Comparative Proteomic Analysis of Proteins in Breast Milk during Different Lactation Periods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Protein Extraction and Digestion
2.3. Nano-Liquid Chromatographic Analysis
2.4. DIA Analysis by Nanospray Electrospray Ionization Mass Spectrometry
2.5. Protein Identification and Quantitative Analysis
2.6. Amino Acids Analysis
2.7. Target Proteins Identification and Quantification
2.8. Bioinformatics Analysis
2.9. Cell Culture and Viability
2.10. Detection of TNF-α
2.11. Western Blotting Analysis
3. Results
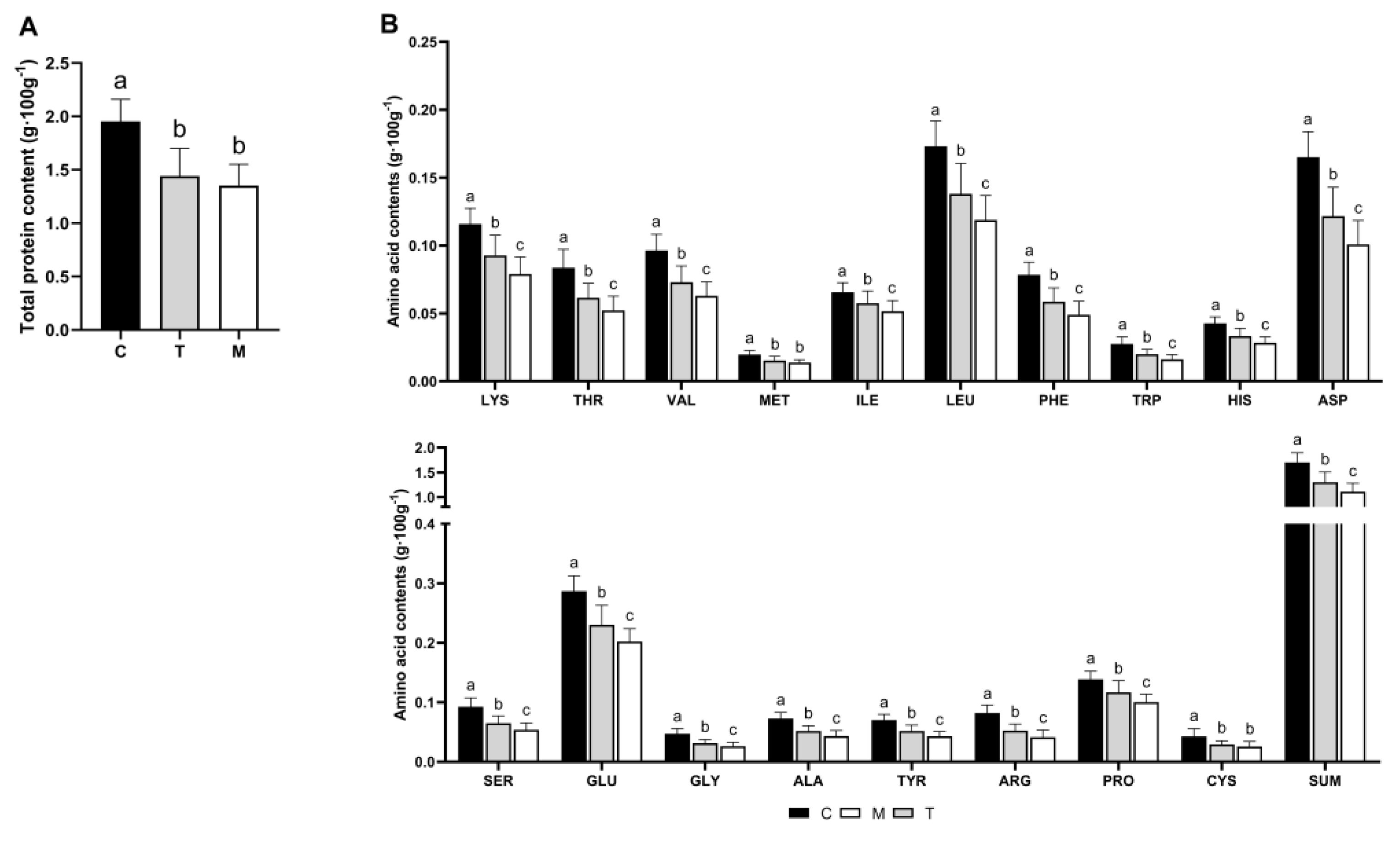
3.1. Comparison of Total Protein and Amino Acid Content of Breast Milk from Different Lactation Periods
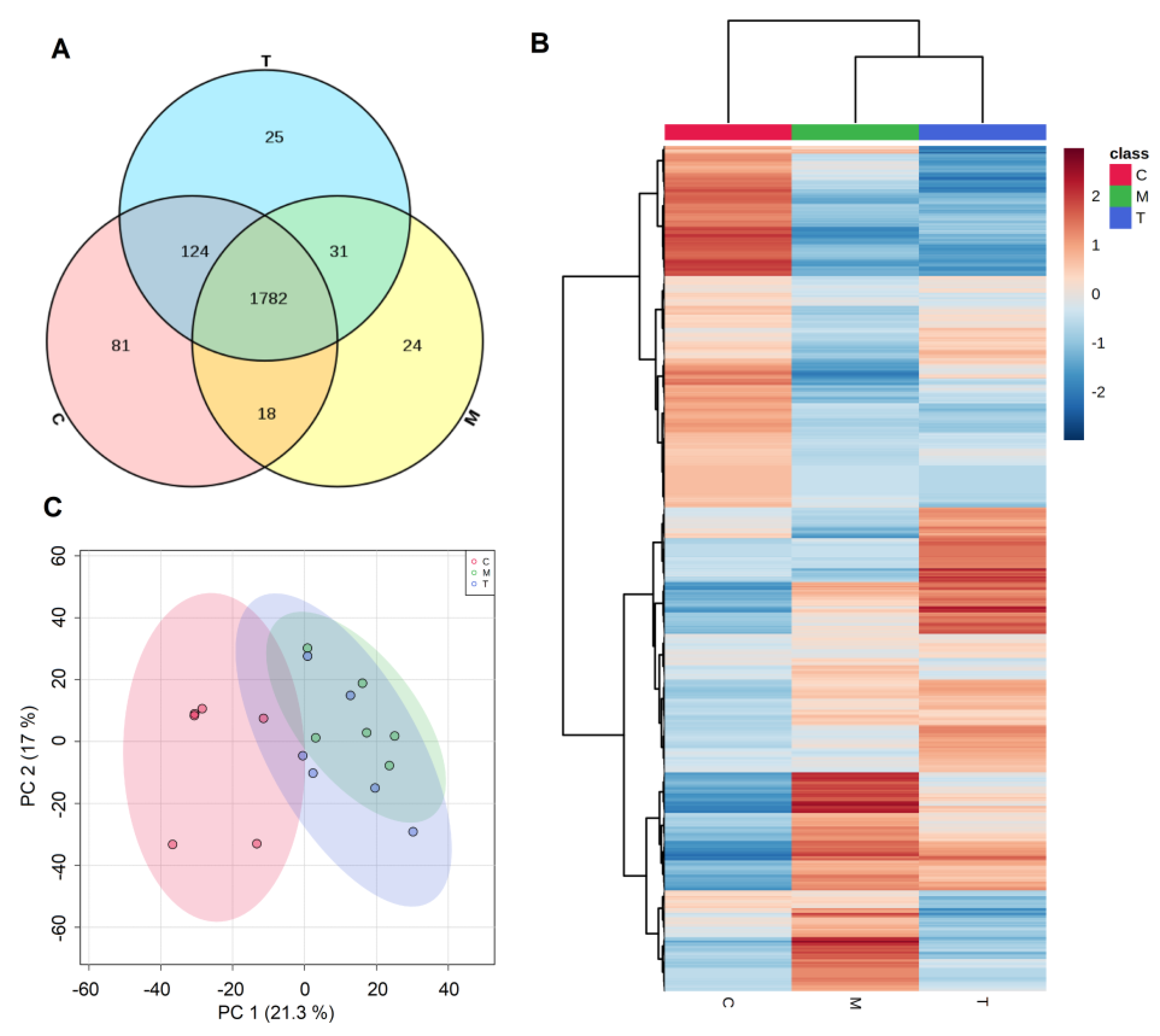
3.2. Identification and Quantification of the Proteome in Breast Milk
3.3. Proteins Differentially Expressed in Different Lactation Periods
3.4. GO Analysis of the Differentially Expressed Proteins in Breast Milk
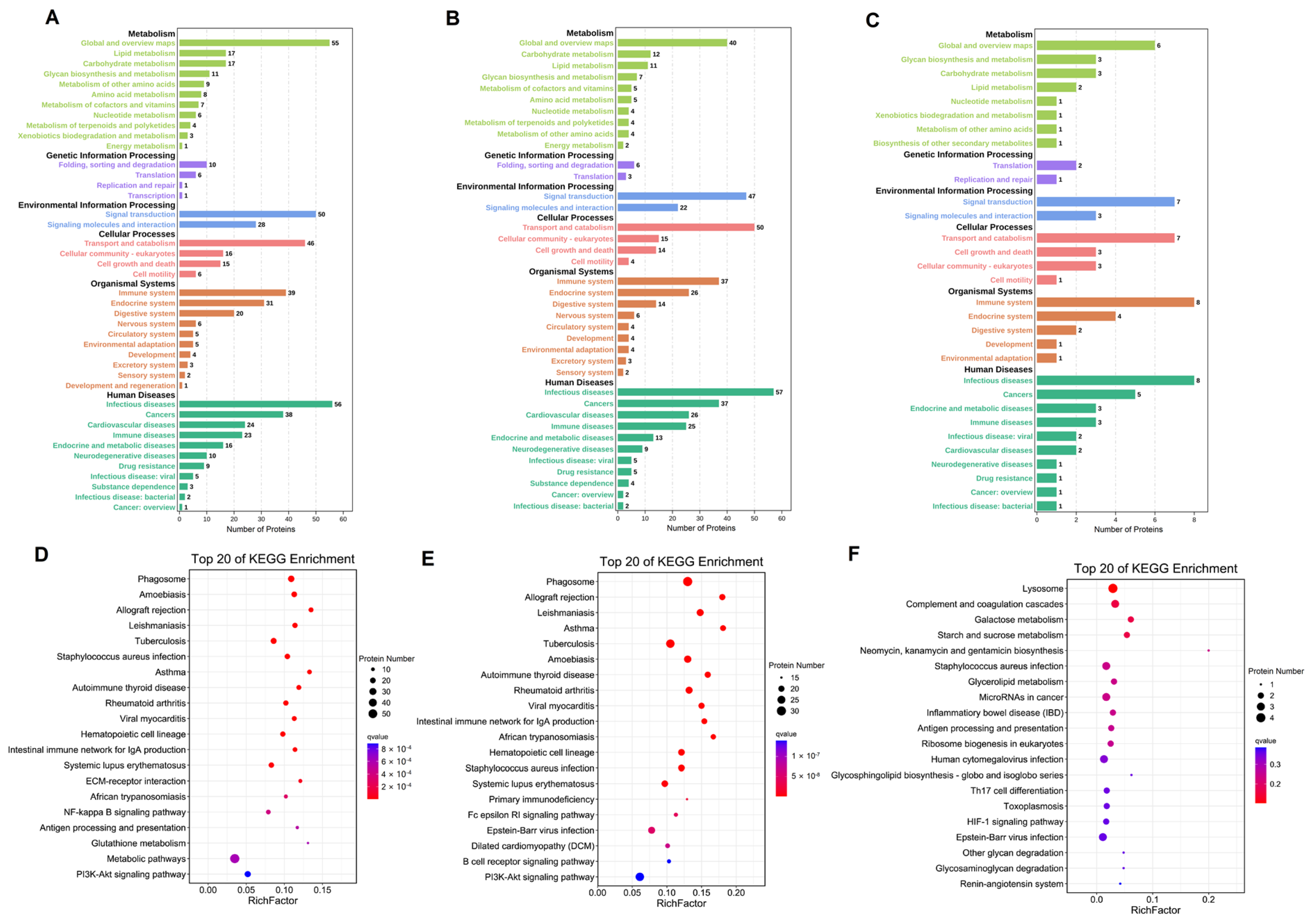
3.5. KEGG Pathway Analysis of the Differentially Expressed Proteins in Breast Milk
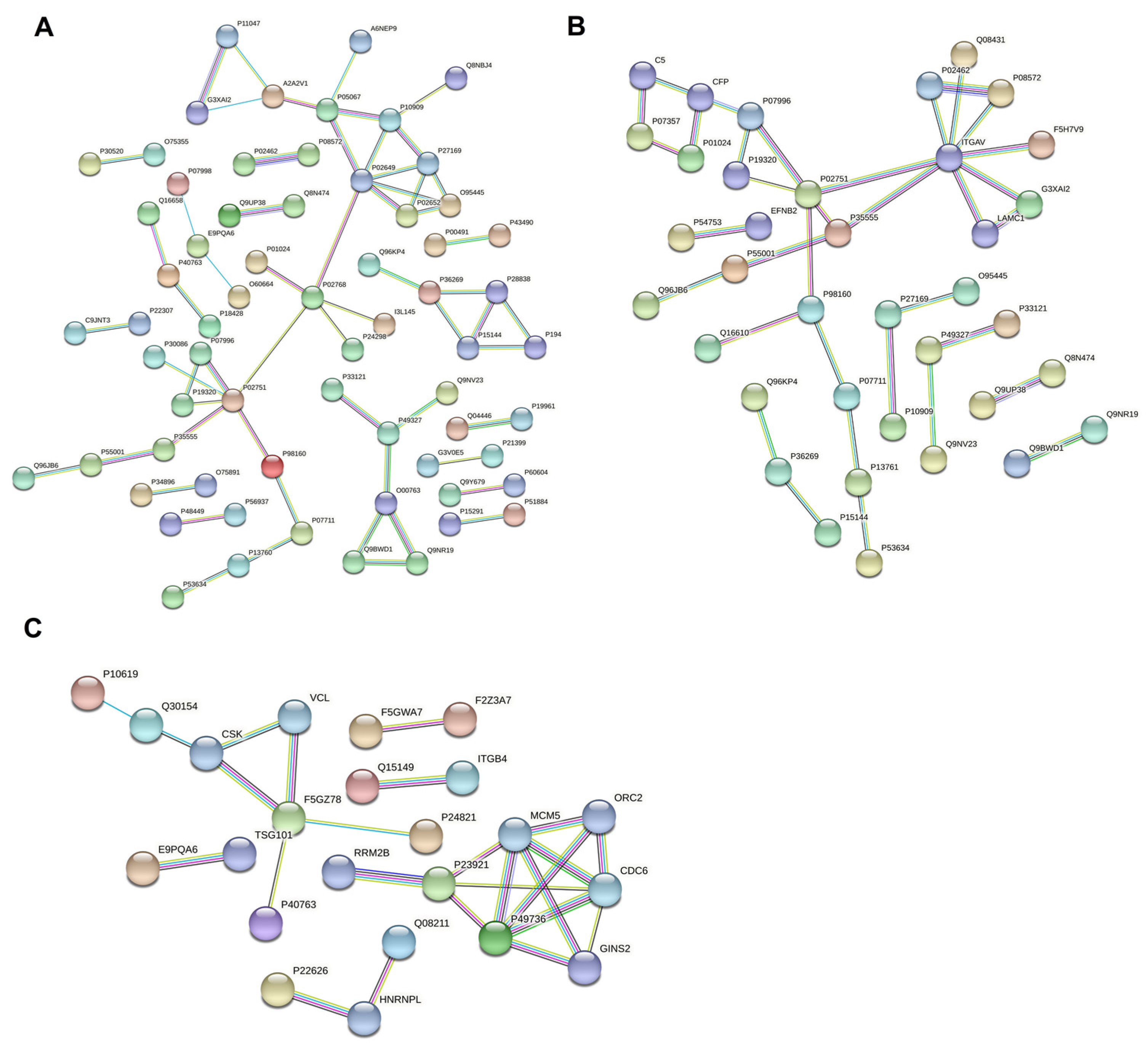
3.6. Protein–Protein Interaction Network Analysis of Differentially Expressed Proteins in Breast Milk
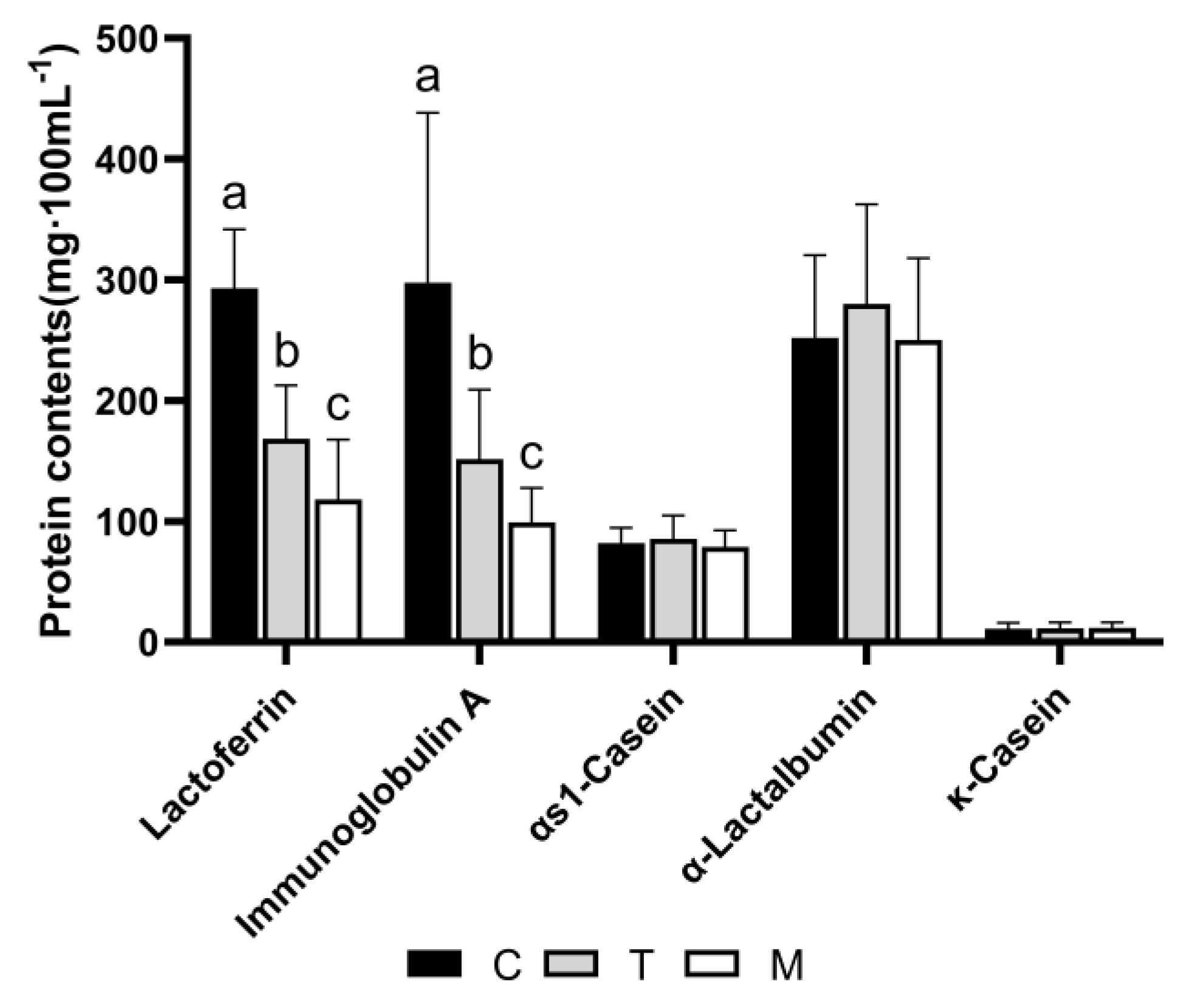
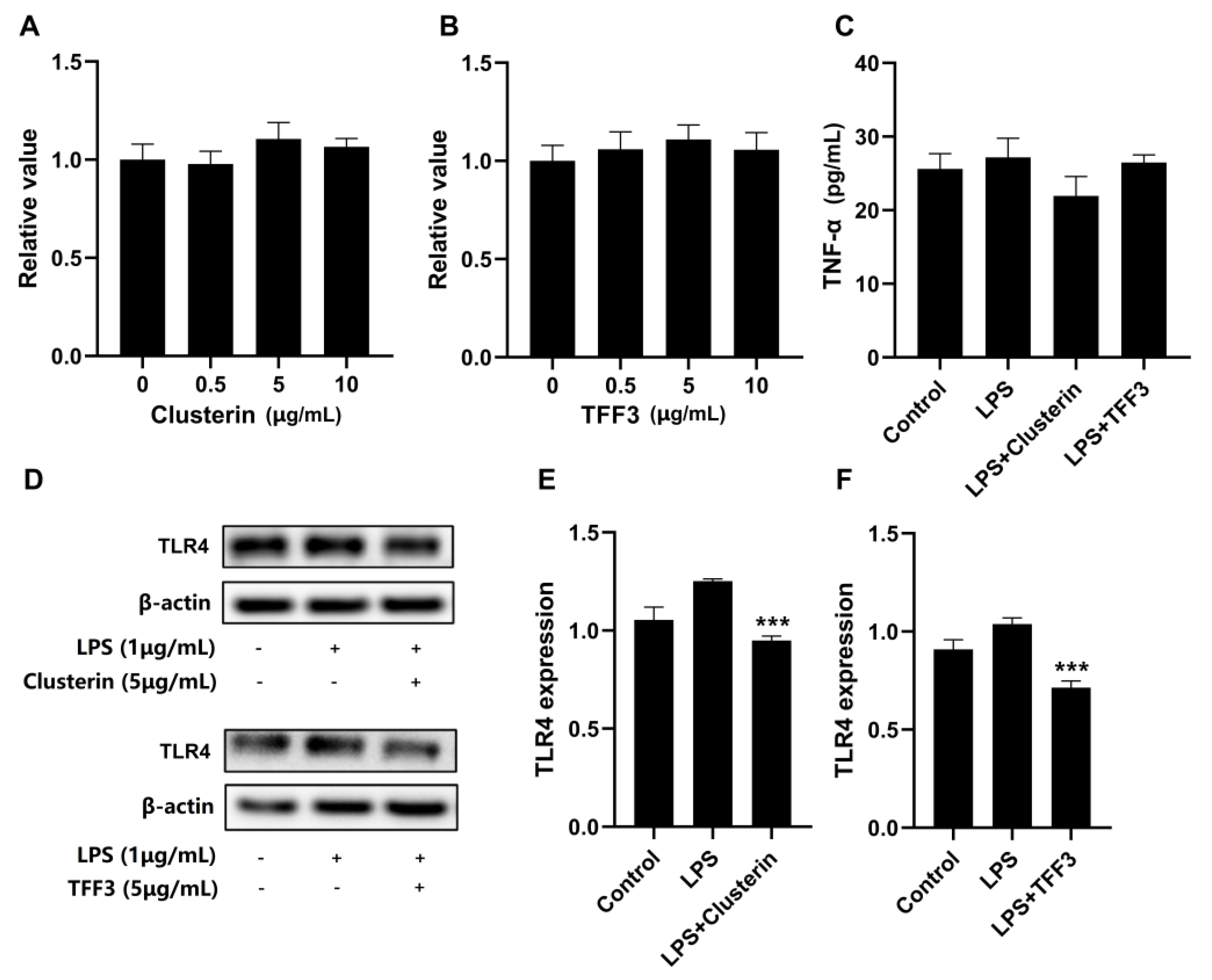
