6.9 ± 6.8) × 10−5 | 3.046 | Decrease | 0.05 (*) | |
| Klebsiella sp. | 0.0 ± 0.0 | (1.5 ± 1.7) × 10−5 | −2.367 | Increase | 0.05 (*) | |
| Klebsiella sp. 8.1T | 0.0 ± 0.0 | (1.2 ± 1.3) × 10−5 | −2.482 | Increase | 0.05 (*) | |
| Klebsiella sp. XW111 | (7.6 ± 2.0) × 10−7 | (7.8 ± 7.7) × 10−5 | −2.651 | Increase | 0.05 (*) | |
| Klebsiella sp. YSI6A | 0.0 ± 0.0 | (2.9 ± 3.2) × 10−5 | −2.445 | Increase | 0.05 (*) | |
| Klebsiella variicola | 0.0 ± 0.0 | (8.7 ± 9.9) × 10−5 | −2.318 | Increase | 0.06 (°) | |
| Lachnospiraceae bacterium DJF RP14 | (2.7 ± 5.7) × 10−4 | (3.9 ± 6.2) × 10−4 | −2.407 | Increase | 0.05 (*) | |
| Lachnospiraceae bacterium DJF VP18k1 | (1.5 ± 1.8) × 10−4 | (3.6 ± 3.2) × 10−4 | −2.768 | Increase | 0.05 (*) | |
| Lactobacillus crispatus | (4.2 ± 3.1) × 10−5 | (1.7 ± 1.6) × 10−4 | −2.467 | Increase | 0.05 (*) | |
| Roseburia faecis | (1.2 ± 1.2) × 10−4 | (2.0 ± 3.4) × 10−5 | 2.587 | Decrease | 0.05 (*) | |
| Roseburia sp. 11SE39 | (1.5 ± 1.4) × 10−4 | (2.1 ± 3.3) × 10−4 | 2.748 | Decrease | 0.05 (*) | |
| Bacterium NLAE–zl–P167 | (1.2 ± 1.2) × 10−5 | (1.6 ± 4.1) × 10−6 | 2.468 | Decrease | 0.05 (*) | |
| Butyrate–producing bacterium PH07BW10 | (2.5 ± 2.6) × 10−3 | (1.3 ± 2.0) × 10−3 | 2.385 | Decrease | 0.05 (*) | |
| Butyrate–producing bacterium SR1/5 | (4.0 ± 3.8) × 10−4 | (8.7 ± 5.9) × 10−4 | −2.597 | Increase | 0.05 (*) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchetti, G.; De Maio, F.; Abeltino, A.; Serantoni, C.; Riente, A.; Santarelli, G.; Sanguinetti, M.; Delogu, G.; Martinoli, R.; Barbaresi, S.; et al. Unraveling the Gut Microbiome–Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology. Nutrients 2023, 15, 3931. https://doi.org/10.3390/nu15183931
Bianchetti G, De Maio F, Abeltino A, Serantoni C, Riente A, Santarelli G, Sanguinetti M, Delogu G, Martinoli R, Barbaresi S, et al. Unraveling the Gut Microbiome–Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology. Nutrients. 2023; 15(18):3931. https://doi.org/10.3390/nu15183931
Chicago/Turabian StyleBianchetti, Giada, Flavio De Maio, Alessio Abeltino, Cassandra Serantoni, Alessia Riente, Giulia Santarelli, Maurizio Sanguinetti, Giovanni Delogu, Roberta Martinoli, Silvia Barbaresi, and et al. 2023. "Unraveling the Gut Microbiome–Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology" Nutrients 15, no. 18: 3931. https://doi.org/10.3390/nu15183931
APA StyleBianchetti, G., De Maio, F., Abeltino, A., Serantoni, C., Riente, A., Santarelli, G., Sanguinetti, M., Delogu, G., Martinoli, R., Barbaresi, S., Spirito, M. D., & Maulucci, G. (2023). Unraveling the Gut Microbiome–Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology. Nutrients, 15(18), 3931. https://doi.org/10.3390/nu15183931











