Impact of Clarified Apple Juices with Different Processing Methods on Gut Microbiota and Metabolomics of Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. CAJ
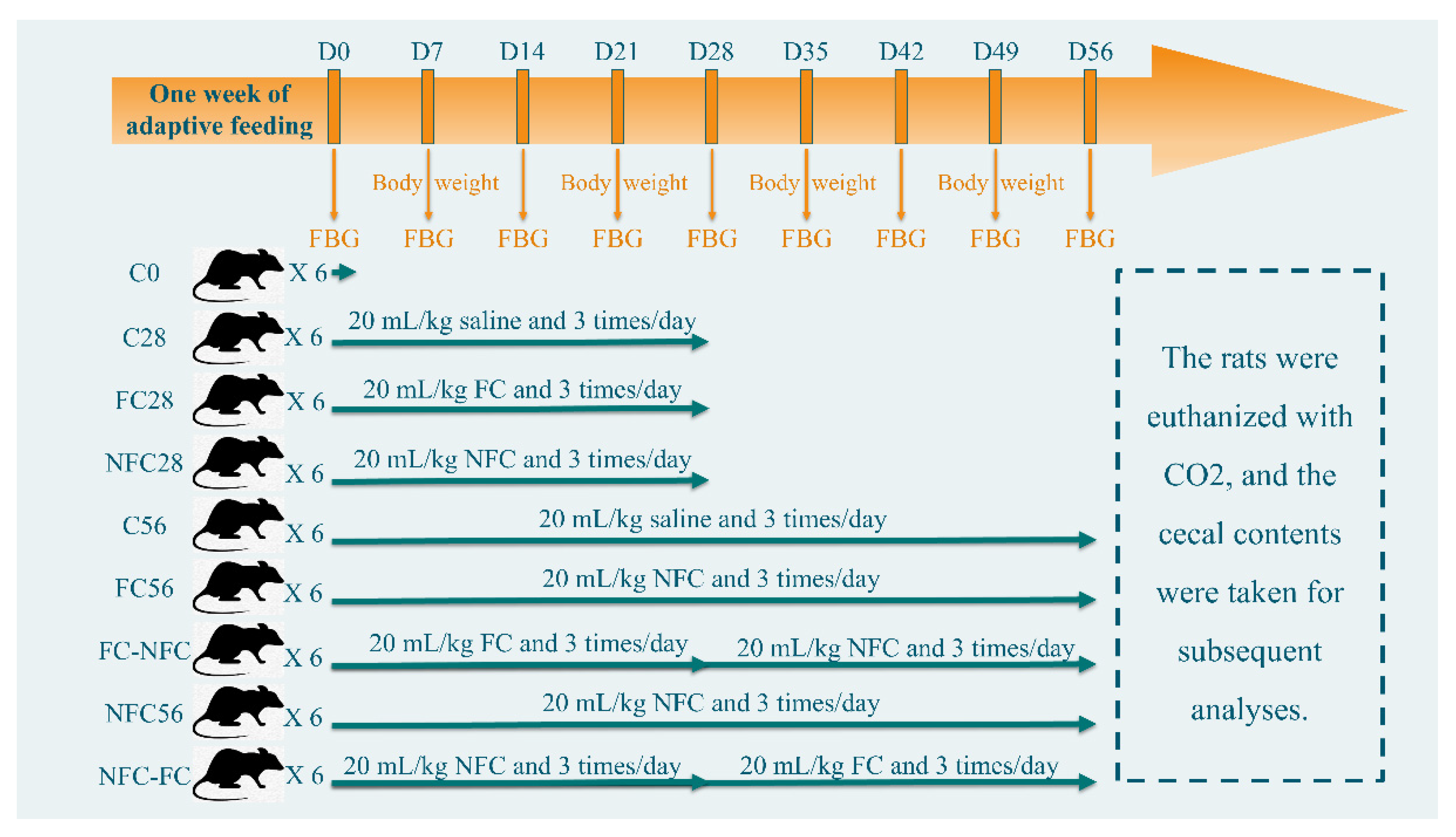
2.2. Animal Intervention
2.3. 16 S rRNA Sequencing
2.4. Untargeted Metabolomics
2.5. CAJ Analyses
2.6. Data Processing, Statistical Analysis, and Visualization
3. Results
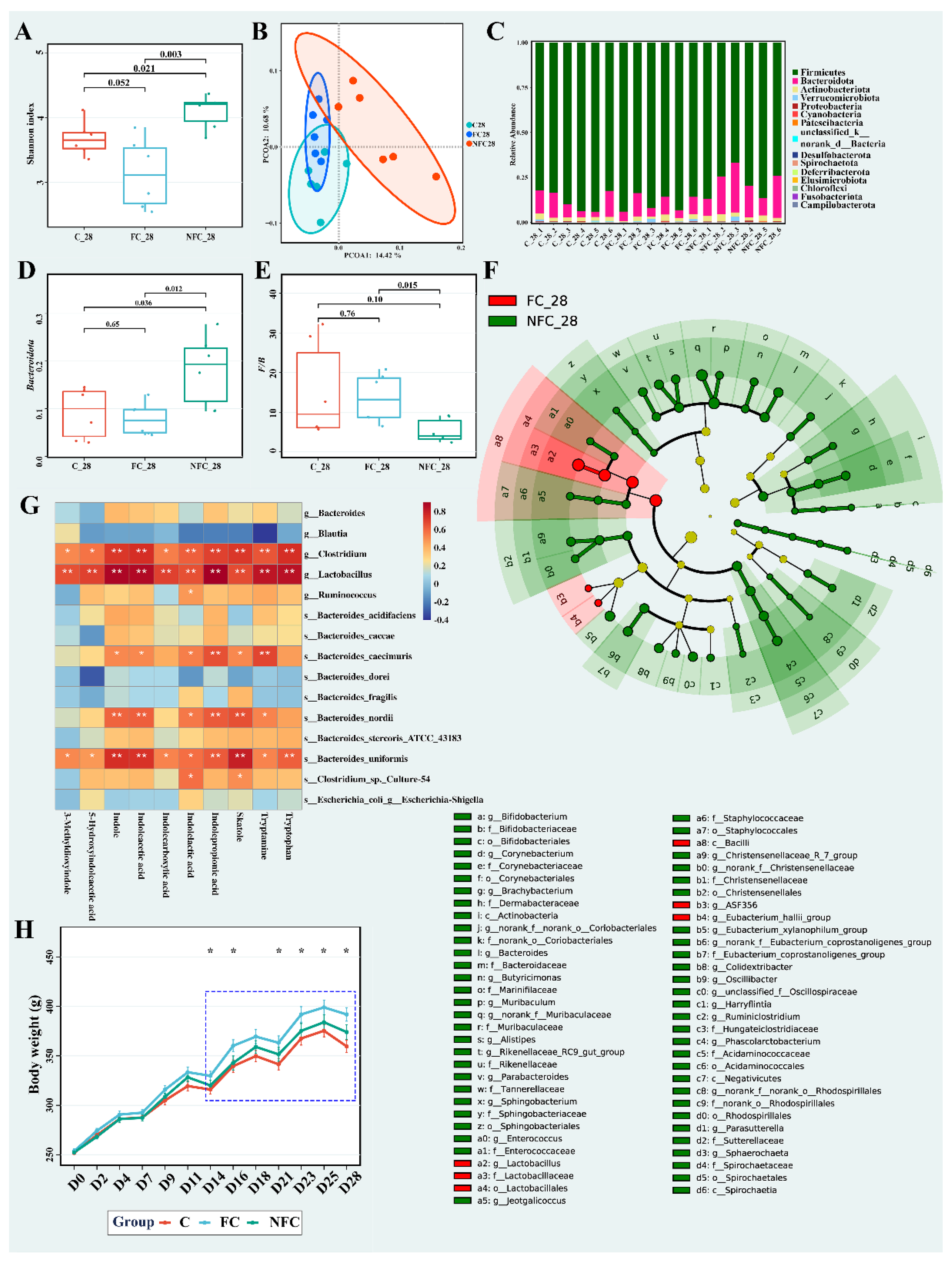
3.1. Continuous Intake of CAJ Has Limited Effect on Body Weight and No Effect on the Intake Amount, Gut Microbiota, and Blood Lipids
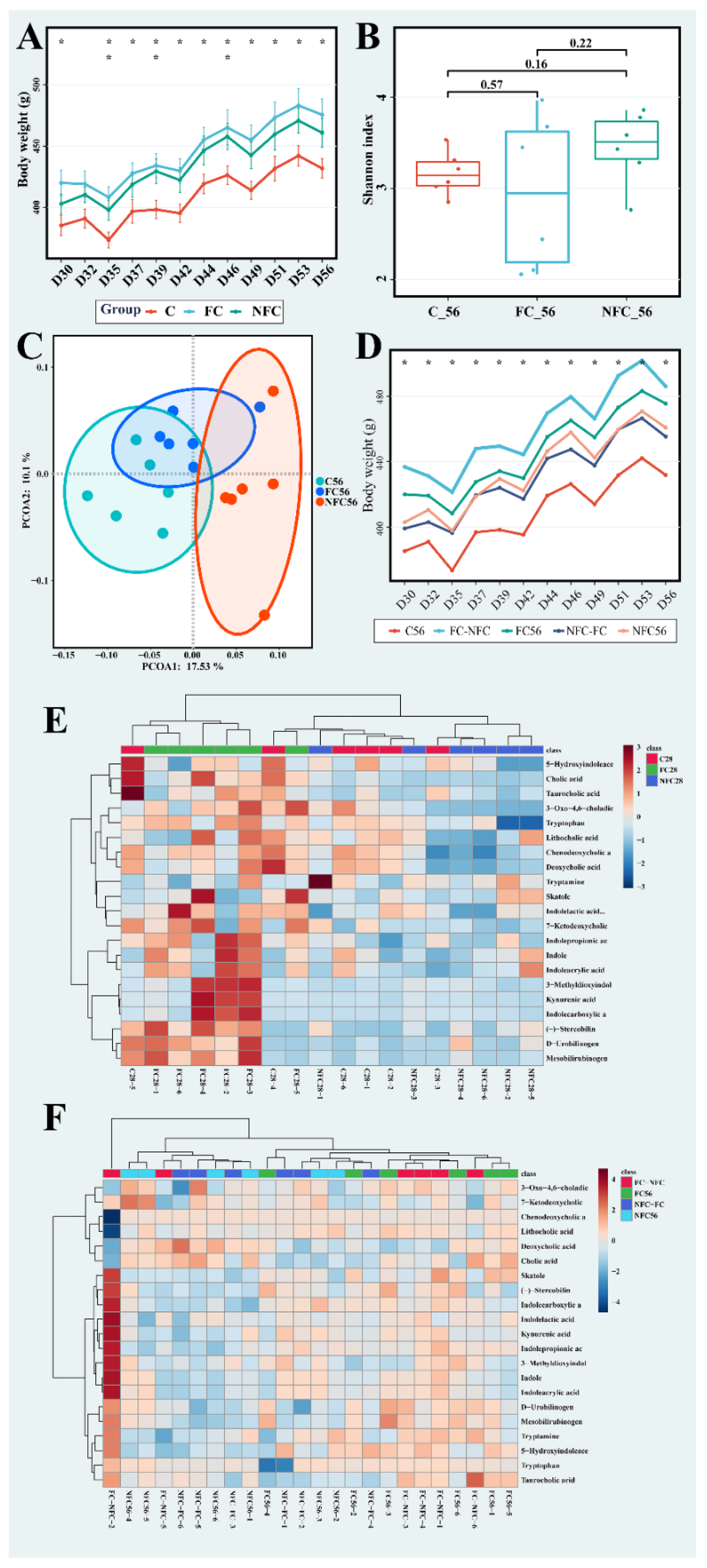
3.2. CAJ with Lower Processing Degree Could Improve Microbiota Diversity and Inhibit the Metabolism of Bile Acids, Bilirubin, and Tryptophan in the Gut
3.3. Latter Intervention of NFC Did Not Show the Same Effect
3.4. The Beneficial Effect of NFC Could Come from Polyphenol Compounds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monteiro, C.A.; Levy, R.B.; Claro, R.M.; Castro, I.R.R.D.; Cannon, G. A new classification of foods based on the extent and purpose of their processing. Cad. Saúde Pública 2010, 26, 2039–2049. [Google Scholar] [CrossRef]
- Monteiro, C.A.; Cannon, G.; Moubarac, J.-C.; Levy, R.B.; Louzada, M.L.C.; Jaime, P.C. The UN Decade of Nutrition, the NOVA food classification and the trouble with ultra-processing. Public Health Nutr. 2018, 21, 5–17. [Google Scholar] [CrossRef]
- Lawrence, M.A.; Baker, P.I. Ultra-processed food and adverse health outcomes. BMJ-Br. Med. J. 2019, 365, l2289. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, T.; El-Nezami, H.; Savidge, T.C. Food ingredients in human health: Ecological and metabolic perspectives implicating gut microbiota function. Trends Food Sci. Technol. 2020, 100, 103–117. [Google Scholar] [CrossRef]
- Hur, J.; Otegbeye, E.; Joh, H.-K.; Nimptsch, K.; Ng, K.; Ogino, S.; Meyerhardt, J.A.; Chan, A.T.; Willett, W.C.; Wu, K.; et al. Sugar-sweetened beverage intake in adulthood and adolescence and risk of early-onset colorectal cancer among women. Gut 2021, 70, 2330–2336. [Google Scholar] [CrossRef]