3.7. Validation of Proteomic Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martin, C.R.; Ling, P.R.; Blackburn, G.L. Review of Infant Feeding: Key Features of Breast Milk and Infant Formula. Nutrients 2016, 8, 279. [Google Scholar] [CrossRef]
- Eidelman, A.I. Breastfeeding and the Use of Human Milk: An Analysis of the American Academy of Pediatrics 2012 Breastfeeding Policy Statement. Breastfeed. Med. 2012, 7, 323–324. [Google Scholar] [CrossRef]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.P.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T.; et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018, 9, 4169. [Google Scholar] [CrossRef]
- Jackson, K.M.; Nazar, A.M. Breastfeeding, the immune response, and long-term health. J. Am. Osteopath. Assoc. 2006, 106, 203–207. [Google Scholar]
- Isaacs, E.B.; Fischl, B.R.; Quinn, B.T.; Chong, W.K.; Gadian, D.G.; Lucas, A. Impact of breast milk on intelligence quotient, brain size, and white matter development. Pediatr. Res. 2010, 67, 357–362. [Google Scholar] [CrossRef]
- Savino, F.; Benetti, S.; Liguori, S.A.; Sorrenti, M.; Cordero di Montezemolo, L. Advances on human milk hormones and protection against obesity. Cell. Mol. Biol. 2013, 59, 89–98. [Google Scholar]
- World Health Organization. Guideline: Delayed Umbilical Cord Clamping for Improved Maternal and Infant Health and Nutrition Outcomes; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156 (Suppl. S2), S8–S15. [Google Scholar] [CrossRef]
