Increased Fat Taste Preference in Progranulin-Deficient Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Progranulin-Deficient and Control Mice
2.2. IntelliCage Description and Setup
2.3. IntelliCage Tasks and Schedules and Analysis of Fat Taste Preference
2.4. Phenomaster Analysis of Fat Taste Preference
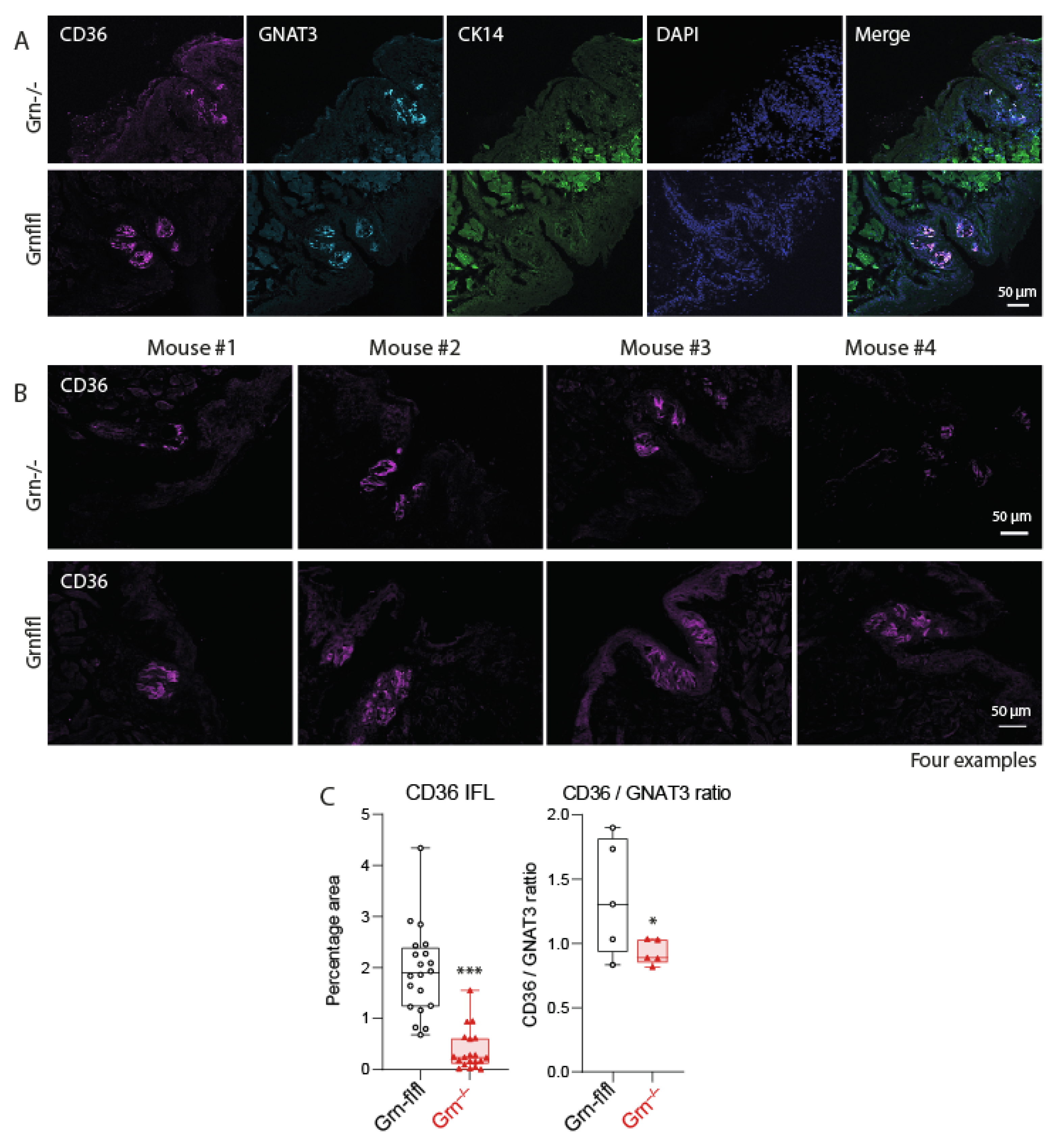
2.5. Immunofluorescence Analysis of CD36 in Taste Buds
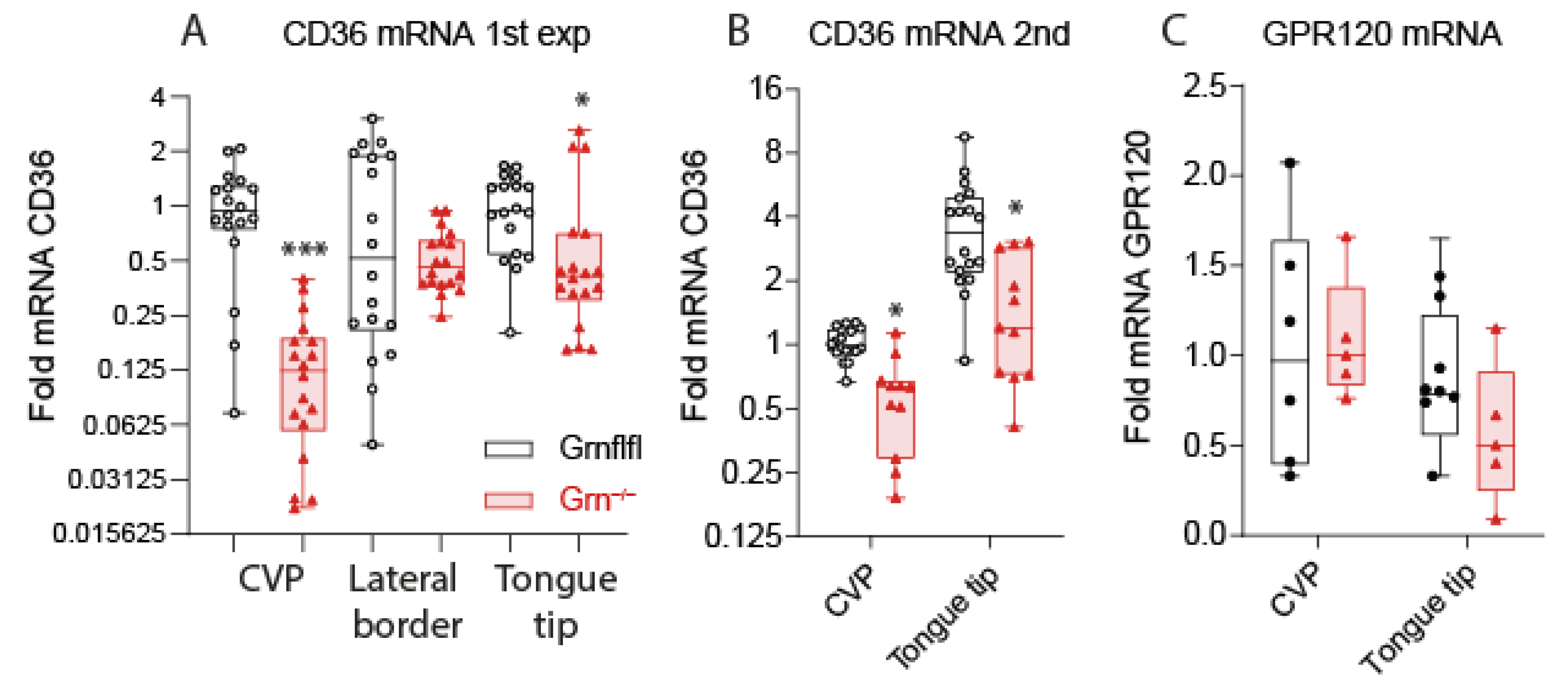
2.6. RNA Extraction and QRT-PCR Analysis of Fat Taste Receptors in the Tongue
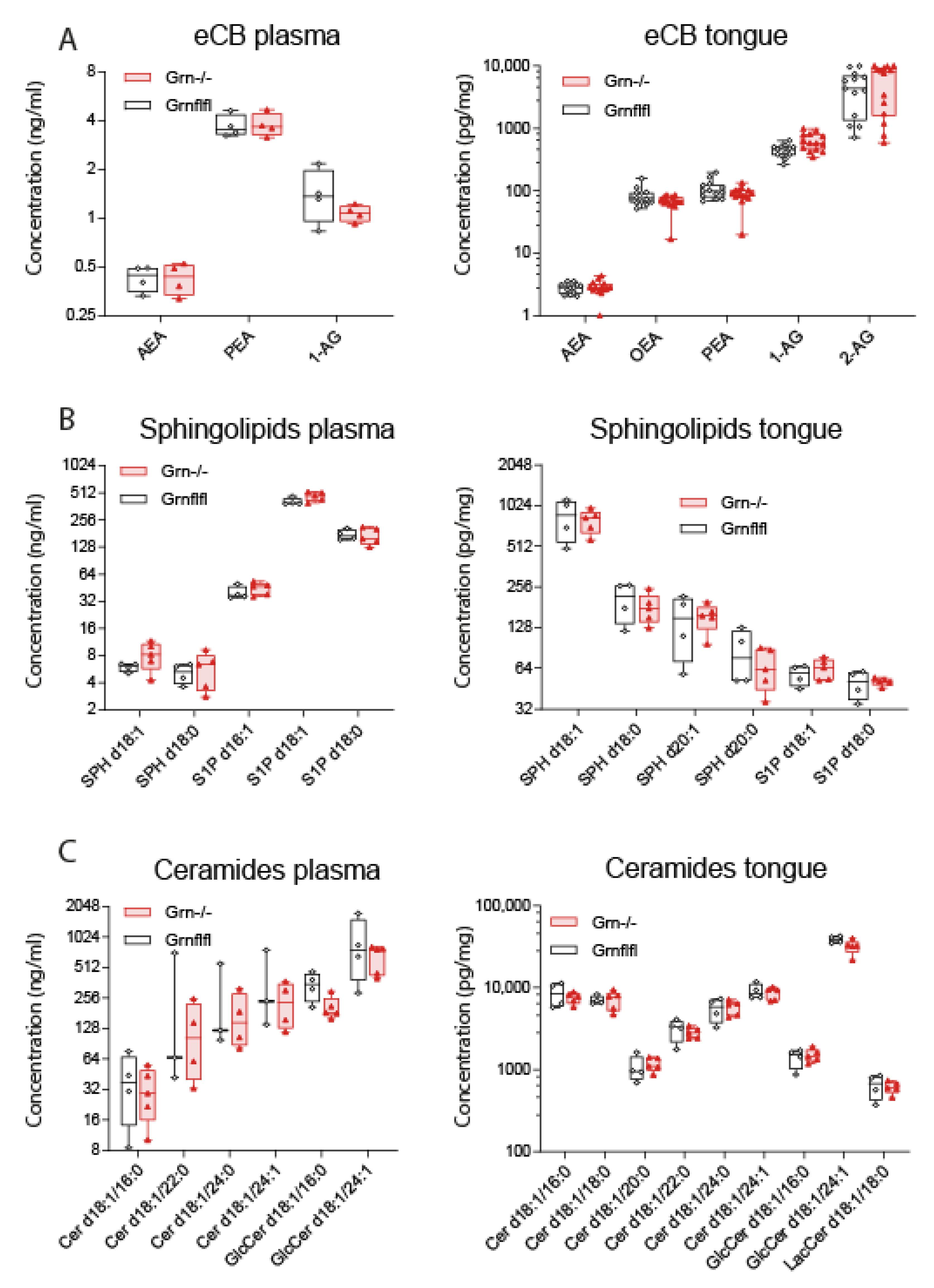
2.7. Analysis of Lipid Signalling Molecules in Tongue Tissue
2.8. Statistical Analyses
3. Results
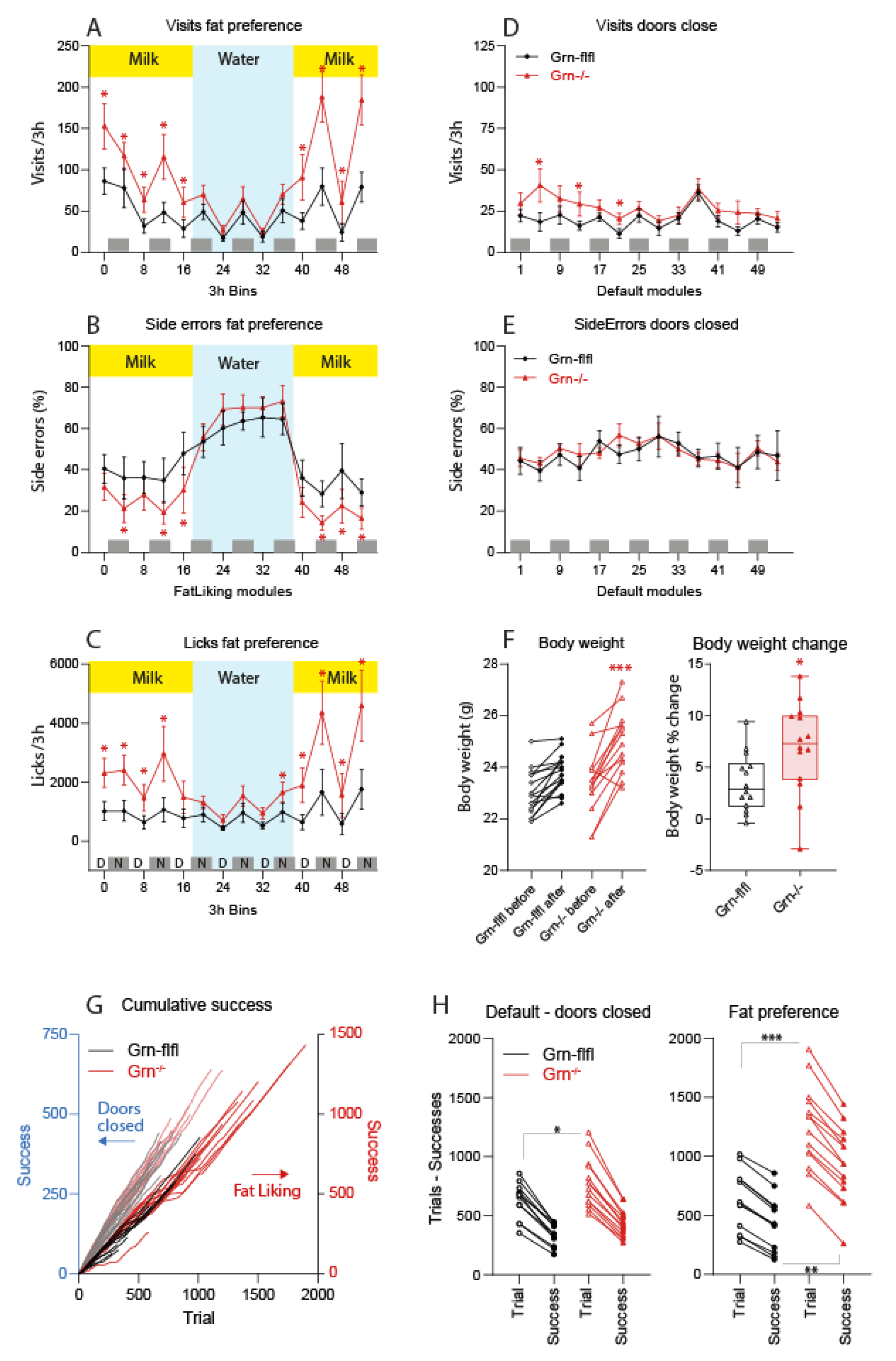
3.1. Fat Milk Taste Preference of Progranulin-Deficient Mice in the IntelliCage
3.2. MCT Fat Taste Preference of Progranulin-Deficient Mice in Phenomaster Cages
3.3. Reduced Expression of CD36 in Taste Buds of Progranulin-Deficient Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Altmann, C.; Hardt, S.; Fischer, C.; Heidler, J.; Lim, H.-Y.; Häussler, A.; Albuquerque, B.; Zimmer, B.; Möser, C.; Behrends, C.; et al. Progranulin overexpression in sensory neurons attenuates neuropathic pain in mice: Role of autophagy. Neurobiol. Dis. 2016, 96, 294–311. [Google Scholar] [CrossRef]
- Altmann, C.; Vasic, V.; Hardt, S.; Heidler, J.; Häussler, A.; Wittig, I.; Schmidt, M.H.H.; Tegeder, I. Progranulin promotes peripheral nerve regeneration and reinnervation: Role of notch signaling. Mol. Neurodegener. 2016, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Lui, H.; Zhang, J.; Makinson, S.R.; Cahill, M.K.; Kelley, K.W.; Huang, H.