- Fardet, A. Minimally processed foods are more satiating and less hyperglycemic than ultra-processed foods: A preliminary study with 98 ready-to-eat foods. Food Funct. 2016, 7, 2338–2346. [Google Scholar] [CrossRef]
- Marty, L.; de Lauzon-Guillain, B.; Labesse, M.; Nicklaus, S. Food choice motives and the nutritional quality of diet during the COVID-19 lockdown in France. Appetite 2021, 157, 105005. [Google Scholar] [CrossRef]
- Bracale, R.; Vaccaro, C.M. Changes in food choice following restrictive measures due to COVID-19. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 1423–1426. [Google Scholar] [CrossRef]
- Licht, T.R.; Hansen, M.; Bergstrom, A.; Poulsen, M.; Krath, B.N.; Markowski, J.; Dragsted, L.O.; Wilcks, A. Effects of apples and specific apple components on the cecal environment of conventional rats: Role of apple pectin. BMC Microbiol. 2010, 10, 13. [Google Scholar] [CrossRef]
- Tomas-Barberan, F.A.; Espin, J.C. Effect of Food Structure and Processing on (Poly)phenol-Gut Microbiota Interactions and the Effects on Human Health. Annu. Rev. Food Sci. Technol. 2019, 10, 221–238. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.J.; Zhang, W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef]
- Miclotte, L.; Van de Wiele, T. Food processing, gut microbiota and the globesity problem. Crit. Rev. Food Sci. Nutr. 2020, 60, 1769–1782. [Google Scholar] [CrossRef]
- Grigor’eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2020, 11, 13. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Flegal, K.M.; Troiano, R.P. Changes in the distribution of body mass index of adults and children in the US population. Int. J. Obes. 2000, 24, 807–818. [Google Scholar] [CrossRef]
