Dietary Inulin Supplementation Affects Specific Plasmalogen Species in the Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice and Diets
2.2. Lipid Extraction and Determination of Fatty Methyl Ester and Dimetyl Acetal Profiles
2.3. Analysis of Phospholipid Molecular Species
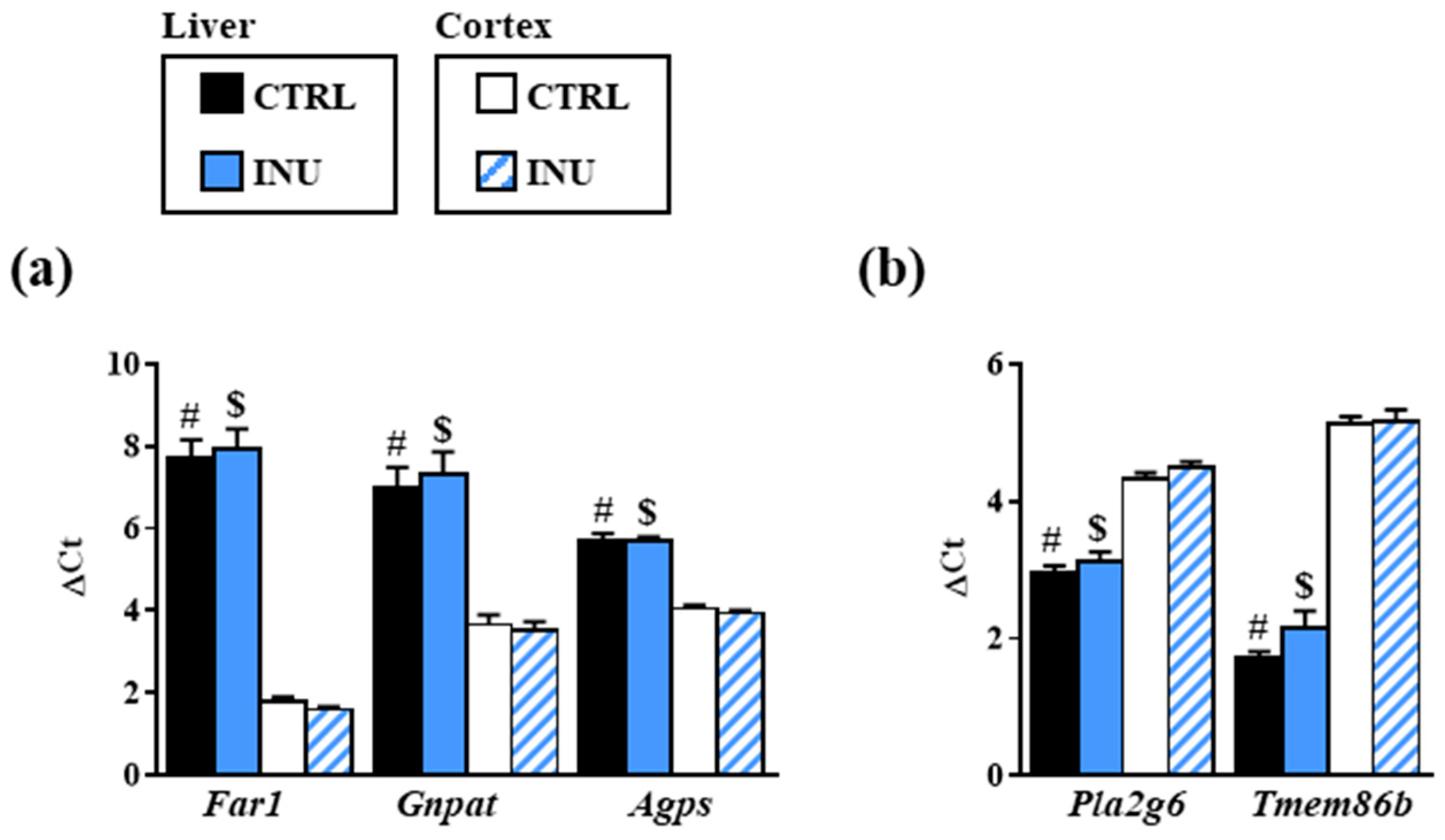
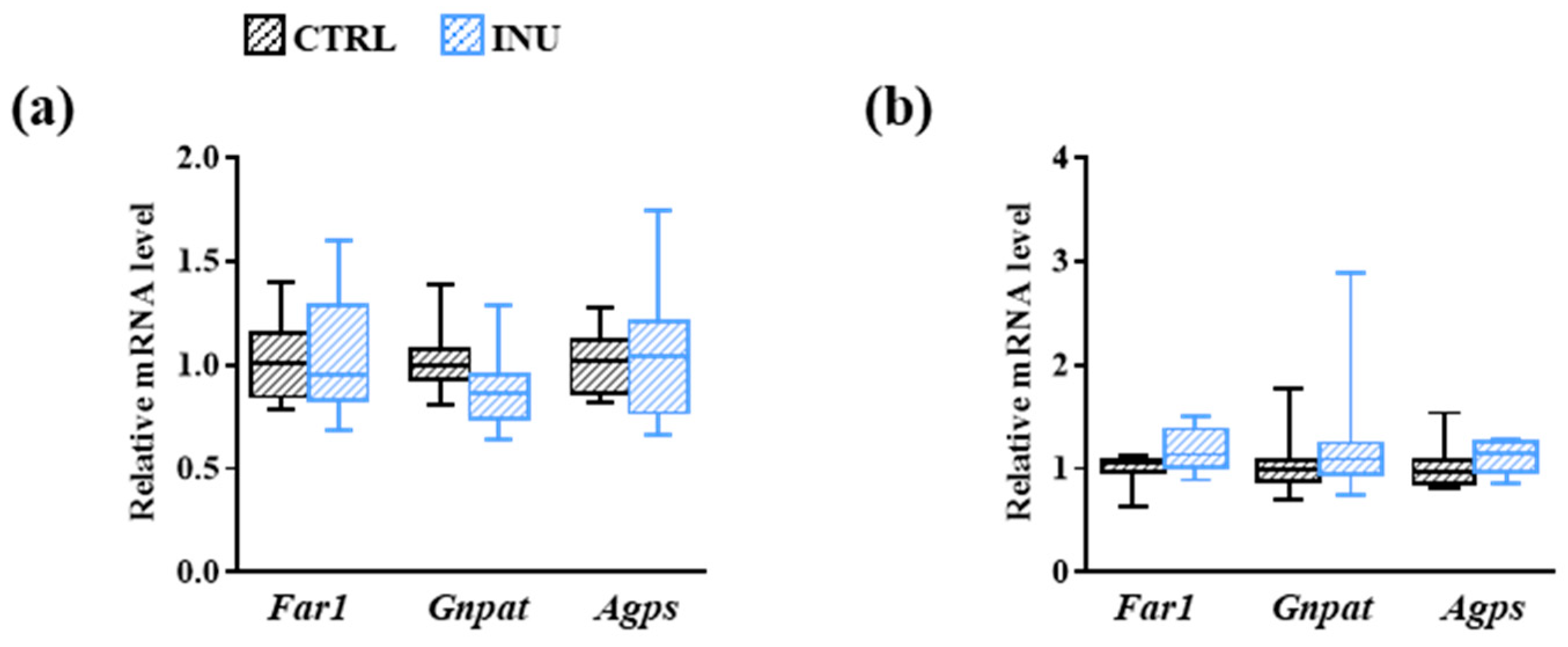
2.4. Gene Expression
2.5. Statistical Analysis
3. Results
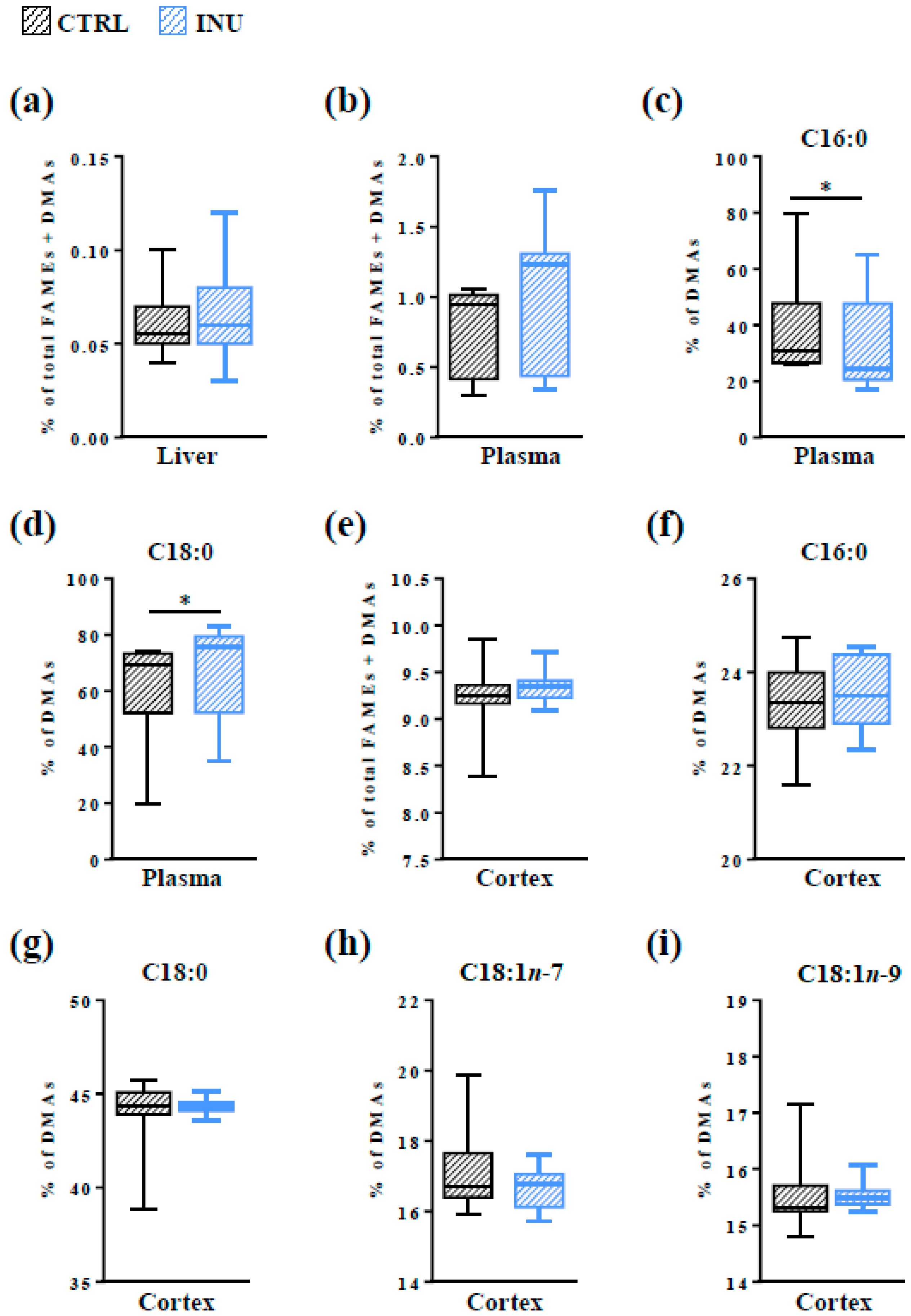
3.1. Effect of Inulin on the Level of Total Pls in the Liver, in the Plasma and in the Cortex
3.2. Impact of Dietary Supplementation with Inulin on the Plasmalogen Content of the Cortex
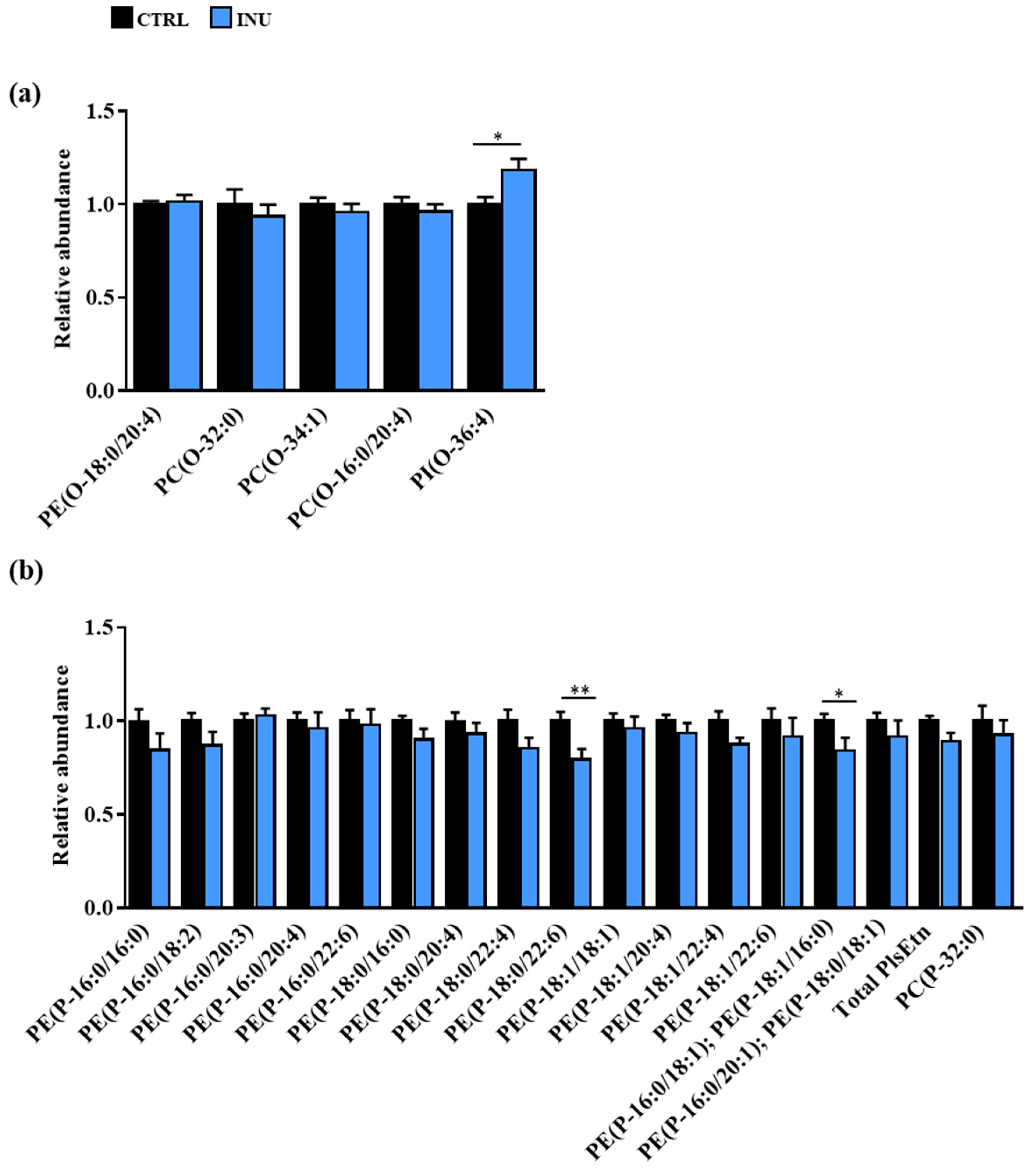
3.2.1. Overview of Plasmalogen Species
3.2.2. Impact of Dietary Inulin Supplementation on the Abundance of Plasmalogen Species in the Liver and the Cortex
3.3. Effect of Inulin on the Expression of Genes Encoding Enzymes Involved in Plasmalogen Biosynthesis
3.4. Modulation of the Fatty Acid Content of the Cortex by the Dietary Intake of Inulin
3.5. Influence of Inulin on the Production of Lyso-Glycerophospholipids in the Cortex
3.6. Influence of Inulin on Oxidative Stress-Related Mechanisms in the Cortex
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Adibhatla, R.M.; Hatcher, J.F. Role of Lipids in Brain Injury and Diseases. Future Lipidol. 2007, 2, 403–422. [Google Scholar] [CrossRef] [Green Version]
- Hussain, G.; Anwar, H.; Rasul, A.; Imran, A.; Qasim, M.; Zafar, S.; Imran, M.; Kamran, S.K.S.; Aziz, N.; Razzaq, A.; et al. Lipids as biomarkers of brain disorders. Crit. Rev. Food Sci. Nutr. 2020, 60, 351–374. [Google Scholar] [CrossRef] [PubMed]
- Sastry, P.S. Lipids of nervous tissue: Composition and metabolism. Prog. Lipid Res. 1985, 24, 69–176. [Google Scholar] [CrossRef]
- Bozek, K.; Wei, Y.; Yan, Z.; Liu, X.; Xiong, J.; Sugimoto, M.; Tomita, M.; Paabo, S.; Sherwood, C.C.; Hof, P.R.; et al. Organization and evolution of brain lipidome revealed by large-scale analysis of human, chimpanzee, macaque, and mouse tissues. Neuron 2015, 85, 695–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzner, D.; Bader, J.M.; Penkert, H.; Bergner, C.G.; Su, M.; Weil, M.T.; Surma, M.A.; Mann, M.; Klose, C.; Simons, M. Cell-Type- and Brain-Region-Resolved Mouse Brain Lipidome. Cell Rep. 2020, 32, 108132. [Google Scholar] [CrossRef]