-Y.; Shang, Y.; Oldham, M.C.; Martens, L.H.; Gao, F.; et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell 2016, 165, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Mackenzie, I.R.; Pickering-Brown, S.; Gass, J.; Rademakers, R.; Lindholm, C.; Snowden, J.; Adamson, J.; Sadovnick, A.D.; Rollinson, S.; et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nat. Cell Biol. 2006, 442, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Cruts, M.; Gijselinck, I.; Van Der Zee, J.; Engelborghs, S.; Wils, H.; Pirici, D.; Rademakers, R.; Vandenberghe, R.; Dermaut, B.; Martin, J.-J.; et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nat. Cell Biol. 2006, 442, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Götzl, J.K.; Mori, K.; Damme, M.; Fellerer, K.; Tahirovic, S.; Kleinberger, G.; Janssens, J.; Van Der Zee, J.; Lang, C.M.; Kremmer, E.; et al. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014, 127, 845–860. [Google Scholar] [CrossRef]
- Liu, C.-J.; Bosch, X. Progranulin: A growth factor, a novel TNFR ligand and a drug target. Pharmacol. Ther. 2012, 133, 124–132. [Google Scholar] [CrossRef]
- Mundra, J.J.; Jian, J.; Bhagat, P.; Liu, C.-J. Progranulin inhibits expression and release of chemokines CXCL9 and CXCL10 in a TNFR1 dependent manner. Sci. Rep. 2016, 6, 21115. [Google Scholar] [CrossRef]
- Pickford, F.; Marcus, J.; Camargo, L.M.; Xiao, Q.; Graham, D.; Mo, J.-R.; Burkhardt, M.; Kulkarni, V.; Crispino, J.; Hering, H.; et al. Progranulin Is a Chemoattractant for Microglia and Stimulates Their Endocytic Activity. Am. J. Pathol. 2011, 178, 284–295. [Google Scholar] [CrossRef]
- Schmitz, K.; Wilken-Schmitz, A.; Vasic, V.; Brunkhorst, R.; Schmidt, M.; Tegeder, I. Progranulin deficiency confers resistance to autoimmune encephalomyelitis in mice. Cell. Mol. Immunol. 2020, 17, 1077–1091. [Google Scholar] [CrossRef]
