Abstract
BACKGROUND: Non-alcoholic fatty liver disease (NAFLD) is a prevalent metabolic disorder. Defects in function/expression of genes/proteins are critical in initiation/progression of NAFLD. Natural products may modulate these genes/proteins. Curcumin improves steatosis, inflammation, and fibrosis progression. Here, bioinformatic tools, gene–drug and gene-disease databases were utilized to explore targets, interactions, and pathways through which curcumin could impact NAFLD. METHODS: Significant curcumin–protein interaction was identified (high-confidence:0.7) in the STITCH database. Identified proteins were investigated to determine association with NAFLD. gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were analyzed for significantly involved targets (p < 0.01). Specificity of obtained targets with NAFLD was estimated and investigated in Tissue/Cells–gene associations (PanglaoDB Augmented 2021, Mouse Gene Atlas) and Disease–gene association-based EnrichR algorithms (Jensen DISEASES, DisGeNET). RESULTS: Two collections were constructed: 227 protein–curcumin interactions and 95 NAFLD-associated genes. By Venn diagram, 14 significant targets were identified, and their biological pathways evaluated. Based on gene ontology, most targets involved stress and lipid metabolism. KEGG revealed chemical carcinogenesis, the AGE-RAGE signaling pathway in diabetic complications and NAFLD as the most common significant pathways. Specificity to diseases database (EnrichR algorithm) revealed specificity for steatosis/steatohepatitis. CONCLUSION: Curcumin may improve, or inhibit, progression of NAFLD through activation/inhibition of NAFLD-related genes.
1. Introduction
In recent decades, non-alcoholic fatty liver disease (NAFLD) has attracted interest from researchers as a highly prevalent metabolic disorder. Metabolic dysfunction can be initiated when there is only 5% fat accumulation in the liver. Furthermore, while an increasing proportion of the population has been diagnosed with NAFLD, scientific studies suggest that many individuals are unaware of the presence of the disease, which complicates the therapy [1]. NAFLD comprises a spectrum of liver disorders, from hepatic steatosis to non-alcoholic steatohepatitis (NASH) and, if unchecked, may progress to fibrosis and cirrhosis [2]. NAFLD is recognized to be closely associated with other metabolic disorders such as hyperlipidemia, obesity, hypertension, cardiovascular disease, and insulin resistance [3,4]. The global prevalence of NAFLD is approximately ~25%, with the highest prevalence in the Middle East. The prevalence of NAFLD in Asia has been determined to be 52.3 per 1000 people years [5]. Despite the health burden it imposes, no specific drugs have been approved for the treatment of NAFLD, though various therapeutic approaches have been proposed. Lifestyle intervention and pharmacological interventions are the mainstays of treatment for patients with NAFLD [6]. A number of studies have identified defects in the function or expression of genes and proteins as critical factors in the initiation and progression of NAFLD [7,8]. Various natural and chemical drugs are being studied to determine whether they modulate these genes and proteins [9,10]. One of the natural products that has drawn much attention for the treatment of metabolic diseases is curcumin.
Curcumin is a bioactive polyphenolic compound, isolated from Curcuma longa Linn, which is endowed with diverse pharmacological activities [11,12,13,14,15,16,17,18,19,20,21,22]. Numerous in-vitro and in-vivo investigations have indicated that curcumin exerts a positive effect at each stage of NAFLD, improving both inflammation and the extent of fat deposition. Curcumin was also shown to inhibit the progression from NAFLD to fibrosis and decrease the risk of liver cancer. More recently, several clinical trials have reported beneficial changes, including a decrease in inflammation, increased antioxidant factors, and improvement in liver histopathology [23,24,25,26,27,28,29,30,31].
Emerging from these studies, therefore, are data supporting the significant clinical effect of curcumin on NAFLD. Although extensive data exist on the benefits of curcumin in NAFLD, studies on the mechanism of action of curcumin and its interaction with various genes have not been evaluated in detail.
Systems biology and bioinformatics, a branch of biology that combines molecular biology and computer science, have been an integral part of research investigations in recent decades. This science focuses on the entire biological process and system as compared to past years where there was a tendency to investigate only a single protein or gene [32].
In this bioinformatics study, we aimed to identify important gene targets for NAFLD and high probability protein targets of interaction for curcumin. Moreover, we have tried to correlate these targets with NAFLD using published literature on curcumin. Figure 1 provides an overview of the investigative strategy undertaken to perform this study.
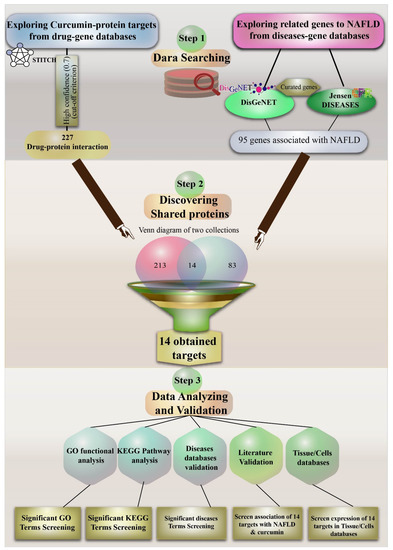
Figure 1.
A comprehensive diagram illustrating the investigative strategy undertaken in the present study. Non-alcoholic fatty liver disease (NAFLD); gene ontology (GO); Kyoto Encyclopedia of Genes and Genomes (KEGG).
2. Methods
Using the bioinformatics approach explores critical genes associated with curcumin and NAFLD in the online databases, then investigates their relations as targets of curcumin for NAFLD.
2.1. Curcumin and Target Search
We first searched interactions of curcumin in the STITCH database (http://stitch.embl.de/ (accessed on 3 October 2021)) to explore essential protein targets. STITCH is a platform for diagnosing interactions between chemicals and proteins. This database currently contains more than 9,640,000 proteins from 2031 organisms [33]. Here, we considered the high confidence cut-off (0.700) and limited the species only to Homo sapiens. We also illustrated the presence of proteins obtained in liver tissue based on the String plugin algorithm (1–5 score) in Cytoscape software (version 3.8.2).
2.2. Exploring Important NAFLD Genes in DISEASES and DisGeNET Databases
Next, we investigated the proteins on curated targets in two databases, the DisGeNET database (https://www.disgenet.org/ (accessed on 3 October 2021)) and the DISEASES database (diseases.jensenlab.org (accessed on 3 October 2021)), to find their association with NAFLD. DisGeNET is a database that contains a collection of genes associated with specific diseases. These data are integrated from a variety of sources, such as expert-curated repositories, the scientific literature and GWAS catalogs. DisGeNET currently covers more than 1700 genes and 24,000 diseases and traits [34]. For association genes with NAFLD, 1058 genes were registered in this database. Curated data contained seven primary resources: UNIPROT, ORPHANET, CTD, GENOMICS ENGLAND, CLINGEN, PSYGENET, and CGI. To achieve a curated dataset from DisGeNET, we used a plugin in the cystoscope to construct curated sources targets for NAFLD. This plugin contains different categories of existing sources in DisGeNET. The most important collection is curated data that are manually achieved. Therefore, we only used curated data by selecting CURATED from Select Source. DISEASES database is a weekly updated database that comprises diseases and gene relations from different resources, including manually curated literature, text mining, cancer mutation data, and genome-wide association research [35]. We extracted the targets from the available resources, including experiments and manually curated literature associated with NAFLD.
2.3. Venn Diagram to Obtain Important Curcumin Interaction Protein Targets in NAFLD
Before identification of common genes between two collections (curated genes-NAFLD and curcumin-STITCH), to eliminate the bias of different forms of gene names, all genes were first converted to UniProt by https://www.uniprot.org/uploadlists/ (accessed on 3 October 2021). For this purpose, we collected all genes and protein targets in different formats (gene symbols) and in the section “Retrieve/ID mapping” and converted to UniProtKB. We finally created a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 3 October 2021)) for these two sets of protein collections to find important targets of curcumin beyond the key recognized targets. A Venn diagram is a suitable method for identifying interesting overlaps between two or more sets.
2.4. Gene Ontology Pathway Enrichment Analyses for Target Proteins of Curcumin
Gene ontology (GO) enrichment is a popular procedure used to interpret genes and stratify them into three major categories: those that contribute to molecular function (MF), biological process (BP) or cellular component (CC) [36]. GO was analyzed for important targets obtained from the Venn diagram using the gene ontology resource with the web address: http://geneontology.org (accessed on 3 October 2021). Additionally, KEGG was analyzed using the Enrichr database with the web address: https://maayanlab.cloud/Enrichr/ (accessed on 3 October 2021). The KEGG pathway is a comprehensive database that maps pathways according to their metabolic inter-relationships [37]. In GO and KEGG analysis, the p-value < 0.01 was considered as the cut-off criterion. We also analyzed the enrichment pathways using the Wikipathway plugin in Cytoscape version 3.8.2. Cytoscape is a powerful software for integrating biomolecular interaction networks. It also has the practical capability to install multiple plugins to analyze high throughput data and different biological processes [38]. A p-value < 0.01 was considered as the cut-off criterion. In this plugin, we selected the Nonalcoholic fatty liver disease–Homo sapiens pathway and highlighted the curcumin protein interactions in this pathway based on their STITCH score. In addition, we estimated the specificity of obtained targets with NAFLD and investigated them in Tissue/Cells–gene associations (PanglaoDB Augmented 2021 and Mouse Gene Atlas) and the Disease–gene associations based EnrichR algorithm (Jensen DISEASES and DisGeNET). EnrichR is a free enrichment analysis website that possesses 180 184 annotated gene sets from 102 gene set libraries. It is a comprehensive online analyzer and engine searching machine to assemble biological knowledge [39]. PanglaoDB is a web server for probing single-cell RNA sequencing data, covering more than 1054 single-cell investigations from more than 4 million cells that derive from a broad range of tissues [40]. Mouse Gene Atlas is an online database resource for mouse research, providing integrated genomic, genetic, and biological information for promoting clinical studies in the field of human diseases [41].
3. Results
3.1. Protein Target Interaction with Curcumin in the STITCH Database
Screening curcumin in the STITCH database with high confidence (0.7) identified 227 protein targets. The Drug–Protein interaction was visualized on Cytoscape (Figure 2A). STITCH scores are indicated by color intensity.