- Bazinet, R.P.; Laye, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Martinez, M.; Mougan, I. Fatty acid composition of human brain phospholipids during normal development. J. Neurochem. 1998, 71, 2528–2533. [Google Scholar] [CrossRef] [Green Version]
- Braverman, N.E.; Moser, A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta 2012, 1822, 1442–1452. [Google Scholar] [CrossRef] [Green Version]
- Das, A.K.; Holmes, R.D.; Wilson, G.N.; Hajra, A.K. Dietary ether lipid incorporation into tissue plasmalogens of humans and rodents. Lipids 1992, 27, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Nishimukai, M.; Wakisaka, T.; Hara, H. Ingestion of plasmalogen markedly increased plasmalogen levels of blood plasma in rats. Lipids 2003, 38, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Su, X.Q.; Wang, J.; Sinclair, A.J. Plasmalogens and Alzheimer’s disease: A review. Lipids Health Dis. 2019, 18, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honsho, M.; Fujiki, Y. Plasmalogen homeostasis-regulation of plasmalogen biosynthesis and its physiological consequence in mammals. FEBS Lett. 2017, 591, 2720–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiesner, P.; Leidl, K.; Boettcher, A.; Schmitz, G.; Liebisch, G. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J. Lipid Res. 2009, 50, 574–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brites, P.; Ferreira, A.S.; da Silva, T.F.; Sousa, V.F.; Malheiro, A.R.; Duran, M.; Waterham, H.R.; Baes, M.; Wanders, R.J. Alkyl-glycerol rescues plasmalogen levels and pathology of ether-phospholipid deficient mice. PLoS ONE 2011, 6, e28539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, P.L.; Smith, T.; Lane, N.; Khan, M.A.; Ehrmantraut, G.; Goodenowe, D.B. Oral bioavailability of the ether lipid plasmalogen precursor, PPI-1011, in the rabbit: A new therapeutic strategy for Alzheimer’s disease. Lipids Health Dis. 2011, 10, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorninger, F.; Forss-Petter, S.; Berger, J. From peroxisomal disorders to common neurodegenerative diseases-the role of ether phospholipids in the nervous system. FEBS Lett. 2017, 591, 2761–2788. [Google Scholar] [CrossRef] [Green Version]
- Almsherqi, Z.A. Potential Role of Plasmalogens in the Modulation of Biomembrane Morphology. Front. Cell Dev. Biol 2021, 9, 673917. [Google Scholar] [CrossRef]