- Chen, X.; Chang, J.; Deng, Q.; Xu, J.; Nguyen, T.A.; Martens, L.H.; Cenik, B.; Taylor, G.; Hudson, K.F.; Chung, J.; et al. Progranulin Does Not Bind Tumor Necrosis Factor (TNF) Receptors and Is Not a Direct Regulator of TNF-Dependent Signaling or Bioactivity in Immune or Neuronal Cells. J. Neurosci. 2013, 33, 9202–9213. [Google Scholar] [CrossRef]
- Tang, W.; Lu, Y.; Tian, Q.-Y.; Zhang, Y.; Guo, F.-J.; Liu, G.-Y.; Syed, N.M.; Lai, Y.; Lin, E.A.; Kong, L.; et al. The Growth Factor Progranulin Binds to TNF Receptors and Is Therapeutic Against Inflammatory Arthritis in Mice. Science 2011, 332, 478–484. [Google Scholar] [CrossRef]
- Wei, F.; Zhang, Y.; Zhao, W.; Yu, X.; Liu, C.-J. Progranulin Facilitates Conversion and Function of Regulatory T Cells under Inflammatory Conditions. PLoS ONE 2014, 9, e112110. [Google Scholar] [CrossRef]
- Amado, D.A.; Rieders, J.M.; Diatta, F.; Hernandez-Con, P.; Singer, A.; Mak, J.T.; Zhang, J.; Lancaster, E.; Davidson, B.L.; Chen-Plotkin, A.S. AAV-Mediated Progranulin Delivery to a Mouse Model of Progranulin Deficiency Causes T Cell-Mediated Toxicity. Mol. Ther. 2019, 27, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Murakoshi, M.; Gohda, T.; Adachi, E.; Ichikawa, S.; Hagiwara, S.; Suzuki, Y. Differential organ-specific inflammatory response to progranulin in high-fat diet-fed mice. Sci. Rep. 2021, 11, 1194. [Google Scholar] [CrossRef]
- Schmid, A.; Hochberg, A.; Kreiß, A.F.; Gehl, J.; Patz, M.; Thomalla, M.; Hanses, F.; Karrasch, T.; Schäffler, A. Role of progranulin in adipose tissue innate immunity. Cytokine 2020, 125, 154796. [Google Scholar] [CrossRef]