- Kato, I.; Horike, K.; Kawada, K.; Htun, Y.; Nishida, T.; Nakamura, S.; Koyano, K.; Konishi, Y.; Kusaka, T. The Trajectory of Expressed Colostrum Volume in the First 48 Hours Postpartum: An Observational Study. Breastfeed. Med. 2022, 17, 52–58. [Google Scholar] [CrossRef]
- Lonnerdal, B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173, S4–S9. [Google Scholar] [CrossRef]
- Arnold, R.R.; Russell, J.E.; Champion, W.J.; Gauthier, J.J. Bactericidal Activity of Human Lactoferrin—Influence of Physical Conditions and Metabolic State of the Target Microorganism. Infect. Immun. 1981, 32, 655–660. [Google Scholar] [CrossRef]
- Lawrence, R.A.; Lawrence, R.A. Breastfeeding, a Guide for the Medical Profession, 2nd ed.; Mosby: St. Louis, MO, USA, 1985; Volume XVI, 601p. [Google Scholar]
- Akhter, H.; Aziz, F.; Ullah, F.R.; Ahsan, M.; Islam, S.N. Immunoglobulins content in colostrum, transitional and mature milk of Bangladeshi mothers: Influence of parity and sociodemographic characteristics. J. Mother Child 2021, 24, 8–15. [Google Scholar] [CrossRef]
- Manoni, M.; di Lorenzo, C.; Ottoboni, M.; Tretola, M.; Pinotti, L. Comparative Proteomics of Milk Fat Globule Membrane (MFGM) Proteome across Species and Lactation Stages and the Potentials of MFGM Fractions in Infant Formula Preparation. Foods 2020, 9, 1251. [Google Scholar] [CrossRef]