- Lessig, J.; Fuchs, B. Plasmalogens in biological systems: Their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Curr. Med. Chem. 2009, 16, 2021–2041. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J.; Han, X.; Chung, K.N.; Gross, R.W. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: A quantitative electrospray ionization/mass spectrometric analysis. Biochemistry 2002, 41, 2075–2088. [Google Scholar] [CrossRef] [PubMed]
- Koivuniemi, A. The biophysical properties of plasmalogens originating from their unique molecular architecture. FEBS Lett. 2017, 591, 2700–2713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorninger, F.; Forss-Petter, S.; Wimmer, I.; Berger, J. Plasmalogens, platelet-activating factor and beyond-Ether lipids in signaling and neurodegeneration. Neurobiol. Dis. 2020, 145, 105061. [Google Scholar] [CrossRef] [PubMed]
- Nagan, N.; Zoeller, R.A. Plasmalogens: Biosynthesis and functions. Prog. Lipid Res. 2001, 40, 199–229. [Google Scholar] [CrossRef]
- Fujino, T.; Yamada, T.; Asada, T.; Tsuboi, Y.; Wakana, C.; Mawatari, S.; Kono, S. Efficacy and Blood Plasmalogen Changes by Oral Administration of Plasmalogen in Patients with Mild Alzheimer’s Disease and Mild Cognitive Impairment: A Multicenter, Randomized, Double-blind, Placebo-controlled Trial. EBioMedicine 2017, 17, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Rowan, S.; Taylor, A. The Role of Microbiota in Retinal Disease. Adv. Exp. Med. Biol. 2018, 1074, 429–435. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Oresic, M.; Seppanen-Laakso, T.; Yetukuri, L.; Backhed, F.; Hanninen, V. Gut microbiota affects lens and retinal lipid composition. Exp. Eye Res. 2009, 89, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Albouery, M.; Buteau, B.; Gregoire, S.; Cherbuy, C.; Pais de Barros, J.P.; Martine, L.; Chain, F.; Cabaret, S.; Berdeaux, O.; Bron, A.M.; et al. Age-Related Changes in the Gut Microbiota Modify Brain Lipid Composition. Front. Cell Infect. Microbiol. 2019, 9, 444. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, J.L.; Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E.C.; Walter, J.; Backhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassaing, B.; Miles-Brown, J.; Pellizzon, M.; Ulman, E.; Ricci, M.; Zhang, L.; Patterson, A.D.; Vijay-Kumar, M.; Gewirtz, A.T. Lack of soluble fiber drives diet-induced adiposity in mice. Am. J. Physiol. Gastrointest Liver Physiol. 