- Matsubara, T.; Mita, A.; Minami, K.; Hosooka, T.; Kitazawa, S.; Takahashi, K.; Tamori, Y.; Yokoi, N.; Watanabe, M.; Matsuo, E.-I.; et al. PGRN is a Key Adipokine Mediating High Fat Diet-Induced Insulin Resistance and Obesity through IL-6 in Adipose Tissue. Cell Metab. 2012, 15, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Xu, L.; Li, H.; Sun, H.; Liu, J.; Wu, S.; Zhou, B. Progranulin causes adipose insulin resistance via increased autophagy resulting from activated oxidative stress and endoplasmic reticulum stress. Lipids Health Dis. 2017, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, H.; Liu, J.; Xu, L.; Guo, Q.; Sun, H.; Wu, S. Progranulin induces adipose insulin resistance and autophagic imbalance via TNFR1 in mice. J. Mol. Endocrinol. 2015, 55, 231–243. [Google Scholar] [CrossRef][Green Version]
- Tanaka, Y.; Takahashi, T.; Tamori, Y. Circulating progranulin level is associated with visceral fat and elevated liver enzymes: Significance of serum progranulin as a useful marker for liver dysfunction. Endocr. J. 2014, 61, 1191–1196. [Google Scholar] [CrossRef]
- Youn, B.-S.; Bang, S.-I.; Klöting, N.; Park, J.W.; Lee, N.; Oh, J.-E.; Pi, K.-B.; Lee, T.H.; Ruschke, K.; Fasshauer, M.; et al. Serum Progranulin Concentrations May Be Associated with Macrophage Infiltration into Omental Adipose Tissue. Diabetes 2008, 58, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Fasshauer, M.; Blüher, M. Adipokines in health and disease. Trends Pharmacol. Sci. 2015, 36, 461–470. [Google Scholar] [CrossRef]
- Li, H.; Zhou, B.; Liu, J.; Li, F.; Li, Y.; Kang, X.; Sun, H.; Wu, S. Administration of progranulin (pgrn) triggers er stress and impairs insulin sensitivity via perk-eif2alpha-dependent manner. Cell Cycle 2015, 14, 1893–1907. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Zhou, B.; Xu, L.; Kang, X.; Yang, W.; Wu, S.; Sun, H. PGRN Induces Impaired Insulin Sensitivity and Defective Autophagy in Hepatic Insulin Resistance. Mol. Endocrinol. 2015, 29, 528–541. [Google Scholar] [CrossRef]