- Yang, M.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Yue, X.; Wu, J. Comparative proteomic exploration of whey proteins in human and bovine colostrum and mature milk using iTRAQ-coupled LC-MS/MS. Int. J. Food Sci. Nutr. 2017, 68, 671–681. [Google Scholar] [CrossRef]
- Elwakiel, M.; Boeren, S.; Hageman, J.A.; Szeto, I.M.; Schols, H.A.; Hettinga, K.A. Variability of Serum Proteins in Chinese and Dutch Human Milk during Lactation. Nutrients 2019, 11, 499. [Google Scholar] [CrossRef]
- Jin, D.P.; Liu, H.; Bu, L.L.; Ke, Q.H.; Li, Z.Y.; Han, W.N.; Zhu, S.Y.; Liu, C.H. Comparative Analysis of Whey Proteins in Human Milk Using a Data-Independent Acquisition Proteomics Approach during the Lactation Period. J. Agric. Food Chem. 2021, 69, 4319–4330. [Google Scholar]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Bobe, G.; Beitz, D.C.; Freeman, A.E.; Lindberg, G.L. Separation and Quantification of Bovine Milk Proteins by Reversed-Phase High-Performance Liquid Chromatography. J. Agric. Food Chem. 1998, 46, 458–463. [Google Scholar] [CrossRef]
- Gura, T. Nature’s first functional food. Science 2014, 345, 747–749. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- El-Hatmi, H.; Jrad, Z.; Salhi, I.; Aguibi, A.; Nadri, A.; Khorchani, T. Comparison of composition and whey protein fractions of human, camel, donkey, goat and cow milk. Mljekarstvo 2015, 65, 159–167. [Google Scholar] [CrossRef]