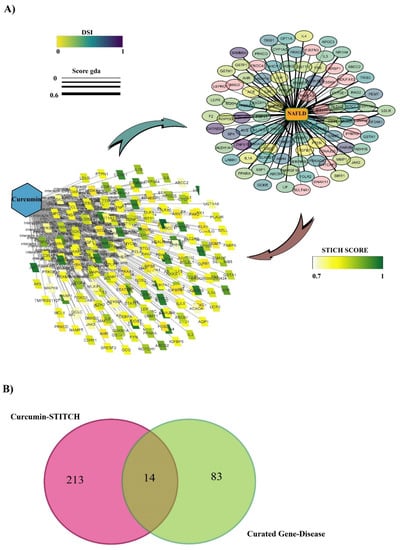
Figure 2.
(A) Curated disease–gene database and curcumin–protein interaction visualized with Cytoscape software. For the curated disease-gene: Color intensity indicates the disease specificity index and the thickness of the edges indicates the genes–disease association score. For the curcumin–protein interaction, color intensity indicates STITCH score. (B) Venn diagram of the two datasets comprising the curated disease–gene database and curcumin-protein interaction.
3.2. Discovering Curated NAFLD Genes
The curated data DisGeNet plugin on Cytoscape and DISEASES database identified 95 genes associated with NAFLD. All the data were visualized with Cytoscape software (Figure 2A). In this figure, the disease specificity index with genes (DSI g) is indicated by color intensity, and the genes–disease association score (gda score) is indicated with the thickness of the edges.
3.3. The Overlap of Curcumin Targets on the STITCH and Curated NAFLD Genes Were Visualized Using a Venn Diagram
A Venn diagram of the two created datasets revealed 14 candidates, CYP1A2, NFE2L2, PPARA, GSTA1, IL1A, LEP, LDLR, CSF2, GSTP1, PRKCE, TGFB1, ABCC2, AHR, and PRKCD (Figure 2B), that may be directly or indirectly affected by curcumin. Direct interaction refers to physical interaction, and indirect interaction refers to functional association. The scoring based on DisGeNET is shown in Table 1. It was noted that the highest relation with NAFLD belongs to TM6SF2 and PNPLA3, with gda scores of 6 and 5, respectively. However, due to the high confidence cut-off, seven candidates showed no relationship, and therefore we excluded them from our study.

Table 1.
The relationship of genes associated with NAFLD that are targets of curcumin (www.disgenet.org (accessed on 3 October 2021)).
3.4. GO and KEGG Enrichment Analyses of Protein Targets of Curcumin
GO analysis of the 14 identified protein targets demonstrated major involvement in response to oxygen-containing compounds, cellular response to lipid and stress, and regulation of lipid metabolic process under biological process (Table 2). This analysis additionally showed that these protein targets were chiefly involved in calcium-independent protein kinase C activity, signaling receptor activator and regulator activity, and transcription coregulator binding under the molecular function category. Furthermore, cellular components included endolysosome, extracellular space, and secretory granule lumen (Table 2).

Table 2.
Gene ontology enrichment analysis via Enrichr for the 14 identified genes with the best score interaction with curcumin.
In KEGG enrichment, we observed several biological pathways. The highest p-value pathways included chemical carcinogenesis, the AGE-RAGE signaling pathway in diabetic complications, fluid shear stress and atherosclerosis, non-alcoholic fatty liver disease and cytokine–cytokine receptor interaction (Table 3).

Table 3.
KEGG pathways for 14 critical genes interact with curcumin.
3.5. Specificity of 14 Obtained Protein Targets with Cells/Liver Tissue and Fatty Liver Disease in Databases
In Cytoscape software, the intensity of the color indicates the number of expressed genes that interacted with curcumin in liver tissue (1–5) (Figure 3). The score of expression of 14 obtained genes is reported in Table 1. Eight genes, including GSTA1, LDLR, PPARA, ABCC2, CYP1A2, PRKCD, GSTP1, and AHR, had high confidence scores (score 4). Additionally, the specificity of all 14 obtained targets in enrichment analysis for liver cells and tissues in two databases, PanglaoDB Augmented 2021 and Mouse Gene Atlas based on the EnrichR algorithm, was confirmed (Figure 3).
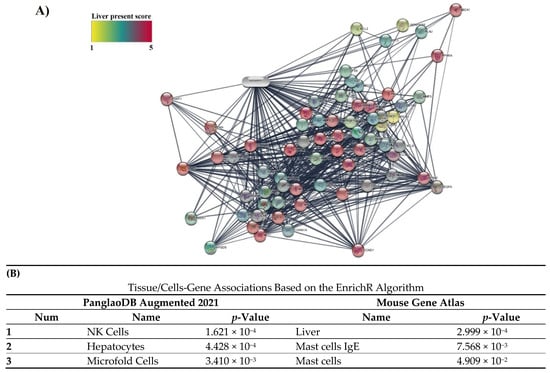
Figure 3.
(A) Amount of expression of protein targets in interaction with curcumin in liver tissue. The intensity of color shows the amount of expression in the liver. (B) Association of 14 obtained protein targets in the interaction of curcumin with Tissue/Cells databases.
Moreover, the specificity of 14 obtained targets with NAFLD was confirmed based on the EnrichR algorithm in two databases, including Jensen DISEASES and DisGeNET. The result showed that NAFLD, and its advanced form NASH, were ranked in the top three in these databases (Table 4).

Table 4.
Association of protein targets obtained in the interaction of curcumin with the indicated disease in two databases.
Furthermore, we visualized all curcumin–protein targets with high confidence (0.7) in the NAFLD pathway in Wikipathway. The more intense color indicates greater interaction (based on the STITCH score) (Figure 4). The pathways are constructed using the Cytoscape with Wikipathway plugin (version 3.3.7).
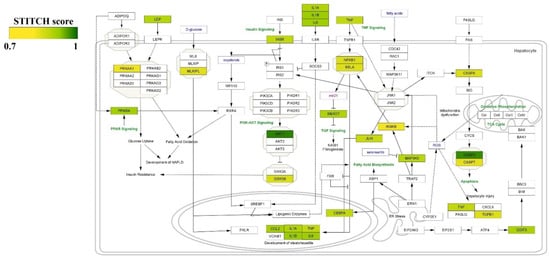
Figure 4.
Visualizing protein interactions with curcumin in the NAFLD pathway with high confidence (0.7). The intensity of color illustrates the strength of interaction of curcumin with the targets.
4. Discussion
As a metabolic disorder, the importance of NAFLD has gained prominence in parallel with the accelerating prevalence of other metabolic syndromes [42]. In addition to causing metabolic dysfunction, NAFLD can progress and increase the risk of transforming to hepatocellular carcinoma if left untreated [43]. Identifying several important susceptibility genes that play a central role in the development and phenotype of NAFLD is a critical and initial step in order to determine suitable drugs for managing NAFLD. Curcumin has been shown to alleviate the pathological dysfunction of NAFLD and could exert these mitigating influences through effects on several important NAFLD-related genes [9,44,45,46]. Here, we investigated and analyzed possible protein targets of curcumin in databases and biological pathways, intending to understand the effects of curcumin in NAFLD in greater depth.
In the present study, we first searched significant predictions of protein interaction with high confidence for curcumin. Then, we probed protein interaction associated with fatty liver disease, determined its impact as an inhibitor, activator, binding, or transcription factor on these genes, and selected the most relevant targets. In doing so, we identified 14 significant targets: CYP1A2, NFE2L2, PPARA, GSTA1, IL1A, LEP, LDLR, CSF2, GSTP1, PRKCE, TGFB1, ABCC2, AHR, and PRKCD, and then evaluated their biological pathways. Most of the targets were involved in response to lipid and stress and lipid metabolism based on gene ontology. KEGG enrichment pathways showed chemical carcinogenesis, the AGE-RAGE signaling pathway in diabetic complications and NAFLD as the most common significant pathways. The targets we found among the authentic disease databases were more specific to fatty liver disease (steatosis and steatohepatitis) than other diseases in these databases. We also investigated the possible presence of these important protein targets in the liver. Those 14 protein targets that we identified were responsible for different stages of NAFLD, including steatosis, steatohepatitis, and fibrosis. We determined that curcumin could impact each of those stages through modulating essential protein targets.
PPARA (peroxisome proliferator-activator receptors-alpha) is a ligand-activated transcription factor, richly expressed in the liver and modulates various genes implicated in the catabolism of fatty acids [47]. Activation of PPARA promotes genes involved in oxidative phosphorylation and fatty acid beta-oxidation [48]. This activation has a protective effect in NAFLD with antioxidant, anti-inflammation, and lipid accumulation inhibition in the liver [49,50,51,52,53,54]. Numerous studies have been designed to activate PPARA for slowing the progression or curing NAFLD, and they demonstrate enhancement in metabolic activity and decrease of steatosis and fibrosis in NAFLD patients and models [48,55,56,57]. One study showed that curcumin activated PPARA and modulated several upstream signaling pathways (AMPK, PI3K/AKT/mTOR), which inhibit oxidative stress and increase autophagic flow in liver cells [58]. Another study reported that, in high-fat diet (HFD) mice, the level of expression of PPARA was reduced, whereas, after treatment with curcumin, the PPARA expression level was restored [59]. Of note, in our study (Table 1), PPARA had the third-highest score for interaction with curcumin and showed a strong relationship to NAFLD (gda score 0.4). This gene is extensively expressed in the liver (4.76), and curcumin was shown to activate PPARA (Table 1).
NFE2L2 (nuclear factor erythroid-derived 2-like 2) is a potent transcription factor in modulating xenobiotics and antioxidants in stress responses. However, oxidative stress causes the progression of NAFLD, and NFE2L2 could promote the reduction of reactive oxygen species (ROS) and amelioration of NAFLD [60]. In NFE2L2-knockout-mice, the ratio of reduced glutathione/oxidized glutathione (GSH/GSSG) decreased and steatohepatitis was severe [61]. Another study showed that NFE2L2 could be involved in cholesterologenic and lipogenic pathways, repressing fat accumulation and oxidative stress in HFD mice. In a study on hepatocyte-specific NFE2L2-upregulation mice with diet-inducing hepatic steatosis, thioredoxin-1, glutathione peroxidase-2, and Nqo1 were increased, and oxidative stress markers decreased. Further experiments also indicated that lipogenesis was inhibited and that the catabolism of lipid was increased in hepatocytes [62]. The protective role of overexpression of NFE2L2 against oxidative stress and reduced hepatic steatosis was demonstrated in mouse hepatic cells from a methionine–choline deficient (MCD) diet-induced NAFLD model [63]. In mice models with lipotoxicity, NFE2L2 was shown to mediate induction of SQTM1, enabling hepatoprotection under lipotoxic conditions [64]. NFE2L2 also was shown to regulate inflammation by repressing JNK (c-Jun N-terminal kinase) and the nuclear factor-kappa B (NF-κB) pathway in HFD diet mice.