- Hardt, S.; Heidler, J.; Albuquerque, B.; Valek, L.; Altmann, C.; Wilken-Schmitz, A.; Schäfer, M.K.; Wittig, I.; Tegeder, I. Loss of synaptic zinc transport in progranulin deficient mice may contribute to progranulin-associated psychopathology and chronic pain. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 2727–2745. [Google Scholar] [CrossRef]
- Kawase, R.; Ohama, T.; Matsuyama, A.; Matsuwaki, T.; Okada, T.; Yamashita, T.; Yuasa-Kawase, M.; Nakaoka, H.; Nakatani, K.; Inagaki, M.; et al. Deletion of progranulin exacerbates atherosclerosis in ApoE knockout mice. Cardiovasc. Res. 2013, 100, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Dumont, M.; Banerjee, R.; Ma, Y.; Li, H.; Lin, M.T.; Beal, M.F.; Nathan, C.; Thomas, B.; Ding, A. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: A mouse model of frontotemporal dementia. FASEB J. 2010, 24, 4639–4647. [Google Scholar] [PubMed]
- Ghoshal, N.; Dearborn, J.; Wozniak, D.; Cairns, N. Core features of frontotemporal dementia recapitulated in progranulin knockout mice. Neurobiol. Dis. 2012, 45, 395–408. [Google Scholar] [CrossRef]
- Jian, J.; Zhao, S.; Tian, Q.-Y.; Liu, H.; Zhao, Y.; Chen, W.-C.; Grunig, G.; Torres, P.A.; Wang, B.C.; Zeng, B.; et al. Association Between Progranulin and Gaucher Disease. EBioMedicine 2016, 11, 127–137. [Google Scholar] [CrossRef]
- Aflaki, E.; Moaven, N.; Borger, D.K.; Lopez, G.; Westbroek, W.; Chae, J.J.; Marugan, J.; Patnaik, S.; Maniwang, E.; Gonzalez, A.N.; et al. Lysosomal storage and impaired autophagy lead to inflammasome activation in gaucher macrophages. Aging Cell 2016, 15, 77–88. [Google Scholar] [CrossRef]