- de Souza, A.Z.; Zambom, A.Z.; Abboud, K.Y.; Reis, S.K.; Tannihao, F.; Guadagnini, D.; Saad, M.J.; Prada, P.O. Oral supplementation with L-glutamine alters gut microbiota of obese and overweight adults: A pilot study. Nutrition 2015, 31, 884–889. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Y.; Tan, S.; Wang, J. Effects of Banana Resistant Starch on the Biochemical Indexes and Intestinal Flora of Obese Rats Induced by a High-Fat Diet and Their Correlation Analysis. Front. Bioeng. Biotechnol. 2021, 9, 575724. [Google Scholar] [CrossRef]
- Lobb, B.; Tremblay, B.J.; Moreno-Hagelsieb, G.; Doxey, A.C. An assessment of genome annotation coverage across the bacterial tree of life. Microb. Genom. 2020, 6, e000341. [Google Scholar] [CrossRef]
- Zhou, S.; Luo, R.; Gong, G.; Wang, Y.; Gesang, Z.; Wang, K.; Xu, Z.; Suolang, S. Characterization of Metagenome-Assembled Genomes and Carbohydrate-Degrading Genes in the Gut Microbiota of Tibetan Pig. Front. Microbiol. 2020, 11, 595066. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Q.; Ma, W.; Tian, F.; Shen, H.; Zhou, M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017, 8, 4644–4656. [Google Scholar] [CrossRef]
- Hamoud, A.R.; Weaver, L.; Stec, D.E.; Hinds, T.D., Jr. Bilirubin in the Liver-Gut Signaling Axis. Trends Endocrinol. Metab. 2018, 29, 140–150. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Gao, K.; Mu, C.L.; Farzi, A.; Zhu, W.Y. Tryptophan Metabolism: A Link between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D. Thermal processing of food reduces gut microbiota diversity of the host and triggers adaptation of the microbiota: Evidence from two vertebrates. Microbiome 2018, 6, 99. [Google Scholar] [CrossRef]
- Kawabata, K.; Yoshioka, Y.; Terao, J. Role of Intestinal Microbiota in the Bioavailability and Physiological Functions of Dietary Polyphenols. Molecules 2019, 24, 370. [Google Scholar] [CrossRef]