Curcumin exerts its anti-diabetic, cardioprotective, hepatoprotective, neuroprotective, and antitumor effects via NFE2L2 signaling pathways [65]. Curcumin activates NFE2L2 signaling pathways in four ways: impacting mediators of NFE2L2, inhibition of KEAP1, impacting NFE2L2 expression, and reducing nuclear translocation of NFE2L2 [65]. In a high-fat and high-fructose diet (HFHFr) mouse model, NFE2L2 was downregulated, while curcumin administration could reverse the abnormal serum biochemical parameters of hepatic steatosis [66]. Another animal study using carbon tetrachloride (CCL4) induced liver damage showed that curcumin’s protective role in reducing inflammation and oxidative stress was mediated through NFE2L2/HO-1 pathways [67]. An in-vitro study showed that curcumin, through activation of NFE2L2, can promote lipocyte activation in stellate cells (HSCs) and repress hepatic fibrosis [68].
In our current study, curcumin demonstrated a robust interaction with NFE2L2 based upon the STITCH score, having the second-highest score after CYP1A2 (high confidence: 0.877), and its function was determined to be through activation of NFE2L2. Similarly, NFE2L2 had the highest score (0.4), together with leptin (LEP) and PPARA, in the gene-diseases association database (Table 1), indicating a significant role in the pathogenesis of NAFLD. For expression in liver tissue, however, it ranked ninth when compared with the other genes.
LEP was shown to be important with regards to NAFLD in our study (gda score: 0.4). LEP is a polypeptide hormone that interacts with its receptor, lepRb [69]. Several studies have investigated the role of leptin in NAFLD. Serum leptin levels are increased in both patients with NAFLD and in animal models of the disease and may be associated with the development of hepatocyte steatosis and its progression through OB-R (leptin receptor) activation of the PI3-K/Akt kinase pathway [70,71]. Moreover, leptin could be considered a surrogate biomarker for diagnosing patients with NASH/NAFLD.
Curcumin reduces the expression and secretion of leptin [72,73,74]. Askari et al. reported that curcumin delivered in a nanoparticle carrier caused a time and dose-dependent decrease in leptin levels [75]. Curcumin inhibits hepatic stellate cell (HSC) activation, while leptin induces HSCs which, in turn, cause progression of NASH to fibrosis. By decreasing phosphorylation of Ob-R, inducing PPAR-gamma, and reducing oxidative stress, curcumin inhibits Ob-R expression and blocks leptin signaling [72]. A recent study indicated that curcumin represses the methionine adenosyltransferase 2A (MAT2A) promoter that promotes leptin expression. MAT2A activity is linked with HSC activation and DNA methylation [76]. According to our STITCH results, curcumin had a significant interaction with LEP (0.82). Additionally, LEP had a robust relationship with NAFLD (gda: 0.4), but its expression was not specific to the liver (3.17).
Low-density apolipoprotein receptor (LDLR) is a mediator for cholesterol uptake in cells and is crucial for the clearance of cholesterol by the liver [77]. LDLR deficient rodents have been used to establish models of NAFLD [78,79]. In those models, elevations in hepatic neutral and hepatic pro-inflammatory oxylipins were observed [80]. An in-vitro study demonstrated that, by inducing fatty acid, the expression of LDLR was upregulated [81]. Curcumin was reported to induce LDLR expression on the cell surface and upregulate its activity in hepatic cells. Moreover, the uptake of LDL is significantly elevated in HepG2 cells following curcumin treatment [82,83], which might contribute to the clearance of cholesterol by the liver. Research on the CCl4 induced cirrhosis rat model also showed upregulation of LDLR expression with curcumin treatment [84]. With a STITCH score of 0.824 and a gda score of 0.37, our results show a strong relation of curcumin with LDLR and of LDLR with NAFLD diseases, respectively. As we show in Table 1, based upon its high score (4.41), the expression of LDLR is specific to the liver.
Cytochrome P450 isozyme 1A2 (CYP1A2) plays an important role in metabolizing drugs and carcinogens [85]. Research on high-fat and high-sucrose (HFHS) diet mice revealed an increase in the expression of CYP1A2 [86]. Additionally, the increased CYP profile, evidenced by increased CYP1A2, normalized after tetrahydro-curcumin administration in a study using the HFHS mouse model [87]. However, a study in humans reported that microsomal CYP1A2 decreased with NAFLD progression [88]. An in-vitro study using the palmitic acid (PA)-induced NAFLD model also indicated that expression of CYP1A2 was inhibited. Several studies have reported that curcumin downregulates the expression of CYP1A2 and, in this way, ameliorates the dysregulation of CYPs in various diseases [89,90,91,92].
In our study, CYP1A2 was identified as one of the most important NAFLD genes inhibited by curcumin (STITCH score: 0.948). According to our dataset, this gene is specifically present in the liver (high confidence: 4.78). However, due to conflicting results in studies about the increase and decrease of CYP1A2 in NAFLD, curcumin administration may benefit or exacerbate the disease, an issue that requires further investigation.
Several studies have shown that IL1α is increased in NASH/NAFLD patients [93,94,95] and is positively correlated with advanced stages of steatosis and steatohepatitis. IL1α was reported to be ~122% and ~300% higher in patients with steatosis and steatohepatitis than in controls [96]. Saberi-Karimian in 2020 reported that curcumin ameliorates inflammatory cytokines like IL1A in serum of patients with NAFLD, and curcumin exhibited anti-steatotic properties [29]. Curcumin was shown to modulate IL1A in a dose and time-dependent manner [97]. An investigation of the effect of curcumin on bone-marrow stromal cells showed that IL1-A was repressed [98]. Our study also reported that IL1A, with a gda score of 0.33, is associated with NAFLD, but its expression in the liver has a low score compared to the other protein target genes (2.78) in Table 1. However, the STITCH score of 0.829 indicates the possibility for curcumin to exhibit potent inhibition of IL1A.
Glutathione s-transferases (GSTs) are catalyzing enzymes that catalase hydrophobic and electrophilic compounds through glutathione reduction. GSTA1 is a member of the alpha class of GSTs and is principally expressed in the liver [99]. GSTA1 polymorphisms are associated with an increased risk of NAFLD [100]. In HepG2 induced lipotoxicity, the expression of GSTA1 was down-regulated [101]. In in-vivo studies, the expression of GSTA1 was decreased in fatty liver mouse models [102,103]. An in-silico docking study revealed that curcumin exhibited a potent affinity to GST isoforms such as GSTA1 [104], and that curcumin could modulate the expression of GSTA1 [105,106]. Here, we showed that curcumin, with a high confidence score of 0.846, influences GSTA1 in both activation and inhibition. Furthermore, with a gda score of 0.31, GSTA1 is a critical protein target in NAFLD and was also shown here to be the most specific protein target in the liver, based on a score of 4.90, in comparison with other identified protein targets.
Glutathione S transferase Pi 1 (GSTP1) is another GST enzyme that plays a vital role in antioxidant defense through detoxifying foreign substances and inactivating byproducts of oxidative stress [107,108]. Research in 2009 by Hori et al. demonstrated for the first time that GSTs could be involved in the progression of NAFLD [108]. Proteomic analysis of GST proteins showed that pi1, mu1, and selenium binding protein-2 were reduced in diet-induced hepatic steatosis [109]. Moreover, several studies have reported that polymorphisms of GSTP1 are common in patients with NAFLD [110,111]. Various studies have investigated the effects of curcumin on GSTP1. An in-vitro investigation by Nishinaka et al. showed that antioxidant response element (ARE) is the primary region where curcumin induces transactivation of the GSTP1 gene [112]. Other research indicates that curcumin might induce apoptosis due to its inhibition of GSTP1 expression at the transcription level [113]. A 2015 study reported that mRNA and protein levels of GSTP1 in curcumin-treated groups were significantly lower than in control groups in human colon carcinoma cells [114]. In our data, curcumin was shown to be a transcriptional regulator of GSTP1 with high confidence (0.794). GSTP1 was present in the liver with a high confidence STITCH-based tissue score (4.64) and a close relationship with NAFLD (gda score: 0.33).
TGF-β is a growth factor and cytokine with profibrotic properties and is the most abundant isoform in the liver. TGF-βis also a key inducer of reactive oxygen species (ROS) [115]. A study in humans reported that profibrotic/scar deposition genes such as TGFB1 are elevated in steatohepatitis, which is purported to be due to the dynamic state of tissue remodeling [116]. In both mouse and human liver, TGFB1 as a pro-fibrogenic marker associated with NAFLD was upregulated [117]. A recent study reported that curcumin, an inhibitor of TGF-beta, also inhibits the transition of hepatocytes to myofibroblasts, a critical step in the progression of liver fibrosis [58]. A 2017 study reported that curcumin inhibited TGFβ1-induced Smad3 phosphorylation by conjunctival fibroblasts [118]. Further research demonstrated that curcumin decreases TGFB1, thereby delivering beneficial effects in resolving fibrosis [119]. High-dose curcumin was reported to significantly decrease the serum level of TGFB1 [120]. As shown here in Table 1, TGFB1 was 1 of the 14 important genes that interacted with curcumin and has a strong association with NAFLD. However, its distribution is not specific to the liver.