- Lopez Alvarez, M.J. Proteins in human milk. Breastfeed. Rev. 2007, 15, 5–16. [Google Scholar]
- Yen, C.C.; Shen, C.J.; Hsu, W.H.; Chang, Y.H.; Lin, H.T.; Chen, H.L.; Chen, C.M. Lactoferrin: An iron-binding antimicrobial protein against Escherichia coli infection. Biometals 2011, 24, 585–594. [Google Scholar] [CrossRef]
- Manzoni, P.; Stolfi, I.; Messner, H.; Cattani, S.; Laforgia, N.; Romeo, M.G.; Bollani, L.; Rinaldi, M.; Gallo, E.; Quercia, M.; et al. Bovine Lactoferrin Prevents Invasive Fungal Infections in Very Low Birth Weight Infants: A Randomized Controlled Trial. Pediatrics 2012, 129, 116–123. [Google Scholar] [CrossRef]
- Montagne, P.; Cuilliere, M.L.; Mole, C.; Bene, M.C.; Faure, G. Changes in lactoferrin and lysozyme levels in human milk during the first twelve weeks of lactation. Bioact. Compon. Hum. Milk 2001, 501, 241–247. [Google Scholar]
- Dawarkadas, A.M.; Saha, K.; Mathur, N.B. A Comparative-Study of Cells and Antimicrobial Proteins in Colostrum of Mothers Delivering Preterm and Full-Term Babies. J. Trop. Pediatr. 1991, 37, 214–219. [Google Scholar]
- Hanson, L.A.; Korotkova, M. The role of breastfeeding in prevention of neonatal infection. Semin. Neonatol. 2002, 7, 275–281. [Google Scholar] [CrossRef]