2015, 309, G528–G541. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albouery, M.; Bretin, A.; Buteau, B.; Gregoire, S.; Martine, L.; Gambert, S.; Bron, A.M.; Acar, N.; Chassaing, B.; Bringer, M.A. Soluble Fiber Inulin Consumption Limits Alterations of the Gut Microbiota and Hepatic Fatty Acid Metabolism Caused by High-Fat Diet. Nutrients 2021, 13, 1037. [Google Scholar] [CrossRef] [PubMed]
- Le Bastard, Q.; Chapelet, G.; Javaudin, F.; Lepelletier, D.; Batard, E.; Montassier, E. The effects of inulin on gut microbial composition: A systematic review of evidence from human studies. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 403–413. [Google Scholar] [CrossRef]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Wang, J.; Sailer, M.; Theis, S.; Verbeke, K.; Raes, J. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 2017, 66, 1968–1974. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qin, S.; Zhai, S.; Gao, Y.; Li, L. Inulin with different degrees of polymerization modulates composition of intestinal microbiota in mice. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Mistry, R.H.; Gu, F.; Schols, H.A.; Verkade, H.J.; Tietge, U.J.F. Effect of the prebiotic fiber inulin on cholesterol metabolism in wildtype mice. Sci. Rep. 2018, 8, 13238. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.J.; Rohde, J.K.; John, C.; Evangelakos, I.; Koop, A.C.; Pertzborn, P.; Todter, K.; Scheja, L.; Heeren, J.; Worthmann, A. Inulin Supplementation Disturbs Hepatic Cholesterol and Bile Acid Metabolism Independent from Housing Temperature. Nutrients 2020, 12, 3200. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.A.; Conceicao, L.L.; Rosa, D.D.; Dias, M.M.; Peluzio Mdo, C. Mechanisms used by inulin-type fructans to improve the lipid profile. Nutr. Hosp. 2014, 31, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Morrison, W.R.; Smith, L.M. Preparation of Fatty Acid Methyl Esters and Dimethylacetals from Lipids with Boron Fluoride--Methanol. J. Lipid Res. 1964, 5, 600–608. [Google Scholar] [CrossRef]
- Bartlett, E.M.; Lewis, D.H. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 1970, 36, 159–167. [Google Scholar] [CrossRef]
- LIPID MAPS® database. Available online: https://www.lipidmaps.org/ (accessed on 22 June 2022).