- Wu, M.; Li, P.; An, Y.; Ren, J.; Yan, D.; Cui, J.; Li, D.; Li, M.; Wang, M.; Zhong, G. Phloretin ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating the gut microbiota. Pharmacol. Res. 2019, 150, 104489. [Google Scholar] [CrossRef]
- Hong, Z.; Piao, M. Effect of Quercetin Monoglycosides on Oxidative Stress and Gut Microbiota Diversity in Mice with Dextran Sodium Sulphate-Induced Colitis. Biomed Res. Int. 2018, 2018, 8343052. [Google Scholar] [CrossRef]
- Luo, J.; Han, L.; Liu, L.; Gao, L.; Xue, B.; Wang, Y.; Ou, S.; Miller, M.; Peng, X. Catechin supplemented in a FOS diet induces weight loss by altering cecal microbiota and gene expression of colonic epithelial cells. Food Funct. 2018, 9, 2962–2969. [Google Scholar] [CrossRef]
- Bhandarkar, N.S.; Brown, L.; Panchal, S.K. Chlorogenic acid attenuates high-carbohydrate, high-fat diet-induced cardiovascular, liver, and metabolic changes in rats. Nutr. Res. 2019, 62, 78–88. [Google Scholar] [CrossRef]
- Wang, Z.; Lam, K.L.; Hu, J.; Ge, S.; Zhou, A.; Zheng, B.; Zeng, S.; Lin, S. Chlorogenic acid alleviates obesity and modulates gut microbiota in high-fat-fed mice. Food Sci. Nutr. 2019, 7, 579–588. [Google Scholar] [CrossRef]
- Williamson, G. Protection against developing type 2 diabetes by coffee consumption: Assessment of the role of chlorogenic acid and metabolites on glycaemic responses. Food Funct. 2020, 11, 4826–4833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Chen, J.; Yi, K.; Peng, L.; Xie, J.; Gou, X.; Peng, T.; Tang, L. Phlorizin ameliorates obesity-associated endotoxemia and insulin resistance in high-fat diet-fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microbes 2020, 12, 1842990. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ge, J.; He, X.; Sheng, Y.; Zheng, S.; Zhang, C.; Xu, W.; Huang, K. Caffeic acid reduces body weight by regulating gut microbiota in diet-induced-obese mice. J. Funct. Foods 2020, 74, 104061. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef]