Colony-stimulating factor 2 (CSF2) has a role in binding to extracellular proteoglycans and modulating biological function [121]. CSF2 is produced by various cells, including fibroblasts, endothelial cells, macrophages, and most inflammatory cells, and, accordingly, as shown here in Table 1, CSF2 is not specific for hepatocytes. CSF2 is an important factor in the progression of liver fibrosis, and anti-CSF2 has been shown to ameliorate liver fibrosis [122]. CSF2 is increased in NASH patients and animal models of NASH [93,123,124,125]. Curcumin was reported to repress CSF2 in a concentration-dependent manner in allergic diseases [126]. Administration of curcumin to cancer patients resulted in a decrease in CSF2 [127]. In line with previous studies, our bioinformatics analysis shows that CSF2 has a strong relationship with NAFLD (gda score 0.3) and is robustly inhibited by curcumin (high confidence-based STITCH score: 0.816).
PRKCE is a member of the protein kinase C family, and has a C2 conserved domain (non-Ca2+ binding) and C1A and C1B domains for binding diacylglycerol (DAG) [128]. Several studies have reported that the accumulation of DAG causes activation of PRKCE, which leads to repression of insulin receptor kinase. This process induces hepatic insulin resistance, the most prevalent complication in NAFLD patients [129,130,131]. Curcumin binds to a specific domain attached to PRKCE and inhibits its activity [132]. Wang et al. in 2016 demonstrated that curcumin represses the accumulation of DAG and PRKCE translocation in the liver, which improves insulin function and inhibition of hepatic gluconeogenesis [133]. In our analysis, PRKCE was one of the protein targets discovered from 95 curated proteins in databases associated with NAFLD. Moreover, its binding activity with curcumin showed high confidence: 0.763. However, its distribution was not specific to the liver, showing less specificity than the other 14 protein targets.
ABCC2 is mainly expressed in significant physiological barriers such as the canalicular membrane in liver cells. It contains multiple binding sites and shows complex transport kinetics [134]. ABCC2 variants have been found to have a role in the susceptibility to and progression of NAFLD [135]. In patients with NASH, the expression of multiple efflux transporters was elevated, and cellular localization of ABCC2 is altered, which affects the elimination of drugs. According to our data, ABCC2, with a gda score of 0.32, has a relationship with NAFLD, and its presence in the liver was second only to GSTA1. An in-vitro study on hepatocellular carcinoma cell lines showed that ABCC2 has a critical role in hypoxia-induced resistance by curcumin [136]. More recent research revealed that curcumin could improve drug resistance, reversing the increased drug resistance of T24 cells treated with gemcitabine [137]. Curcumin may exert its effect by modulating the expression of ABCC2 [138]. However, as we show in Table 1, its STITCH score showed a low connection compared with most of the protein obtained (high confidence: 0.727).
Aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor with multifunctional activity and may benefit or adversely affect NAFLD [139]. Activation of AHR might positively improve NAFLD and may also have an anti-inflammatory effect in the disease [140]. AHR is highly expressed in the liver (4.40) and, with a gda score of 0.33 is highly associated with NAFLD. As a chemopreventative agent in cancer, curcumin has been shown to decrease AHR levels [141]. Curcumin has been shown to repress 3-methylcholanthrene binding (an AHR agonist) to the receptor; curcumin can bind to the AHR as a ligand and inhibit its transformation via phosphorylation, possibly acting through PKC [142]. However, several contradictory reports indicate that curcumin might decrease or increase AHR. Our data showed that curcumin was less associated with AHR than other targets with a STITCH score of 0.700 (Table 1).
Protein kinase c delta (PRKCD) is a member of the lipid-activated PKC family, which can be activated with DAG but not Ca2+ [143]. PRKCD has been reported to have a role in the progression of NAFLD [144]. PRKCD modulates SERCA (Sarco/endoplasmic reticulum calcium ATPase) activity in the human liver cell line (LO-2) induced with palmitic acid, an in-vitro model for steatosis [143]. SERCA is an important component in Ca2+ homeostasis [145]. Triglyceride (TG) accumulation was associated with dysregulation of PKCD and ER stress [143]. Deletion of PLDD has been shown to cause a reduction in lipogenesis by inducing important transcription factors in the liver in a HFD mouse model [146,147]. Deletion of PRKCD caused a decrease in TG accumulation and lipid droplet accumulation [143]. Various studies indicate that curcumin can stimulate PKCD phosphorylation, subsequently inducing downstream signaling pathways [148,149,150]. In our analysis, PRKCD was 1 of the 14 obtained protein targets (gda score: 0.31) with low association (STITCH score: 0.7) with curcumin in comparison with the other targets.
5. Conclusions
In summary, we surveyed the important NAFLD genes and their potential in targeting curcumin, together with their biological pathways. In this study, we identified 14 genes in NAFLD that are likely to be the target of curcumin and observed that curcumin induces or inhibits them. According to our results, this activity of curcumin was in line with improving NAFLD based on literature. We believe that the ongoing clinical trials investigating the effect of curcumin on fatty liver could yield positive results in the future, enhancing the therapeutic status of curcumin in this metabolic disorder.
Author Contributions
Conceptualization, A.M. and A.S.; methodology, A.M.; investigation, A.M.; writing—original draft preparation, A.M.; writing—review and editing, A.E.B., M.B., M.M. and A.S. All authors have read and agreed to the published version of the manuscript.
Funding
No funding has been received for this research.
Data Availability Statement
All data pertaining to this study will be made available upon reasonable request to the corresponding author.
Conflicts of Interest
Muhammed Majeed is the founder of Sami-Sabinsa group of companies.
References
- Murag, S.; Ahmed, A.; Kim, D. Recent Epidemiology of Nonalcoholic Fatty Liver Disease. Gut Liver 2021, 15, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Manne, V.; Handa, P.; Kowdley, K.V. Pathophysiology of Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis. Clin. Liver Dis. 2018, 22, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Mansour-Ghanaei, F.; Joukar, F.; Mobaraki, S.N.; Mavaddati, S.; Hassanipour, S.; Sepehrimanesh, M. Prevalence of non-alcoholic fatty liver disease in patients with diabetes mellitus, hyperlipidemia, obesity and polycystic ovary syndrome: A cross-sectional study in north of Iran. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1591–1596. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Byrne, C.D.; Tilg, H. NAFLD and increased risk of cardiovascular disease: Clinical associations, pathophysiological mechanisms and pharmacological implications. Gut 2020, 69, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Mundi, M.S.; Velapati, S.; Patel, J.; Kellogg, T.A.; Abu Dayyeh, B.K.; Hurt, R.T. Evolution of NAFLD and Its Management. Nutr. Clin. Pract. 2020, 35, 72–84. [Google Scholar] [CrossRef]
- Sookoian, S.; Pirola, C.J. Precision medicine in nonalcoholic fatty liver disease: New therapeutic insights from genetics and systems biology. Clin. Mol. Hepatol. 2020, 26, 461–475. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Valenti, L.; Miele, L.; Feldstein, A.E.; Alkhouri, N. NAFLD in children: New genes, new diagnostic modalities and new drugs. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 517–530. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, T.; Li, J.; Wang, S.; Qiu, F.; Yu, H.; Zhang, Y.; Wang, T. Effects of Natural Products on Fructose-Induced Nonalcoholic Fatty Liver Disease (NAFLD). Nutrients 2017, 9, 96. [Google Scholar] [CrossRef]
- Reimer, K.C.; Wree, A.; Roderburg, C.; Tacke, F. New drugs for NAFLD: Lessons from basic models to the clinic. Hepatol. Int. 2020, 14, 8–23. [Google Scholar] [CrossRef]
- Afshari, A.R.; Jalili-Nik, M.; Abbasinezhad-Moud, F.; Javid, H.; Karimi, M.; Mollazadeh, H.; Jamialahmadi, T.; Sathyapalan, T.; Sahebkar, A. Anti-tumor Effects of Curcuminoids in Glioblastoma Multiforme: An Updated Literature Review. Curr. Med. Chem. 2021, 28, 8116–8138. [Google Scholar] [CrossRef] [PubMed]