- Igarashi, M.; Ma, K.; Gao, F.; Kim, H.W.; Rapoport, S.I.; Rao, J.S. Disturbed choline plasmalogen and phospholipid fatty acid concentrations in Alzheimer’s disease prefrontal cortex. J. Alzheimers Dis. 2011, 24, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Farooqui, A.A.; Horrocks, L.A. Lyso-Glycerophospholipids. In Glycerophospholipids in the Brain; Springer: New York, NY, USA, 2007. [Google Scholar] [CrossRef]
- Jenkins, C.M.; Yang, K.; Liu, G.; Moon, S.H.; Dilthey, B.G.; Gross, R.W. Cytochrome c is an oxidative stress-activated plasmalogenase that cleaves plasmenylcholine and plasmenylethanolamine at the sn-1 vinyl ether linkage. J. Biol. Chem. 2018, 293, 8693–8709. [Google Scholar] [CrossRef] [Green Version]
- Weisser, M.; Vieth, M.; Stolte, M.; Riederer, P.; Pfeuffer, R.; Leblhuber, F.; Spiteller, G. Dramatic increase of alpha-hydroxyaldehydes derived from plasmalogens in the aged human brain. Chem. Phys. Lipids 1997, 90, 135–142. [Google Scholar] [CrossRef]
- Carrie, I.; Clement, M.; de Javel, D.; Frances, H.; Bourre, J.M. Specific phospholipid fatty acid composition of brain regions in mice. Effects of n-3 polyunsaturated fatty acid deficiency and phospholipid supplementation. J. Lipid Res. 2000, 41, 465–472. [Google Scholar] [PubMed]
- Chianese, R.; Coccurello, R.; Viggiano, A.; Scafuro, M.; Fiore, M.; Coppola, G.; Operto, F.F.; Fasano, S.; Laye, S.; Pierantoni, R.; et al. Impact of Dietary Fats on Brain Functions. Curr. Neuropharmacol. 2018, 16, 1059–1085. [Google Scholar] [CrossRef]
- Giles, C.; Takechi, R.; Mellett, N.A.; Meikle, P.J.; Dhaliwal, S.; Mamo, J.C. The Effects of Long-Term Saturated Fat Enriched Diets on the Brain Lipidome. PLoS ONE 2016, 11, e0166964. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Park, S.M.; Kim, I.Y.; Sung, H.; Seong, J.K.; Moon, M.H. High-fat diet-induced lipidome perturbations in the cortex, hippocampus, hypothalamus, and olfactory bulb of mice. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 980–990. [Google Scholar] [CrossRef]
- Naoe, S.; Tsugawa, H.; Takahashi, M.; Ikeda, K.; Arita, M. Characterization of Lipid Profiles after Dietary Intake of Polyunsaturated Fatty Acids Using Integrated Untargeted and Targeted Lipidomics. Metabolites 2019, 9, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velagapudi, V.R.; Hezaveh, R.; Reigstad, C.S.; Gopalacharyulu, P.; Yetukuri, L.; Islam, S.; Felin, J.; Perkins, R.; Boren, J.; Oresic, M.; et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010, 51, 1101–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindt, A.; Liebisch, G.; Clavel, T.; Haller, D.; Hormannsperger, G.; Yoon, H.; Kolmeder, D.; Sigruener, A.; Krautbauer, S.; Seeliger, C.; et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat. Commun. 2018, 9, 3760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e455. [Google Scholar] [CrossRef] [PubMed]