- Molinaro, A.; Wahlstrom, A.; Marschall, H.U. Role of Bile Acids in Metabolic Control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef]
- Wei, M.; Huang, F.; Zhao, L.; Zhang, Y.; Yang, W.; Wang, S.; Li, M.; Han, X.; Ge, K.; Qu, C.; et al. A dysregulated bile acid-gut microbiota axis contributes to obesity susceptibility. EBioMedicine 2020, 55, 102766. [Google Scholar] [CrossRef]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Bou Sleiman, M.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 2020, 159, 956–968.e8. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.; Li, D.; Cui, Y.; Li, X. Apple polyphenols extract alleviated dextran sulfate sodium-induced ulcerative colitis in C57BL/6 male mice by restoring bile acid metabolism disorder and gut microbiota dysbiosis. Phytother. Res. 2021, 35, 1468–1485. [Google Scholar] [CrossRef]
- Lin, L.; Huang, Z.; Gao, Y.; Yan, X.; Xing, J.; Hang, W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 2011, 10, 1396–1405. [Google Scholar] [CrossRef]
- Aprikian, O.; Duclos, V.; Guyot, S.; Besson, C.; Manach, C.; Bernalier, A.; Morand, C.; Rémésy, C.; Demigné, C. Apple Pectin and a Polyphenol-Rich Apple Concentrate Are More Effective Together than Separately on Cecal Fermentations and Plasma Lipids in Rats. J. Nutr. 2003, 133, 1860–1865. [Google Scholar] [CrossRef] [Green Version]
- Lustig, R.H.; Schmidt, L.A.; Brindis, C.D. The toxic truth about sugar. Nature 2012, 482, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, L.; Wang, Y.; Tang, H. Age-related topographical metabolic signatures for the rat gastrointestinal contents. J. Proteome Res. 2012, 11, 1397–1411. [Google Scholar] [CrossRef]
- Xie, G.; Zhong, W.; Zheng, X.; Li, Q.; Qiu, Y.; Li, H.; Chen, H.; Zhou, Z.; Jia, W. Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J. Proteome Res. 2013, 12, 3297–3306. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Z.; Strashnov, I.; Liao, X. Use of information dependent acquisition mass spectra and sequential window acquisition of all theoretical fragment-ion mass spectra for fruit juices metabolomics and authentication. Metabolomics 2020, 16, 81. [Google Scholar] [CrossRef]