- Takahashi, T.; Yoshida, Y.; Hatano, S.; Sugita-Konishi, Y.; Igimi, S.; Yajima, M.; Kojima, T.; Kanno, T.; Yonekubo, A.; Yajima, T.; et al. Reactivity of secretory IgA antibodies in breast milk from 107 Japanese mothers to 20 environmental antigens. Biol. Neonate 2002, 82, 238–242. [Google Scholar] [CrossRef]
- Rizzi, F.; Caccamo, A.E.; Belloni, L.; Bettuzzi, S. Clusterin Is a Short Half-Life, Poly-Ubiquitinated Protein, Which Controls the Fate of Prostate Cancer Cells. J. Cell. Physiol. 2009, 219, 314–323. [Google Scholar] [CrossRef]
- Santilli, G.; Aronow, B.J.; Sala, A. Essential requirement of apolipoprotein J (clusterin) signaling for I kappa B expression and regulation of NF-kappa B activity. J. Biol. Chem. 2003, 278, 38214–38219. [Google Scholar] [CrossRef]
- Ren, Q.; Zhou, Y.; Zhang, W.; Tian, Y.; Sun, H.; Zhao, X.; Xu, Y.; Jiang, S. Longitudinal changes in the bioactive proteins in human milk of the Chinese population: A systematic review. Food Sci. Nutr. 2021, 9, 25–35. [Google Scholar] [CrossRef]
- Levy, O. Innate immunity of the newborn: Basic mechanisms and clinical correlates. Nat. Rev. Immunol. 2007, 7, 379–390. [Google Scholar] [CrossRef]
- Riskin, A.; Almog, M.; Peri, R.; Halasz, K.; Srugo, I.; Kessel, A. Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 2012, 71, 220–225. [Google Scholar] [CrossRef]