- Joffre, C.; Gregoire, S.; De Smedt, V.; Acar, N.; Bretillon, L.; Nadjar, A.; Laye, S. Modulation of brain PUFA content in different experimental models of mice. Prostaglandins Leukot. Essent. Fat. Acids 2016, 114, 1–10. [Google Scholar] [CrossRef]
- Dorninger, F.; Brodde, A.; Braverman, N.E.; Moser, A.B.; Just, W.W.; Forss-Petter, S.; Brugger, B.; Berger, J. Homeostasis of phospholipids-The level of phosphatidylethanolamine tightly adapts to changes in ethanolamine plasmalogens. Biochim. Biophys. Acta 2015, 1851, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Mallick, R.; Basak, S.; Duttaroy, A.K. Docosahexaenoic acid,22:6n-3: Its roles in the structure and function of the brain. Int. J. Dev. Neurosci. 2019, 79, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Avallone, R.; Vitale, G.; Bertolotti, M. Omega-3 Fatty Acids and Neurodegenerative Diseases: New Evidence in Clinical Trials. Int. J. Mol. Sci. 2019, 20, 4256. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Holtzman, D.M.; McKeel, D.W., Jr. Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: Molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001, 77, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Hoving, L.R.; de Vries, M.R.; de Jong, R.C.M.; Katiraei, S.; Pronk, A.; Quax, P.H.A.; van Harmelen, V.; Willems van Dijk, K. The Prebiotic Inulin Aggravates Accelerated Atherosclerosis in Hypercholesterolemic APOE*3-Leiden Mice. Nutrients 2018, 10, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.W.F.; Houben, T.; Katiraei, S.; Dijk, W.; Boutens, L.; van der Bolt, N.; Wang, Z.; Brown, J.M.; Hazen, S.L.; Mandard, S.; et al. Modulation of the gut microbiota impacts nonalcoholic fatty liver disease: A potential role for bile acids. J. Lipid Res. 2017, 58, 1399–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, J.P.; Zou, J.; Kumar, M.V.; Pellizzon, M.; Ulman, E.; Ricci, M.; Gewirtz, A.T.; Chassaing, B. Supplementation of Low- and High-fat Diets with Fermentable Fiber Exacerbates Severity of DSS-induced Acute Colitis. Inflamm. Bowel. Dis. 2017, 23, 1133–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D., Jr.; Zhang, L.; Wang, W.B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694.e622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murga-Garrido, S.M.; Hong, Q.; Cross, T.L.; Hutchison, E.R.; Han, J.; Thomas, S.P.; Vivas, E.I.; Denu, J.; Ceschin, D.G.; Tang, Z.Z.; et al. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome 2021, 9, 117. [Google Scholar] [CrossRef]

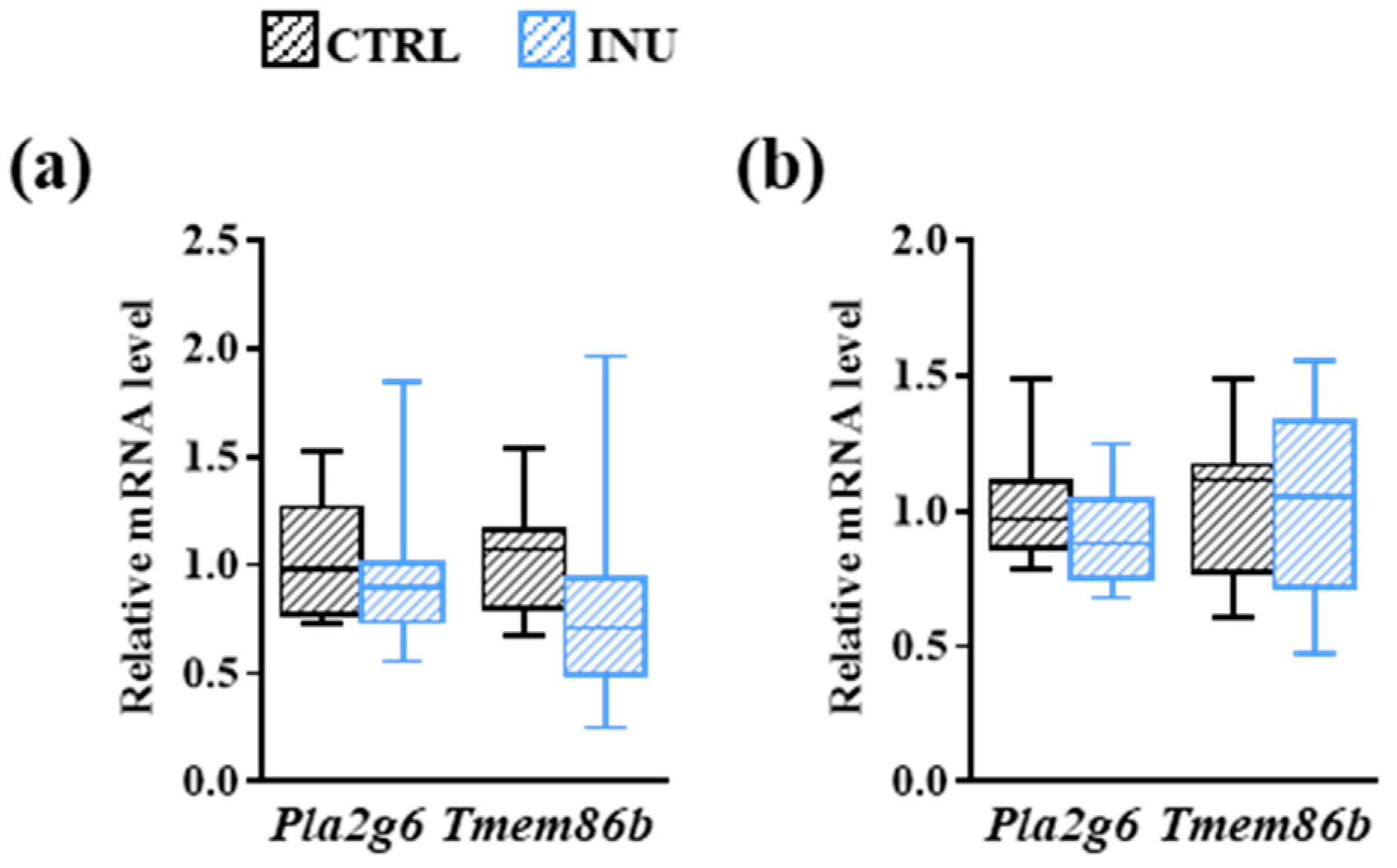
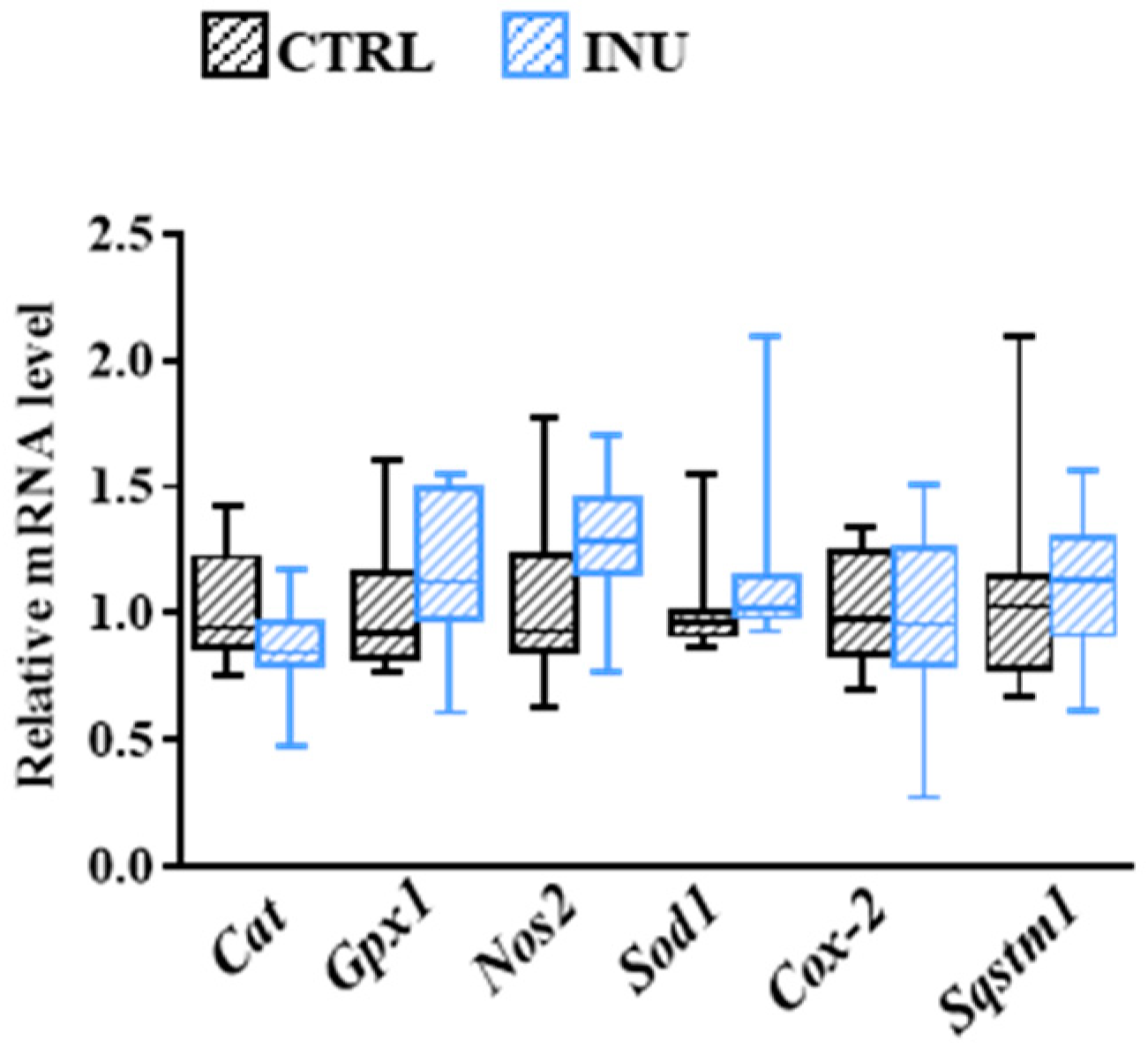
| Genes (ID) | Sense (5′-3′) | Antisense (5′-3′) |
|---|---|---|
| Hprt (15452) | CAGTCCCAGCGTCGTGATTA | TGGCCTCCCATCTCCTTCAT |
| Far1 (67420) | GCTCGGAAGCATCTCAACAAG | GTGCTGGATGCTCGGAAGTAT |
| Gnpat (14712) | TCACCGCAGCTACATTGACT | GCAGCTCACTGACCACTCTC |
| Agps (228061) | GTGCAGGGTGACACAGACTT | CCATGGTGATGTGACAGGCT |