- Zhang, Y.; Bilbao, A.; Bruderer, T.; Luban, J.; Strambio-De-Castillia, C.; Lisacek, F.; Hopfgartner, G.; Varesio, E. The Use of Variable Q1 Isolation Windows Improves Selectivity in LC-SWATH-MS Acquisition. J. Proteome Res. 2015, 14, 4359–4371. [Google Scholar] [CrossRef]
- Sabina, B.; Ivana, B.; Verheij, E.R.; Raymond, R.; Sunil, K.; Macdonald, I.A.; Ben, V.O.; Smilde, A.K. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 2006, 78, 567–574. [Google Scholar]
- Wang, B.; Wang, X.; Bei, J.; Xu, L.; Zhang, X.; Xu, Z. Development and Validation of an Analytical Method for the Quantification of Arabinose, Galactose, Glucose, Sucrose, Fructose, and Maltose in Fruits, Vegetables, and Their Products. Food Anal. Methods 2021, 14, 1227–1238. [Google Scholar] [CrossRef]
- Wei-de, S.; Li-jun, D.; Yao-hua, G.; Chuan-jie, Z.; Xiang-ying, Y.; Ming-qin, S. Simultaneous determination of twenty amino acids in fruit juice by ion chromatography with integrated pulsed amperometric detection. China Food Addit. 2019, 30, 132–138. [Google Scholar]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Wohlgemuth, G.; Mehta, S.S.; Mejia, R.F.; Neumann, S.; Pedrosa, D.; Pluskal, T.; Schymanski, E.L.; Willighagen, E.L.; Wilson, M.; Wishart, D.S.; et al. SPLASH, a hashed identifier for mass spectra. Nat. Biotechnol. 2016, 34, 1099–1101. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef]

| Index | FC | NFC |
|---|---|---|
| Total phenol (GAE mg/100 mL) | 1073.32 ± 72.33 b | 2169.03 ± 116.84 a |
| pH | 3.57 ± 0.02 b | 3.63 ± 0.02 a |
| Sugar content (°Bx) | 10.83 ± 1.41 a | 10.49 ± 0.11 a |
| Glucose (g/100 mL) | 7.81 ± 0.92 a | 6.13 ± 1.05 b |
| Fructose (g/100 mL) | 4.24 ± 0.79 a | 2.85 ± 0.74 b |
| Sucrose (g/100 mL) | 0.16 ± 0.06 a | 0.10 ± 0.02 a |