- Gorabi, A.M.; Kiaie, N.; Hajighasemi, S.; Jamialahmadi, T.; Majeed, M.; Sahebkar, A. The Effect of Curcumin on the Differentiation of Mesenchymal Stem Cells into Mesodermal Lineage. Molecules 2019, 24, 4029. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, M.; Bianconi, V.; Ávila-Rodriguez, M.F.; Barreto, G.E.; Jamialahmadi, T.; Pirro, M.; Sahebkar, A. Curcumin: A phytochemical modulator of estrogens and androgens in tumors of the reproductive system. Pharmacol. Res. 2020, 156, 104765. [Google Scholar] [CrossRef] [PubMed]
- Alidadi, M.; Jamialahmadi, T.; Cicero, A.F.; Bianconi, V.; Pirro, M.; Banach, M.; Sahebkar, A. The potential role of plant-derived natural products in improving arterial stiffness: A review of dietary intervention studies. Trends Food Sci. Technol. 2020, 99, 426–440. [Google Scholar] [CrossRef]
- Farhood, B.; Mortezaee, K.; Goradel, N.H.; Khanlarkhani, N.; Salehi, E.; Nashtaei, M.S.; Najafi, M.; Sahebkar, A. Curcumin as an anti-inflammatory agent: Implications to radiotherapy and chemotherapy. J. Cell. Physiol. 2019, 234, 5728–5740. [Google Scholar] [CrossRef] [PubMed]
- Ganjali, S.; Blesso, C.N.; Banach, M.; Pirro, M.; Majeed, M.; Sahebkar, A. Effects of curcumin on HDL functionality. Pharmacol. Res. 2017, 119, 208–218. [Google Scholar] [CrossRef]
- Mortezaee, K.; Salehi, E.; Mirtavoos-Mahyari, H.; Motevaseli, E.; Najafi, M.; Farhood, B.; Rosengren, R.J.; Sahebkar, A. Mechanisms of apoptosis modulation by curcumin: Implications for cancer therapy. J. Cell. Physiol. 2019, 234, 12537–12550. [Google Scholar] [CrossRef]
- Panahi, Y.; Ghanei, M.; Bashiri, S.; Hajihashemi, A.; Sahebkar, A. Short-term Curcuminoid Supplementation for Chronic Pulmonary Complications due to Sulfur Mustard Intoxication: Positive Results of a Randomized Double-blind Placebo-controlled Trial. Drug Res. 2014, 65, 567–573. [Google Scholar] [CrossRef]
- Parsamanesh, N.; Moossavi, M.; Bahrami, A.; Butler, A.E.; Sahebkar, A. Therapeutic potential of curcumin in diabetic complications. Pharmacol. Res. 2018, 136, 181–193. [Google Scholar] [CrossRef]
- Sahebkar, A.; Henrotin, Y. Analgesic Efficacy and Safety of Curcuminoids in Clinical Practice: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Pain Med. 2016, 17, 1192–1202. [Google Scholar] [CrossRef]
- Shakeri, A.; Cicero, A.F.G.; Panahi, Y.; Mohajeri, M.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell. Physiol. 2019, 234, 5643–5654. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Harsha, C.; Parama, D.; Girisa, S.; Daimary, U.D.; Mao, X.; Kunnumakkara, A.B. Current clinical developments in curcumin-based therapeutics for cancer and chronic diseases. Phytotherapy Res. 2021, 35, 6768–6801. [Google Scholar] [CrossRef] [PubMed]
- Amato, A.; Caldara, G.-F.; Nuzzo, D.; Baldassano, S.; Picone, P.; Rizzo, M.; Mulè, F.; Di Carlo, M. NAFLD and Atherosclerosis Are Prevented by a Natural Dietary Supplement Containing Curcumin, Silymarin, Guggul, Chlorogenic Acid and Inulin in Mice Fed a High-Fat Diet. Nutrients 2017, 9, 492. [Google Scholar] [CrossRef] [PubMed]
- Cicero, A.F.G.; Sahebkar, A.; Fogacci, F.; Bove, M.; Giovannini, M.; Borghi, C. Effects of phytosomal curcumin on anthropometric parameters, insulin resistance, cortisolemia and non-alcoholic fatty liver disease indices: A double-blind, placebo-controlled clinical trial. Eur. J. Nutr. 2020, 59, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Hariri, M.; Gholami, A.; Mirhafez, S.R.; Bidkhori, M.; Sahebkar, A. A pilot study of the effect of curcumin on epigenetic changes and DNA damage among patients with non-alcoholic fatty liver disease: A randomized, double-blind, placebo-controlled, clinical trial. Complement. Ther. Med. 2020, 51, 102447. [Google Scholar] [CrossRef] [PubMed]
- Jalali, M.; Mahmoodi, M.; Mosallanezhad, Z.; Jalali, R.; Imanieh, M.H.; Moosavian, S.P. The effects of curcumin supplementation on liver function, metabolic profile and body composition in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis of randomized controlled trials. Complement. Ther. Med. 2020, 48, 102283. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, I.A.; Farrell, G.C.; Sempoux, C.; Peña, A.D.; Horsmans, Y. Curcumin inhibits NF-κB activation and reduces the severity of experimental steatohepatitis in mice. J. Hepatol. 2004, 41, 926–934. [Google Scholar] [CrossRef]
- Rahmani, S.; Asgary, S.; Askari, G.; Keshvari, M.; Hatamipour, M.; Feizi, A.; Sahebkar, A. Treatment of Non-alcoholic Fatty Liver Disease with Curcumin: A Randomized Placebo-controlled Trial. Phytother. Res. 2016, 30, 1540–1548. [Google Scholar] [CrossRef]
- Saberi-Karimian, M.; Keshvari, M.; Ghayour-Mobarhan, M.; Salehizadeh, L.; Rahmani, S.; Behnam, B.; Jamialahmadi, T.; Asgary, S.; Sahebkar, A. Effects of curcuminoids on inflammatory status in patients with non-alcoholic fatty liver disease: A randomized controlled trial. Complement. Ther. Med. 2020, 49, 102322. [Google Scholar] [CrossRef]
- Vizzutti, F.; Provenzano, A.; Galastri, S.; Milani, S.; Delogu, W.; Novo, E.; Caligiuri, A.; Zamara, E.; Arena, U.; Laffi, G.; et al. Curcumin limits the fibrogenic evolution of experimental steatohepatitis. Lab. Investig. 2010, 90, 104–115. [Google Scholar] [CrossRef]
- Bagherniya, M.; Nobili, V.; Blesso, C.N.; Sahebkar, A. Medicinal plants and bioactive natural compounds in the treatment of non-alcoholic fatty liver disease: A clinical review. Pharmacol. Res. 2018, 130, 213–240. [Google Scholar] [CrossRef] [PubMed]
- Likić, V.A.; McConville, M.J.; Lithgow, T.; Bacic, A. Systems Biology: The Next Frontier for Bioinformatics. Adv. Bioinform. 2010, 2010, 268925. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Szklarczyk, D.; Franceschini, A.; Campillos, M.; von Mering, C.; Jensen, L.J.; Beyer, A.; Bork, P. STITCH 2: An interaction network database for small molecules and proteins. Nucleic Acids Res. 2010, 38, D552–D556. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Pletscher-Frankild, S.; Pallejà, A.; Tsafou, K.; Binder, J.X.; Jensen, L.J. DISEASES: Text mining and data integration of disease–gene associations. Methods 2015, 74, 83–89. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. gene ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Franzén, O.; Gan, L.-M.; Björkegren, J.L.M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef]
- Eppig, J.T.; Richardson, J.E.; Kadin, J.A.; Ringwald, M.; Blake, J.A.; Bult, C.J. Mouse Genome Informatics (MGI): Reflecting on 25 years. Mamm. Genome 2015, 26, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62, S47–S64. [Google Scholar] [CrossRef] [PubMed]
- Margini, C.; Dufour, J.F. The story of HCC in NAFLD: From epidemiology, across pathogenesis, to prevention and treatment. Liver Int. J. 2016, 36, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, R.P.; Moore, M.; Moore, A.N.; Healy, J.C.; Roberts, M.D.; Rector, R.S.; Martin, J.S. Curcumin supplementation mitigates NASH development and progression in female Wistar rats. Physiol. Rep. 2018, 6, e13789. [Google Scholar] [CrossRef] [PubMed]
- Ferramosca, A.; Di Giacomo, M.; Zara, V. Antioxidant dietary approach in treatment of fatty liver: New insights and updates. World J. Gastroenterol. 2017, 23, 4146–4157. [Google Scholar] [CrossRef]
- Gheibi, S.; Ghaleh, H.E.G.; Motlagh, B.M.; Azarbayjani, A.F.; Zarei, L. Therapeutic effects of curcumin and ursodexycholic acid on non-alcoholic fatty liver disease. Biomed. Pharmacother. 2019, 115, 108938. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Stienstra, R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie 2017, 136, 75–84. [Google Scholar] [CrossRef]
- Lakhia, R.; Yheskel, M.; Flaten, A.; Quittner-Strom, E.B.; Holland, W.L.; Patel, V. PPARα agonist fenofibrate enhances fatty acid β-oxidation and attenuates polycystic kidney and liver disease in mice. Am. J. Physiol. Physiol. 2018, 314, F122–F131. [Google Scholar] [CrossRef]
- Karahashi, M.; Hoshina, M.; Yamazaki, T.; Sakamoto, T.; Mitsumoto, A.; Kawashima, Y.; Kudo, N. Fibrates Reduce Triacylglycerol Content by Upregulating Adipose Triglyceride Lipase in the Liver of Rats. J. Pharmacol. Sci. 2013, 123, 356–370. [Google Scholar] [CrossRef]
- Pan, S.-Y.; Yu, Q.; Zhang, Y.; Wang, X.-Y.; Sun, N.; Yu, Z.-L.; Ko, K.-M. Dietary Fructus Schisandrae extracts and fenofibrate regulate the serum/hepatic lipid-profile in normal and hypercholesterolemic mice, with attention to hepatotoxicity. Lipids Health Dis. 2012, 11, 120. [Google Scholar] [CrossRef]
- Pan, S.-Y.; Jia, Z.-H.; Zhang, Y.; Yu, Q.; Wang, X.-Y.; Sun, N.; Zhu, P.-L.; Yu, Z.-L.; Ko, K.-M. A Novel Mouse Model of Combined Hyperlipidemia Associated with Steatosis and Liver Injury by a Single-Dose Intragastric Administration of Schisandrin B/Cholesterol/Bile Salts Mixture. J. Pharmacol. Sci. 2013, 123, 110–119. [Google Scholar] [CrossRef]