- Aflaki, E.; Stubblefield, B.K.; Maniwang, E.; Lopez, G.; Moaven, N.; Goldin, E.; Marugan, J.; Patnaik, S.; Dutra, A.; Southall, N.; et al. Macrophage Models of Gaucher Disease for Evaluating Disease Pathogenesis and Candidate Drugs. Sci. Transl. Med. 2014, 6, 240ra73. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, I.; Kögel, D. When lipid homeostasis runs havoc: Lipotoxicity links lysosomal dysfunction to autophagy. Matrix Biol. 2021, 100–101, 99–117. [Google Scholar] [CrossRef]
- Jian, J.; Tian, Q.Y.; Hettinghouse, A.; Zhao, S.; Liu, H.; Wei, J.; Grunig, G.; Zhang, W.; Setchell, K.D.; Sun, Y.; et al. Progranulin recruits hsp70 to beta-glucocerebrosidase and is therapeutic against gaucher disease. EBioMedicine 2016, 13, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Kao, A.W.; Eisenhut, R.J.; Martens, L.H.; Nakamura, A.; Huang, A.; Bagley, J.A.; Zhou, P.; de Luis, A.; Neukomm, L.J.; Cabello, J.; et al. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4441–4446. [Google Scholar] [CrossRef]
- Valdez, C.; Ysselstein, D.; Young, T.J.; Zheng, J.; Krainc, D. Progranulin mutations result in impaired processing of prosaposin and reduced glucocerebrosidase activity. Hum. Mol. Genet. 2020, 29, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Paushter, D.H.; Du, H.; Feng, T.; Hu, F. The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol. 2018, 136, 1–17. [Google Scholar] [CrossRef]
- Tanaka, Y.; Suzuki, G.; Matsuwaki, T.; Hosokawa, M.; Serrano, G.; Beach, T.G.; Yamanouchi, K.; Hasegawa, M.; Nishihara, M. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 2017, 26, 969–988. [Google Scholar] [CrossRef]
- Pepino, M.Y.; Love-Gregory, L.; Klein, S.; Abumrad, N.A. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 2012, 53, 561–566. [Google Scholar] [CrossRef]