- Martin, E.; Palmic, N.; Sanquer, S.; Lenoir, C.; Hauck, F.; Mongellaz, C.; Fabrega, S.; Nitschke, P.; Degli Esposti, M.; Schwartzentruber, J.; et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 2014, 510, 288–292. [Google Scholar] [CrossRef]
- Mayer, S.; Raulf, M.K.; Lepenies, B. C-type lectins: Their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 2017, 147, 223–237. [Google Scholar]
- Cai, Y.; Winn, M.E.; Zehmer, J.K.; Gillette, W.K.; Lubkowski, J.T.; Pilon, A.L.; Kimura, S. Preclinical evaluation of human secretoglobin 3A2 in mouse models of lung development and fibrosis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2014, 306, L10–L22. [Google Scholar]
- Kimura, S.; Yokoyama, S.; Pilon, A.L.; Kurotani, R. Emerging role of an immunomodulatory protein secretoglobin 3A2 in human diseases. Pharmacol. Ther. 2022, 236, 108112. [Google Scholar]
- Brunner, M.; Millon-Fremillon, A.; Chevalier, G.; Nakchbandi, I.A.; Mosher, D.; Block, M.R.; Albiges-Rizo, C.; Bouvard, D. Osteoblast mineralization requires beta1 integrin/ICAP-1-dependent fibronectin deposition. J. Cell Biol. 2011, 194, 307–322. [Google Scholar] [CrossRef]
- Oertel, M.; Graness, A.; Thim, L.; Buhling, F.; Kalbacher, H.; Hoffmann, W. Trefoil factor family-peptides promote migration of human bronchial epithelial cells—Synergistic effect with epidermal growth factor. Am. J. Respir. Cell Mol. Biol. 2001, 25, 418–424. [Google Scholar]