- Naderali, E.K.; Fatani, S.; Itua, I.; Wong, C. The effects of diet-induced obesity on hepatocyte insulin signaling pathways and induction of non-alcoholic liver damage. Int. J. Gen. Med. 2011, 4, 211–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hong, X.Z.; Da Li, L.; Wu, L.M. Effects of Fenofibrate and Xuezhikang on Highfat Diet-induced Nonalcoholic Fatty Liver Disease. Clin. Exp. Pharmacol. Physiol. 2007, 34, 27–35. [Google Scholar] [CrossRef]
- Shiri-Sverdlov, R.; Wouters, K.; van Gorp, P.; Gijbels, M.J.; Noel, B.; Buffat, L.; Staels, B.; Maeda, N.; van Bilsen, M.; Hofker, M.H. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. J. Hepatol. 2006, 44, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.R.; Giri, S.R.; Bhoi, B.; Trivedi, C.; Rath, A.; Rathod, R.; Ranvir, R.; Kadam, S.; Patel, H.; Swain, P.; et al. Dual PPARα/γ agonist saroglitazar improves liver histopathology and biochemistry in experimental NASH models. Liver Int. 2018, 38, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Hamed, A.M.; El-Kharashi, O.A.; Boctor, S.S.; Abd-Elaziz, L.F. Potential involvement of PPAR α activation in diminishing the hepatoprotective effect of fenofibrate in NAFLD: Accuracy of non- invasive panel in determining the stage of liver fibrosis in rats. Biomed. Pharmacother. 2017, 85, 68–78. [Google Scholar] [CrossRef]
- Seo, Y.S.; Kim, J.H.; Jo, N.Y.; Choi, K.M.; Baik, S.H.; Park, J.-J.; Kim, J.S.; Byun, K.S.; Bak, Y.-T.; Lee, C.H.; et al. PPAR agonists treatment is effective in a nonalcoholic fatty liver disease animal model by modulating fatty-acid metabolic enzymes. J. Gastroenterol. Hepatol. 2006, 23, 102–109. [Google Scholar] [CrossRef]
- Kong, D.; Zhang, Z.; Chen, L.; Huang, W.; Zhang, F.; Wang, L.; Wang, Y.; Cao, P.; Zheng, S. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020, 36, 101600. [Google Scholar] [CrossRef]
- Zeng, K.; Tian, L.; Patel, R.; Shao, W.; Song, Z.; Manuel, J.; Ma, X.; McGilvray, I.; Weng, J.; Jin, T.; et al. Diet polyphenol curcumin stimulates hepatic Fgf21 production and restores its sensitivity in high fat diet fed male mice. Endocrinology 2017, 158, 277–292. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Fu, J.; Sun, J.; Liu, D.; Chen, C.; Wang, H.; Hou, Y.; Xu, Y.; Pi, J. Is Nrf2-ARE a potential target in NAFLD mitigation? Curr. Opin. Toxicol. 2019, 13, 35–44. [Google Scholar] [CrossRef]
- Chowdhry, S.; Nazmy, M.H.; Meakin, P.; Dinkova-Kostova, A.; Walsh, S.V.; Tsujita, T.; Dillon, J.; Ashford, M.; Hayes, J.D. Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic. Biol. Med. 2010, 48, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Ramadori, P.; Drescher, H.; Erschfeld, S.; Schumacher, F.; Berger, C.; Fragoulis, A.; Schenkel, J.; Kensler, T.; Wruck, C.J.; Trautwein, C.; et al. Hepatocyte-specific Keap1 deletion reduces liver steatosis but not inflammation during non-alcoholic steatohepatitis development. Free Radic. Biol. Med. 2016, 91, 114–126. [Google Scholar] [CrossRef]
- Lee, L.-Y.; Köhler, U.A.; Zhang, L.; Roenneburg, E.; Werner, S.; Johnson, J.A.; Foley, D.P. Activation of the Nrf2-ARE pathway in hepatocytes protects against steatosis in nutritionally induced non-alcoholic steatohepatitis in mice. Toxicol. Sci. 2014, 142, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Park, J.S.; Lee, Y.S.; Han, J.; Lee, D.-K.; Kwon, S.W.; Han, D.H.; Lee, Y.-H.; Bae, S.H. SQSTM1/p62 activates NFE2L2/NRF2 via ULK1-mediated autophagic KEAP1 degradation and protects mouse liver from lipotoxicity. Autophagy 2020, 16, 1949–1973. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Ahmadi, Z.; Mohamamdinejad, R.; Farkhondeh, T.; Samarghandian, S. Curcumin Activates the Nrf2 Pathway and Induces Cellular Protection Against Oxidative Injury. Curr. Mol. Med. 2020, 20, 116–133. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhang, Y.; Zhang, X.; Aa, J.; Wang, G.; Xie, Y. Curcumin regulates endogenous and exogenous metabolism via Nrf2-FXR-LXR pathway in NAFLD mice. Biomed. Pharmacother. 2018, 105, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Dai, C.; Liu, Q.; Li, J.; Qiu, J. Curcumin Attenuates on Carbon Tetrachloride-Induced Acute Liver Injury in Mice via Modulation of the Nrf2/HO-1 and TGF-β1/Smad3 Pathway. Molecules 2018, 23, 215. [Google Scholar] [CrossRef]
- Lu, C.; Xu, W.; Zheng, S. Nrf2 activation is required for curcumin to induce lipocyte phenotype in hepatic stellate cells. Biomed. Pharmacother. 2017, 95, 1–10. [Google Scholar] [CrossRef]
- Swellam, M.; Hamdy, N. Association of nonalcoholic fatty liver disease with a single nucleotide polymorphism on the gene encoding leptin receptor. IUBMB Life 2012, 64, 180–186. [Google Scholar] [CrossRef]
- Xu, D.; Huang, X.-D.; Luo, H.-S.; Yuan, J.-P.; Zhang, H.; Wu, J. Impaired activation of phosphatidylinositol 3-kinase by leptin in NAFLD: A novel mechanism of hepatic leptin resistance. World Chin. J. Dig. 2012, 20, 3095–3100. [Google Scholar] [CrossRef]
- Xu, D.; Huang, X.-D.; Yuan, J.-P.; Wu, J.; Fan, Y.; Luo, H.-S.; Yang, Y.-H. Impaired Activation of Phosphatidylinositol 3-Kinase by Leptin is a Novel Mechanism of Hepatic Leptin Resistance in NAFLD. Hepatogastroenterology 2011, 58, 1703–1707. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zheng, S.; Chen, A. Curcumin Eliminates Leptin’s Effects on Hepatic Stellate Cell Activation via Interrupting Leptin Signaling. Endocrinology 2009, 150, 3011–3020. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.-H.; Liao, J.; Zhang, J.-Y.; Hao, X.-H.; Liang, C.; Wang, L.-H.; Xue, H.; Zhang, K.; Yan, G.-T. Localized leptin release may be an important mechanism of curcumin action after acute ischemic injuries. J. Trauma Acute Care Surg. 2013, 74, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Nejati-Koshki, K.; Akbarzadeh, A.; Pourhassan-Moghaddam, M. Curcumin inhibits leptin gene expression and secretion in breast cancer cells by estrogen receptors. Cancer Cell Int. 2014, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Askari, S.; Salehi, R.; Zarghami, N.; Akbarzadeh, A.; Rahmati-Yamchi, M. The anticancer effects of biodegradable nanomagnetic dual natural components on the leptin gene expression in lung cancer. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1753–1763. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, H.; Zhou, Y.; Xu, F. Curcumin Affects Leptin-Induced Expression of Methionine Adenosyltransferase 2A in Hepatic Stellate Cells by Inhibition of JNK Signaling. Pharmacology 2021, 106, 426–434. [Google Scholar] [CrossRef]
- Floeth, M.; Elges, S.; Gerss, J.; Schwöppe, C.; Kessler, T.; Herold, T.; Wardelmann, E.; Berdel, W.E.; Lenz, G.; Mikesch, J.; et al. Low-density lipoprotein receptor (LDLR) is an independent adverse prognostic factor in acute myeloid leukaemia. Br. J. Haematol. 2021, 192, 494–503. [Google Scholar] [CrossRef]
- Álvarez-Amor, L.; Sierra, A.L.; Cárdenas, A.; López-Bermudo, L.; López-Beas, J.; Andújar, E.; Pérez-Alegre, M.; Gallego-Durán, R.; Varela, L.M.; Martin-Montalvo, A.; et al. Extra virgin olive oil improved body weight and insulin sensitivity in high fat diet-induced obese LDLr−/−.Leiden mice without attenuation of steatohepatitis. Sci. Rep. 2021, 11, 8250. [Google Scholar] [CrossRef]
- Schoemaker, M.H.; Kleemann, R.; Morrison, M.C.; Verheij, J.; Salic, K.; Van Tol, E.A.F.; Kooistra, T.; Wielinga, P.Y. A casein hydrolysate based formulation attenuates obesity and associated non-alcoholic fatty liver disease and atherosclerosis in LDLr-/-.Leiden mice. PLoS ONE 2017, 12, e0180648. [Google Scholar] [CrossRef]
- Li, L.; Zhang, G.-F.; Lee, K.; Lopez, R.; Previs, S.F.; Willard, B.; McCullough, A.; Kasumov, T. A Western diet induced NAFLD in LDLR−/− mice is associated with reduced hepatic glutathione synthesis. Free Radic. Biol. Med. 2016, 96, 13–21. [Google Scholar] [CrossRef]
- He, Y.; Yang, W.; Gan, L.; Liu, S.; Ni, Q.; Bi, Y.; Han, T.; Liu, Q.; Chen, H.; Hu, Y.; et al. Silencing HIF-1α aggravates non-alcoholic fatty liver disease in vitro through inhibiting PPAR-α/ANGPTL4 singling pathway. Gastroenterol. Hepatol. 2020, 44, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.-H.; Chen, P.-K.; Chen, P.-Y.; Wu, M.-J.; Ho, C.-T.; Yen, J.-H. Curcumin enhances cell-surface LDLR level and promotes LDL uptake through downregulation of PCSK9 gene expression in HepG2 cells. Mol. Nutr. Food Res. 2014, 58, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.B.; Wo, X.D.; Fan, C.L.; Yu, Y.; Qian, Y.; Huang, X.L.; Ling, X. Effect of curcumin on the expression of low density lipoprotein receptor in HepG2 cell line. Chin. Pharm. J. 2007, 42, 572–575. [Google Scholar]
- Cai, Y.; Lu, D.; Zou, Y.; Zhou, C.; Liu, H.; Tu, C.; Li, F.; Zhang, S. Curcumin Protects Against Intestinal Origin Endotoxemia in Rat Liver Cirrhosis by Targeting PCSK9. J. Food Sci. 2017, 82, 772–780. [Google Scholar] [CrossRef]