| Malic acid (mg/100 mL) | 186.16 ± 2.86 a | 187.10 ± 0.31 a |
| Tartaric acid (mg/100 mL) | 21.33 ± 0.08 a | 18.80 ± 0.15 b |
| Asparagine (mg/100 mL) | 4.57 ± 0.39 a | 10.82 ± 6.58 a |
| Alanine (mg/100 mL) | 4.43 ± 0.39 a | 10.73 ± 6.61 a |
| Serine (mg/100 mL) | 0.69 ± 0.09 a | 1.66 ± 1.08 a |
| Arginine (mg/100 mL) | 0.14 ± 0.02 a | 0.31 ± 0.19 a |
| Glutamine (mg/100 mL) | 0.07 ± 0.01 a | 0.15 ± 0.10 a |
| Class or Name | Formula | C (%) | FC (%) | NFC (%) |
|---|---|---|---|---|
| Bile acids | ||||
| Cholic acid | C24H40O5 | 43.32 a | 47.92 a | 8.77 b |
| Deoxycholic acid | C24H40O4 | 48.24 a | 38.52 a | 13.24 b |
| Chenodeoxycholic acid | C24H40O4 | 41.40 a | 39.77 a | 18.83 b |
| Taurocholic acid | C26H45NO7S | 48.19 a | 37.35 a | 14.45 b |
| Lithocholic acid | C24H40O3 | 35.38 a | 35.79 a | 28.83 a |
| 7-Ketodeoxycholic acid | C24H38O5 | 28.33 a,b | 49.52 a | 22.14 b |
| 3-Oxo-4,6-choladienoic acid | C24H34O3 | 34.36 a,b | 43.91 a | 21.73 b |
| Bilirubinoids | ||||
| Mesobilirubinogen | C33H44N4O6 | 22.30 b | 60.23 a | 17.47 b |
| D-Urobilinogen | C33H42N4O6 | 21.14 a | 57.88 a | 20.97 b |
| (−)-Stercobilin | C33H46N4O6 | 20.85 b | 49.25 a | 29.90 b |
| Tryptophan and its metabolites | ||||
| Tryptophan | C11H12N2O2 | 35.46 b | 43.08 a | 21.46 c |
| Indole | C8H7N | 29.30 b | 39.01 a | 31.69 a,b |
| Tryptamine | C10H12N2 | 27.75 a | 29.21 a | 43.04 a |
| Indolecarboxylic acid | C9H7NO2 | 13.77 b | 72.01 a | 14.22 b |
| Indolelactic acid | C10H9NO2 | 34.41 a | 45.44 a | 20.15 b |
| Indolepropionic acid | C11H11NO2 | 29.88 b | 42.13 a | 27.99 b |
| Indoleacrylic acid | C11H9NO2 | 30.13 b | 38.90 a | 30.97 a,b |
| Skatole | C9H9N | 27.00 a | 39.77 a | 33.23 a |
| 3-Methyldioxyindole | C9H9NO2 | 13.77 b | 72.01 a | 14.22 b |
| 5-Hydroxyindoleacetic acid | C10H9NO3 | 49.36 a | 28.60 a,b | 22.05 b |
| Kynurenic acid | C10H7NO3 | 18.87 b | 63.20 a | 17.94 b |