| Pla2g6 (53357) | AAAGTCCCCTCAAGTGCCTG | ACAGTCCACGACCATCTTGC |
| Tmem86b (68255) | TGGGGTGCTGTGCTCTTTAC | CACTAGGCGGGCAAAAGGTA |
| Cat (12359) | CAACAGCTTCAGCGCACCAG | GGCCGGCAATGTTCTCACAC |
| Gpx1 (14775) | GGAATGCCTTGCCAACACCC | GTCGATGGTACGAAAGCGGC |
| Nos2 (18126) | AGAGCCACAGTCCTCTTTGC | ACCACCAGCAGTAGTTGCTC |
| Sod1 (20655) | GATGAAAGCGGTGTGCGTGC | TGGACGTGGAACCCATGCTG |
| Cox-2 (19225) | TTGCATTCTTTGCCCAGCAC | TTAAGTCCACTCCATGGCCC |
| Sqstm1 (18412) | TAAAAGCTGGGCTCTCGGCG | CGTGAACGACGCCATAACCG |
| Glycerophospholipids | Relative Abundance (%) |
|---|---|
| Ethanolamine glycerophospholipids | |
| Alkyl-glycerophospholipids | |
| PE(O-18:0/20:4) | 0.619 ± 0.011 |
| Plasmalogens | |
| PE(P-16:0/16:0) | 0.178 ± 0.011 |
| PE(P-16:0/18:2) | 0.233 ± 0.010 |
| PE(P-16:0/20:3) | 0.365 ± 0.014 |
| PE(P-16:0/20:4) | 1.546 ± 0.071 |
| PE(P-16:0/22:6) | 5.189 ± 0.299 |
| PE(P-18:0/16:0) | 0.715 ± 0.021 |
| PE(P-18:0/20:4) | 5.001 ± 0.230 |
| PE(P-18:0/22:4) | 2.084 ± 0.126 |
| PE(P-18:0/22:6) | 10.857 ± 0.530 |
| PE(P-18:1/18:1) | 4.206 ± 0.172 |
| PE(P-18:1/20:4) | 3.240 ± 0.109 |
| PE(P-18:1/22:4) | 2.250 ± 0.117 |
| PE(P-18:1/22:6) | 2.757 ± 0.187 |
| PE(P-16:0/18:1); PE(P-18:1/16:0) * | 3.855 ± 0.147 |
| PE(P-16:0/20:1); PE(P-18:0/18:1) * | 3.681 ± 0.164 |
| Total PlsEtn | 46.155 ± 1.303 |
| Choline glycerophospholipids | |
| Alkyl-glycerophospholipids | |
| PC(O-32:0) | 0.134 ± 0.011 |
| PC(O-34:1) | 0.361 ± 0.013 |
| PC(O-16:0/20:4) | 0.108 ± 0.004 |
| Plasmalogens | |
| PC(P-32:0) | 0.125 ± 0.010 |
| Inositol glycerophospholipids | |
| Alkyl-glycerophospholipids | |
| PI(O-16:0/20:4) | 0.109 ± 0.004 |
| Fatty Acids | CTRL | INU |
|---|---|---|
| Saturated fatty acids (SFAs) | ||
| C14:0 | 0.143 ± 0.004 | 0.136 ± 0.004 |
| C15:0 *** | 0.044 ± 0.001 | 0.059 ± 0.003 |
| C16:0 | 22.033 ± 0.153 | 22.014 ± 0.167 |
| C17:0 **** | 0.146 ± 0.002 | 0.181 ± 0.006 |
| C18:0 | 21.454 ± 0.081 | 21.471 ± 0.059 |
| C20:0 | 0.273 ± 0.009 | 0.274 ± 0.006 |
| C22:0 | 0.183 ± 0.006 | 0.181 ± 0.009 |
| C24:0 | 0.207 ± 0.008 | 0.219 ± 0.017 |
| Total | 44.482 ± 0.170 | 44.535 ± 0.159 |
| Monounsaturated fatty acids (MUFAs) | ||
| C16:1n-7 ** | 0.735 ± 0.016 | 0.671 ± 0.022 |
| C18:1n-7 | 4.102 ± 0.042 | 4.038 ± 0.033 |
| C20:1n-7 | 0.384 ± 0.013 | 0.364 ± 0.011 |
| C16:1n-9 | 0.173 ± 0.002 | 0.167 ± 0.002 |
| C18:1n-9 | 17.532 ± 0.170 | 17.534 ± 0.119 |
| C20:1n-9 | 1.548 ± 0.057 | 1.574 ± 0.049 |
| C22:1n-9 | 0.152 ± 0.005 | 0.148 ± 0.005 |
| C24:1n-9 | 0.450 ± 0.019 | 0.454 ± 0.030 |
| Total n-7 MUFAs (p = 0.0572) | 5.221 ± 0.066 | 5.073 ± 0.056 |
| Total n-9 MUFAs | 19.854 ± 0.239 | 19.878 ± 0.188 |
| Total MUFAs | 25.076 ± 0.271 | 24.950 ± 0.216 |
| Polyunsaturated fatty acids (PUFAs) | ||
| C20:5n-3 | 0.062 ± 0.002 | 0.061 ± 0.002 |
| C22:5n-3 * | 0.168 ± 0.002 | 0.157 ± 0.003 |
| C22:6n-3 | 15.360 ± 0.172 | 15.328 ± 0.111 |