- Corral, P.A.; Botello, J.F.; Xing, C. Design, synthesis, and enzymatic characterization of quinazoline-based CYP1A2 inhibitors. Bioorganic Med. Chem. Lett. 2020, 30, 126719. [Google Scholar] [CrossRef]
- Chiba, T.; Noji, K.; Shinozaki, S.; Suzuki, S.; Umegaki, K.; Shimokado, K. Diet-induced non-alcoholic fatty liver disease affects expression of major cytochrome P450 genes in a mouse model. J. Pharm. Pharmacol. 2016, 68, 1567–1576. [Google Scholar] [CrossRef]
- Jearapong, N.; Chatuphonprasert, W.; Jarukamjorn, K. Effect of tetrahydrocurcumin on the profiles of drug-metabolizing enzymes induced by a high fat and high fructose diet in mice. Chem.-Biol. Interact. 2015, 239, 67–75. [Google Scholar] [CrossRef]
- Fisher, C.D.; Lickteig, A.J.; Augustine, L.M.; Ranger-Moore, J.; Jackson, J.P.; Ferguson, S.S.; Cherrington, N.J. Hepatic Cytochrome P450 Enzyme Alterations in Humans with Progressive Stages of Nonalcoholic Fatty Liver Disease. Drug Metab. Dispos. 2009, 37, 2087–2094. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, W.-H.; Chen, B.-L.; Fan, L.; Han, Y.; Wang, G.; Hu, D.-L.; Tan, Z.-R.; Zhou, G.; Cao, S.; et al. Plant Polyphenol Curcumin Significantly Affects CYPIA2 and CYP2A6 Activity in Healthy, Male Chinese Volunteers. Ann. Pharmacother. 2010, 44, 1038–1045. [Google Scholar] [CrossRef]
- Abd-Elhakim, Y.M.; Moustafa, G.G.; El-Sharkawy, N.I.; Hussein, M.M.; Ghoneim, M.H.; El Deib, M.M. The ameliorative effect of curcumin on hepatic CYP1A1 and CYP1A2 genes dysregulation and hepatorenal damage induced by fenitrothion oral intoxication in male rats. Pestic. Biochem. Physiol. 2021, 179, 104959. [Google Scholar] [CrossRef]
- Wu, R.; Cui, X.; Dong, W.; Zhou, M.; Simms, H.H.; Wang, P. Suppression of hepatocyte CYP1A2 expression by Kupffer cells via AhR pathway: The central role of proinflammatory cytokines. Int. J. Mol. Med. 2006, 18, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Appiahopong, R.; Commandeur, J.; Vanvugtlussenburg, B.; Vermeulen, N.P. Inhibition of human recombinant cytochrome P450s by curcumin and curcumin decomposition products. Toxicology 2007, 235, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Gianotti, T.F.; Rosselli, M.S.; Burgueño, A.L.; Castaño, G.O.; Pirola, C.J. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis 2011, 218, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, H.R.H.; El-Nablaway, M.; Othman, B.H.; Abdalla, A.M.; El Nashar, E.M.; Abd-Elmonem, M.M.; El-Gamal, R. Can Dasatinib Ameliorate the Hepatic changes, Induced by Long Term Western Diet, in Mice? Ann. Anat. Anat. Anz. 2021, 234, 151626. [Google Scholar] [CrossRef] [PubMed]
- Mikuriya, Y.; Tashiro, H.; Kuroda, S.; Nambu, J.; Kobayashi, T.; Amano, H.; Tanaka, Y.; Ohdan, H. Fatty liver creates a pro-metastatic microenvironment for hepatocellular carcinoma through activation of hepatic stellate cells. Int. J. Cancer 2014, 136, E3–E13. [Google Scholar] [CrossRef]
- Poniachik, J.; Csendes, A.; Díaz, J.C.; Rojas, J.; Burdiles, P.; Maluenda, F.; Smok, G.; Rodrigo, R.; Videla, L.A. Increased production of IL-1α and TNF-α in lipopolysaccharide-stimulated blood from obese patients with non-alcoholic fatty liver disease. Cytokine 2006, 33, 252–257. [Google Scholar] [CrossRef]
- Xu, Y.X.; Pindolia, K.; Janakiraman, N.; Noth, C.J.; Chapman, R.A.; Gautam, S.C. Curcumin, a compound with anti-inflammatory and anti-oxidant properties, down-regulates chemokine expression in bone marrow stromal cells. Exp. Hematol. 1997, 25, 413–422. [Google Scholar]
- Xu, Y.X.; Pindolia, K.R.; Janakiraman, N.; Chapman, R.A.; Gautam, S.C. Curcumin inhibits IL1α and TNFα induction of AP-1 and NF-kB DNA-binding activity in bone marrow stromal cells. Hematopathol. Mol. Hematol. 1997, 11, 49–62. [Google Scholar]
- Michaud, V.; Tran, M.; Pronovost, B.; Bouchard, P.; Bilodeau, S.; Alain, K.; Vadnais, B.; Franco, M.; Bélanger, F.; Turgeon, J. Impact of GSTA1 Polymorphisms on Busulfan Oral Clearance in Adult Patients Undergoing Hematopoietic Stem Cell Transplantation. Pharmaceutics 2019, 11, 440. [Google Scholar] [CrossRef]
- Oniki, K.; Hori, M.; Saruwatari, J.; Morita, K.; Kajiwara, A.; Sakata, M.; Mihara, S.; Ogata, Y.; Nakagawa, K. Interactive effects of smoking and glutathione S-transferase polymorphisms on the development of non-alcoholic fatty liver disease. Toxicol. Lett. 2013, 220, 143–149. [Google Scholar] [CrossRef]
- Jin, X.-L.; Wang, K.; Li, Q.; Tian, W.-L.; Xue, X.-F.; Wu, L.-M.; Hu, F.-L. Antioxidant and anti-inflammatory effects of Chinese propolis during palmitic acid-induced lipotoxicity in cultured hepatocytes. J. Funct. Foods 2017, 34, 216–223. [Google Scholar] [CrossRef]
- Miyata, M.; Funaki, A.; Fukuhara, C.; Sumiya, Y.; Sugiura, Y. Taurine attenuates hepatic steatosis in a genetic model of fatty liver disease. J. Toxicol. Sci. 2020, 45, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Matsushita, K.; Shindo, R.; Shimokawa, Y.; Sugiura, Y.; Yamashita, M. Selenoneine Ameliorates Hepatocellular Injury and Hepatic Steatosis in a Mouse Model of NAFLD. Nutrients 2020, 12, 1898. [Google Scholar] [CrossRef] [PubMed]
- Robin, S.K.D.; Ansari, M.; Uppugunduri, C.R.S. Spectrophotometric Screening for Potential Inhibitors of Cytosolic Glutathione S-Transferases. J. Vis. Exp. 2020, e61347. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Ellis, B.; Sharma, A. Role of alpha class glutathione transferases (GSTs) in chemoprevention: GSTA1 and A4 overexpressing human leukemia (HL60) cells resist sulforaphane and curcumin induced toxicity. Phytother. Res. 2011, 25, 563–568. [Google Scholar] [CrossRef]
- Odenthal, J.; van Heumen, B.W.H.; Roelofs, H.M.J.; Morsche, R.H.M.T.; Marian, B.; Nagengast, F.M.; Peters, W.H.M. The Influence of Curcumin, Quercetin, and Eicosapentaenoic Acid on the Expression of Phase II Detoxification Enzymes in the Intestinal Cell Lines HT-29, Caco-2, HuTu 80, and LT97. Nutr. Cancer 2012, 64, 856–863. [Google Scholar] [CrossRef]
- Qadri, Q.; Sameer, A.; Shah, Z.; Hamid, A.; Alam, S.; Manzoor, S.; Siddiqi, M. Genetic polymorphism of the glutathione-S-transferase P1 gene (GSTP1) and susceptibility to prostate cancer in the Kashmiri population. Genet. Mol. Res. 2011, 10, 3038–3045. [Google Scholar] [CrossRef]
- Hori, M.; Oniki, K.; Nakagawa, T.; Takata, K.; Mihara, S.; Marubayashi, T.; Nakagawa, K. Association between combinations of glutathione-S-transferaseM1, T1 and P1 genotypes and non-alcoholic fatty liver disease. Liver Int. 2009, 29, 164–168. [Google Scholar] [CrossRef]
- Kirpich, I.A.; Gobejishvili, L.N.; Homme, M.B.; Waigel, S.; Cave, M.; Arteel, G.; Barve, S.S.; McClain, C.J.; Deaciuc, I.V. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J. Nutr. Biochem. 2011, 22, 38–45. [Google Scholar] [CrossRef]
- Prysyazhnyuk, V.; Rossokha, Z.; Gorovenko, N. Variation in particular biochemical indicators, cytokine and adipokine profiles of the blood, and the structural and functional parameters of the liver in patients with nonalcoholic fatty liver disease and different genotypes by the polymorphic locus A313G of the GSTP1 gene. Cytol. Genet. 2017, 51, 455–461. [Google Scholar] [CrossRef]
- Hashemi, M.; Eskandari, E.; Fazaeli, A.; Bahari, A.; Hashemzehi, N.-A.; Shafieipour, S.; Taheri, M.; Moazeni-Roodi, A.; Zakeri, Z.; Bakhshipour, A.; et al. Association of Genetic Polymorphisms of Glutathione-S-Transferase Genes (GSTT1, GSTM1, and GSTP1) and Susceptibility to Nonalcoholic Fatty Liver Disease in Zahedan, Southeast Iran. DNA Cell Biol. 2012, 31, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Nishinaka, T.; Ichijo, Y.; Ito, M.; Kimura, M.; Katsuyama, M.; Iwata, K.; Miura, T.; Terada, T.; Yabe-Nishimura, C. Curcumin activates human glutathione S-transferase P1 expression through antioxidant response element. Toxicol. Lett. 2007, 170, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Duvoix, A.; Morceau, F.; Delhalle, S.; Schmitz, M.; Schnekenburger, M.; Galteau, M.-M.; Dicato, M.; Diederich, M. Induction of apoptosis by curcumin: Mediation by glutathione S-transferase P1-1 inhibition. Biochem. Pharmacol. 2003, 66, 1475–1483. [Google Scholar] [CrossRef]
- Li, H.; Li, L. Relationship of GSTP1 lower expression and multidrug resistance reversing of curcumin on human colon carcinoma cells. Zhonghua Yi Xue Za Zhi 2015, 95, 2478–2482. [Google Scholar]
- Carrillo-Gálvez, A.B.; Gálvez-Peisl, S.; González-Correa, J.E.; Haro-Carrillo, M.; Ayllón, V.; Carmona-Sáez, P.; Ramos-Mejía, V.; Galindo-Moreno, P.; Cara, F.E.; Granados-Principal, S.; et al. GARP is a key molecule for mesenchymal stromal cell responses to TGF-β and fundamental to control mitochondrial ROS levels. STEM CELLS Transl. Med. 2020, 9, 636–650. [Google Scholar] [CrossRef]