| Name | Fold Change (NFC/FC) | p Value | VIP |
|---|---|---|---|
| Chlorogenic acid | 2.21 | 2.96 × 10−7 | 1.19 |
| 5-Methoxysalicylic acid | 46.46 | 5.28 × 10−17 | 2.16 |
| p-Coumaric acid | 18.84 | 3.82 × 10−12 | 2.19 |
| Caffeic acid | 3.32 | 4.94 × 10−6 | 1.44 |
| Ferulic acid | 31.44 | 5.56 × 10−14 | 1.92 |
| Phloretin | 10.21 | 7.02 × 10−7 | 1.53 |
| (+)-Catechin | 16.76 | 1.71 × 10−16 | 2.24 |
| (−)-Epicatechin | 50.53 | 1.04 × 10−16 | 2.40 |
| (−)-gallocatechin | 2.92 | 7.55 × 10−19 | 1.85 |
| Phlorizin | 69.98 | 2.21 × 10−16 | 2.20 |
| Isoquercitrin | 59.83 | 3.96 × 10−11 | 2.09 |
| Rutin | 27.15 | 4.01 × 10−12 | 2.22 |
| Naringenin | 8.52 | 1.84 × 10−9 | 1.42 |
| Eriodictyol | 30.07 | 3.76 × 10−12 | 1.70 |
| (+−)-Taxifolin | 8.05 | 6.21 × 10−13 | 1.64 |
| Quercetin-3-O-galactoside/Hyperoside | 50.38 | 2.19 × 10−12 | 2.07 |
| Procyanidin B1 | 102.51 | 2.84 × 10−16 | 2.51 |
| p-Coumaraldehyde | 67.65 | 2.98 × 10−19 | 2.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Yang, S.; Wang, K.; Lu, A.; Wang, X.; Xu, Z. Impact of Clarified Apple Juices with Different Processing Methods on Gut Microbiota and Metabolomics of Rats. Nutrients 2022, 14, 3488. https://doi.org/10.3390/nu14173488
Xu L, Yang S, Wang K, Lu A, Wang X, Xu Z. Impact of Clarified Apple Juices with Different Processing Methods on Gut Microbiota and Metabolomics of Rats. Nutrients. 2022; 14(17):3488. https://doi.org/10.3390/nu14173488
Chicago/Turabian StyleXu, Lei, Shini Yang, Kewen Wang, Anjing Lu, Xue Wang, and Zhenzhen Xu. 2022. "Impact of Clarified Apple Juices with Different Processing Methods on Gut Microbiota and Metabolomics of Rats" Nutrients 14, no. 17: 3488. https://doi.org/10.3390/nu14173488
APA StyleXu, L., Yang, S., Wang, K., Lu, A., Wang, X., & Xu, Z. (2022). Impact of Clarified Apple Juices with Different Processing Methods on Gut Microbiota and Metabolomics of Rats. Nutrients, 14(17), 3488. https://doi.org/10.3390/nu14173488