| C18:2n-6 ** | 0.660 ± 0.022 | 0.570 ± 0.014 |
| C20:2n-6 | 0.094 ± 0.005 | 0.088 ± 0.005 |
| C20:3n-6 **** | 0.463 ± 0.005 | 0.411 ± 0.010 |
| C20:4n-6 | 10.598 ± 0.072 | 10.768 ± 0.074 |
| C22:4n-6 | 2.551 ± 0.024 | 2.590 ± 0.020 |
| C22:5n-6 * | 0.303 ± 0.006 | 0.351 ± 0.023 |
| C20:3n-9 ** | 0.127 ± 0.003 | 0.141 ± 0.004 |
| Total n-3 PUFAs | 15.590 ± 0.171 | 15.546 ± 0.111 |
| Total n-6 PUFAs | 14.668 ± 0.083 | 14.778 ± 0.091 |
| Total PUFAs | 30.386 ± 0.249 | 30.466 ± 0.163 |
| n-6 PUFAs/n-3 PUFAs | 0.942 ± 0.006 | 0.951 ± 0.008 |
| CTRL | INU | |
|---|---|---|
| a LPE 14:0 * | 0.274 ± 0.053 | 0.259 ± 0.131 |
| LPE 16:0 (p = 0.0576) | 5.485 ± 0.553 | 4.438 ± 0.462 |
| LPE 18:0 | 7.184 ± 1.712 | 5.552 ± 1.782 |
| LPE 20:0 | 0.653 ± 0.044 | 0.522 ± 0.068 |
| LPE 22:0 | 0.034 ± 0.004 | 0.032 ± 0.005 |
| LPE 14:1 | 0.016 ± 0.004 | 0.026 ± 0.010 |
| LPE 16:1 | 0.712 ± 0.051 | 0.652 ± 0.043 |
| LPE 18:1 | 21.223 ± 0.722 | 21.005 ± 0.914 |
| LPE 19:1 | 0.113 ± 0.009 | 0.121 ± 0.012 |
| LPE 20:1 | 8.779 ± 0.396 | 8.129 ± 0.732 |
| LPE 22:1 | 0.760 ± 0.046 | 0.695 ± 0.077 |
| LPE 18:2 | 2.624 ± 0.502 | 2.463 ± 0.440 |
| LPE 20:2 | 0.500 ± 0.049 | 0.513 ± 0.037 |
| LPE 22:2 | 0.134 ± 0.011 | 0.127 ± 0.014 |
| LPE 18:3 | 0.116 ± 0.013 | 0.136 ± 0.025 |
| LPE 20:3 | 0.907 ± 0.078 | 0.938 ± 0.052 |
| LPE 20:4 | 13.377 ± 0.520 | 14.498 ± 0.589 |
| LPE 22:4 | 7.068 ± 0.830 | 8.058 ± 0.715 |
| LPE 20:5 | 0.060 ± 0.008 | 0.059 ± 0.007 |
| LPE 22:5 | 0.626 ± 0.045 | 0.750 ± 0.063 |
| LPE 22:6 | 29.357 ± 1.145 | 31.032 ± 1.146 |
| b LPEs/PEs | 0.807 ± 0.273 | 1.096 ± 0.487 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bizeau, J.-B.; Albouery, M.; Grégoire, S.; Buteau, B.; Martine, L.; Crépin, M.; Bron, A.M.; Berdeaux, O.; Acar, N.; Chassaing, B.; et al. Dietary Inulin Supplementation Affects Specific Plasmalogen Species in the Brain. Nutrients 2022, 14, 3097. https://doi.org/10.3390/nu14153097
Bizeau J-B, Albouery M, Grégoire S, Buteau B, Martine L, Crépin M, Bron AM, Berdeaux O, Acar N, Chassaing B, et al. Dietary Inulin Supplementation Affects Specific Plasmalogen Species in the Brain. Nutrients. 2022; 14(15):3097. https://doi.org/10.3390/nu14153097
Chicago/Turabian StyleBizeau, Jean-Baptiste, Mayssa Albouery, Stéphane Grégoire, Bénédicte Buteau, Lucy Martine, Marine Crépin, Alain M. Bron, Olivier Berdeaux, Niyazi Acar, Benoit Chassaing, and et al. 2022. "Dietary Inulin Supplementation Affects Specific Plasmalogen Species in the Brain" Nutrients 14, no. 15: 3097. https://doi.org/10.3390/nu14153097
APA StyleBizeau, J.-B., Albouery, M., Grégoire, S., Buteau, B., Martine, L., Crépin, M., Bron, A. M., Berdeaux, O., Acar, N., Chassaing, B., & Bringer, M.-A. (2022). Dietary Inulin Supplementation Affects Specific Plasmalogen Species in the Brain. Nutrients, 14(15), 3097. https://doi.org/10.3390/nu14153097