- Subudhi, S.; Drescher, H.K.; Dichtel, L.E.; Bartsch, L.M.; Chung, R.T.; Hutter, M.M.; Gee, D.W.; Meireles, O.R.; Witkowski, E.R.; Gelrud, L.; et al. Distinct Hepatic Gene-Expression Patterns of NAFLD in Patients with Obesity. Hepatol. Commun. 2021, 6, 77–89. [Google Scholar] [CrossRef]
- Tang, M.; Jia, H.; Chen, S.; Yang, B.; Patpur, B.K.; Song, W.; Chang, Y.; Li, J.; Yang, C. Significance of MR/OPN/HMGB1 axis in NAFLD-associated hepatic fibrogenesis. Life Sci. 2021, 264, 118619. [Google Scholar] [CrossRef]
- Brown, K.D.; Shah, M.H.; Liu, G.-S.; Chan, E.C.; Crowston, J.G.; Peshavariya, H.M. Transforming Growth Factor ?1–Induced NADPH Oxidase-4 Expression and Fibrotic Response in Conjunctival Fibroblasts. Investig. Opthalmol. Vis. Sci. 2017, 58, 3011–3017. [Google Scholar] [CrossRef]
- Fu, X.-Y.; Zhang, D.-W.; Li, Y.-D.; Jian-Zhao, N.; Tang, Y.-Q.; Niu, J.-Z.; Li, Y. Curcumin Treatment Suppresses CCR7 Expression and the Differentiation and Migration of Human Circulating Fibrocytes. Cell. Physiol. Biochem. 2015, 35, 489–498. [Google Scholar] [CrossRef]
- Li, H.; Liu, B.; Li, P.; Feng, L.; Ma, H.; Xuan, S.; Cao, Y. Inhibitory effects of curcumin on inflammatory cytokines in rats with paraquat poisoning. Chin. J. Ind. Hyg. Occup. Dis. 2015, 33, 689–692. [Google Scholar]
- Gordon, M.Y.; Riley, G.P.; Watt, S.M.; Greaves, M.F. Compartmentalization of a haematopoietic growth factor (GM-CSF) by glycosaminoglycans in the bone marrow microenvironment. Nature 1987, 326, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Tan-Garcia, A.; Lai, F.; Yeong, J.P.S.; Irac, S.E.; Ng, P.Y.; Msallam, R.; Lim, J.C.T.; Wai, L.-E.; Tham, C.Y.; Choo, S.P.; et al. Liver fibrosis and CD206+ macrophage accumulation are suppressed by anti-GM-CSF therapy. JHEP Rep. 2020, 2, 100062. [Google Scholar] [CrossRef]
- Xin, X.; Jin, Y.; Wang, X.; Cai, B.; An, Z.; Hu, Y.-Y.; Feng, Q. A Combination of Geniposide and Chlorogenic Acid Combination Ameliorates Nonalcoholic Steatohepatitis in Mice by Inhibiting Kupffer Cell Activation. BioMed Res. Int. 2021, 2021, 6615881. [Google Scholar] [CrossRef] [PubMed]
- de Almeida-Souza, C.B.; Antunes, M.M.; Carbonera, F.; Godoy, G.; da Silva, M.A.; Masi, L.N.; Visentainer, J.V.; Curi, R.; Bazotte, R.B. A High-Fat Diet Induces Lower Systemic Inflammation than a High-Carbohydrate Diet in Mice. Metab. Syndr. Relat. Disord. 2021, 19, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.C.; Kleemann, R.; Van Koppen, A.; Hanemaaijer, R.; Verschuren, L. Key Inflammatory Processes in Human NASH Are Reflected in Ldlr−/−.Leiden Mice: A Translational Gene Profiling Study. Front. Physiol. 2018, 9, 132. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hashimoto, S.; Horie, T. Curcumin inhibition of Dermatophagoides farinea-induced interleukin-5 (IL-5) and granulocyte macrophage-colony stimulating factor (GM-CSF) production by lymphocytes from bronchial asthmatics. Biochem. Pharmacol. 1997, 54, 819–824. [Google Scholar] [CrossRef]
- Latimer, B.; Ekshyyan, O.; Nathan, N.; Moore-Medlin, T.; Rong, X.; Ma, X.; Khandelwal, A.; Christy, H.T.; Abreo, F.; McClure, G.; et al. Enhanced Systemic Bioavailability of Curcumin Through Transmucosal Administration of a Novel Microgranular Formulation. Anticancer Res. 2015, 35, 6411–6418. [Google Scholar]
- Mellor, H.; Parker, P. The extended protein kinase C superfamily. Biochem. J. 1998, 332, 281–292. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Shulman, G.I. Diacylglycerol Activation of Protein Kinase Cε and Hepatic Insulin Resistance. Cell Metab. 2012, 15, 574–584. [Google Scholar] [CrossRef]
- ter Horst, K.W.; Gilijamse, P.W.; Versteeg, R.I.; Ackermans, M.T.; Nederveen, A.J.; la Fleur, S.E.; Romijn, J.A.; Nieuwdorp, M.; Zhang, D.; Samuel, V.T.; et al. Hepatic Diacylglycerol-Associated Protein Kinase Cε Translocation Links Hepatic Steatosis to Hepatic Insulin Resistance in Humans. Cell Rep. 2017, 19, 1997–2004. [Google Scholar] [CrossRef]
- Birkenfeld, A.L.; Shulman, G.I. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 Diabetes. Hepatology 2013, 59, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Pany, S.; You, Y.; Das, J. Curcumin Inhibits Protein Kinase Cα Activity by Binding to Its C1 Domain. Biochemistry 2016, 55, 6327–6336. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, B.; Huang, F.; Liu, B.; Xie, Y. Curcumin inhibits lipolysis via suppression of ER stress in adipose tissue and prevents hepatic insulin resistance. J. Lipid Res. 2016, 57, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Jemnitz, K.; Heredi-Szabo, K.; Janossy, J.; Ioja, E.; Vereczkey, L.; Krajcsi, P. ABCC2/Abcc2: A multispecific transporter with dominant excretory functions. Drug Metab. Rev. 2010, 42, 402–436. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Castaño, G.; Gianotti, T.F.; Gemma, C.; Pirola, C.J. Polymorphisms of MRP2 (ABCC2) are associated with susceptibility to nonalcoholic fatty liver disease. J. Nutr. Biochem. 2009, 20, 765–770. [Google Scholar] [CrossRef]
- Sakulterdkiat, T.; Srisomsap, C.; Udomsangpetch, R.; Svasti, J.; Lirdprapamongkol, K. Curcumin resistance induced by hypoxia in HepG2 cells is mediated by multidrug-resistance-associated proteins. Anticancer Res. 2012, 32, 5337–5342. [Google Scholar]
- Cho, C.; Yang, C.; Wu, C.; Ho, J.; Yu, C.; Wu, S.; Yu, D. The modulation study of multiple drug resistance in bladder cancer by curcumin and resveratrol. Oncol. Lett. 2019, 18, 6869–6876. [Google Scholar] [CrossRef]
- Cho, C.; Yu, C.; Wu, C.; Ho, J.; Yang, C.; Yu, D. Decreased drug resistance of bladder cancer using phytochemicals treatment. Kaohsiung J. Med Sci. 2021, 37, 128–135. [Google Scholar] [CrossRef]
- Bock, K.W. Modulation of aryl hydrocarbon receptor (AHR) and the NAD+-consuming enzyme CD38: Searches of therapeutic options for nonalcoholic fatty liver disease (NAFLD). Biochem. Pharmacol. 2020, 175, 113905. [Google Scholar] [CrossRef]
- Bock, K.W. Aryl hydrocarbon receptor (AHR)-mediated inflammation and resolution: Non-genomic and genomic signaling. Biochem. Pharmacol. 2020, 182, 114220. [Google Scholar] [CrossRef]
- Choi, H.; Chun, Y.-S.; Shin, Y.J.; Ye, S.K.; Kim, M.-S.; Park, J.-W. Curcumin attenuates cytochrome P450 induction in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin by ROS-dependently degrading AhR and ARNT. Cancer Sci. 2008, 99, 2518–2524. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Yoshida, K.-I.; Ashida, H. Curcumin suppresses the transformation of an aryl hydrocarbon receptor through its phosphorylation. Arch. Biochem. Biophys. 2007, 466, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Li, Y.; Kuang, Y.; Cui, H.; Yang, Y.; Sun, W.; Liu, K.; Chen, N.; Yan, Q.; Wen, L. PKCδ silencing alleviates saturated fatty acid induced ER stress by enhancing SERCA activity. Biosci. Rep. 2017, 37, BSR20170869. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Song, X.; Yu, C. Identification of genes in hepatocellular carcinoma induced by non-alcoholic fatty liver disease. Cancer Biomarkers 2020, 29, 69–78. [Google Scholar] [CrossRef]
- Park, S.W.; Zhou, Y.; Lee, J.; Lee, J.; Ozcan, U. Sarco(endo)plasmic reticulum Ca 2+ -ATPase 2b is a major regulator of endoplasmic reticulum stress and glucose homeostasis in obesity. Proc. Natl. Acad. Sci. USA 2010, 107, 19320–19325. [Google Scholar] [CrossRef] [PubMed]
- Frangioudakis, G.; Burchfield, J.G.; Narasimhan, S.; Cooney, G.J.; Leitges, M.; Biden, T.J.; Schmitz-Peiffer, C. Diverse roles for protein kinase C δ and protein kinase C ε in the generation of high-fat-diet-induced glucose intolerance in mice: Regulation of lipogenesis by protein kinase C δ. Diabetologia 2009, 52, 2616–2620. [Google Scholar] [CrossRef]
- Bezy, O.; Tran, T.T.; Pihlajamäki, J.; Suzuki, R.; Emanuelli, B.; Winnay, J.; Mori, M.; Haas, J.; Biddinger, S.B.; Leitges, M.; et al. PKCδ regulates hepatic insulin sensitivity and hepatosteatosis in mice and humans. J. Clin. Investig. 2011, 121, 2504–2517. [Google Scholar] [CrossRef]
- Rushworth, S.A.; Ogborne, R.M.; Charalambos, C.A.; O’Connell, M.A. Role of protein kinase C δ in curcumin-induced antioxidant response element-mediated gene expression in human monocytes. Biochem. Biophys. Res. Commun. 2006, 341, 1007–1016. [Google Scholar] [CrossRef]
- Kunwar, A.; Narang, H.; Priyadarsini, K.I.; Krishna, M.; Pandey, R.; Sainis, K. Delayed activation of PKCδ and NFκB and higher radioprotection in splenic lymphocytes by copper (II)–Curcumin (1:1) complex as compared to curcumin. J. Cell. Biochem. 2007, 102, 1214–1224. [Google Scholar] [CrossRef]
- Hsu, H.-Y.; Chu, L.-C.; Hua, K.-F.; Chao, L.K. Heme oxygenase-1 mediates the anti-inflammatory effect of Curcumin within LPS-stimulated human monocytes. J. Cell. Physiol. 2008, 215, 603–612. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).