- Mashimo, H.; Wu, D.C.; Podolsky, D.K.; Fishman, M.C. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 1996, 274, 262–265. [Google Scholar]
- Cook, G.A.; Familari, M.; Thim, L.; Giraud, A.S. The trefoil peptides TFF2 and TFF3 are expressed in rat lymphoid tissues and participate in the immune response. FEBS Lett. 1999, 456, 155–159. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, X.; Mi, L.; Li, C.; Zhang, Y.; Bi, R.; Pang, J.; Li, Y. Comparative Proteomic Analysis of Proteins in Breast Milk during Different Lactation Periods. Nutrients 2022, 14, 3648. https://doi.org/10.3390/nu14173648
Zhang Y, Zhang X, Mi L, Li C, Zhang Y, Bi R, Pang J, Li Y. Comparative Proteomic Analysis of Proteins in Breast Milk during Different Lactation Periods. Nutrients. 2022; 14(17):3648. https://doi.org/10.3390/nu14173648
Chicago/Turabian StyleZhang, Yifan, Xiaoxu Zhang, Lijuan Mi, Chuangang Li, Yiran Zhang, Ran Bi, Jinzhu Pang, and Yixuan Li. 2022. "Comparative Proteomic Analysis of Proteins in Breast Milk during Different Lactation Periods" Nutrients 14, no. 17: 3648. https://doi.org/10.3390/nu14173648
APA StyleZhang, Y., Zhang, X., Mi, L., Li, C., Zhang, Y., Bi, R., Pang, J., & Li, Y. (2022). Comparative Proteomic Analysis of Proteins in Breast Milk during Different Lactation Periods. Nutrients, 14(17), 3648. https://doi.org/10.3390/nu14173648