- Keller, K.L.; Liang, L.C.; Sakimura, J.; May, D.; van Belle, C.; Breen, C.; Driggin, E.; Tepper, B.J.; Lanzano, P.C.; Deng, L.; et al. Common Variants in the CD36 Gene Are Associated with Oral Fat Perception, Fat Preferences, and Obesity in African Americans. Obesity 2012, 20, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Degrace-Passilly, P.; Besnard, P. CD36 and taste of fat. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 107–111. [Google Scholar] [CrossRef]
- Chen, C.S.; Bench, E.M.; Allerton, T.D.; Schreiber, A.L.; Arceneaux, K.P., 3rd; Primeaux, S.D. Preference for linoleic acid in obesity-prone and obesity-resistant rats is attenuated by the reduction of cd36 on the tongue. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R1346–R1355. [Google Scholar] [CrossRef]
- Ozdener, M.H.; Subramaniam, S.; Sundaresan, S.; Sery, O.; Hashimoto, T.; Asakawa, Y.; Besnard, P.; Abumrad, N.A.; Khan, N.A. CD36- and GPR120-Mediated Ca2+ Signaling in Human Taste Bud Cells Mediates Differential Responses to Fatty Acids and Is Altered in Obese Mice. Gastroenterology 2014, 146, 995–1005.e5. [Google Scholar] [CrossRef]
- Braymer, H.D.; Zachary, H.; Schreiber, A.L.; Primeaux, S.D. Lingual CD36 and nutritional status differentially regulate fat preference in obesity-prone and obesity-resistant rats. Physiol. Behav. 2017, 174, 120–127. [Google Scholar] [CrossRef]
- Sollai, G.; Melis, M.; Mastinu, M.; Pani, D.; Cosseddu, P.; Bonfiglio, A.; Crnjar, R.; Tepper, B.J.; Barbarossa, I.T. Human Tongue Electrophysiological Response to Oleic Acid and Its Associations with PROP Taster Status and the CD36 Polymorphism (rs1761667). Nutrients 2019, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Melis, M.; Sollai, G.; Muroni, P.; Crnjar, R.; Barbarossa, I.T. Associations between Orosensory Perception of Oleic Acid, the Common Single Nucleotide Polymorphisms (rs1761667 and rs1527483) in the CD36 Gene, and 6-n-Propylthiouracil (PROP) Tasting. Nutrients 2015, 7, 2068–2084. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Wang, Y.; Zhong, S.; Wang, M.; Lin, P.; Li, H.; Liu, Z. Cd36 is a candidate lipid sensor involved in the sensory detection of fatty acid in zebrafish. Physiol. Behav. 2017, 182, 34–39. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Zhou, L.-H.; Ban, X.; Liu, D.-X.; Jiang, W.; Liu, X.-M. Decreased expression of CD36 in circumvallate taste buds of high-fat diet induced obese rats. Acta Histochem. 2011, 113, 663–667. [Google Scholar] [CrossRef]
- Simons, P.J.; Kummer, J.A.; Luiken, J.J.; Boon, L. Apical CD36 immunolocalization in human and porcine taste buds from circumvallate and foliate papillae. Acta Histochem. 2011, 113, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, A.; Liu, D.; Nowson, C.; Duesing, K.; Archer, N.; Bowe, S.; Keast, R. A low-fat diet up-regulates expression of fatty acid taste receptor gene FFAR4 in fungiform papillae in humans: A co-twin randomised controlled trial. Br. J. Nutr. 2019, 122, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Costanzo, A.; Evans, M.D.M.; Archer, N.S.; Nowson, C.; Duesing, K.; Keast, R. Expression of the candidate fat taste receptors in human fungiform papillae and the association with fat taste function. Br. J. Nutr. 2018, 120, 64–73. [Google Scholar] [CrossRef]
- Tucker, R.M.; Mattes, R.D.; Running, C.A. Mechanisms and effects of “fat taste” in humans. Biofactors 2014, 40, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Cong, W.-N.; Daimon, C.M.; Wang, R.; Tschöp, M.H.; Sévigny, J.; Martin, B.; Maudsley, S. Altered Lipid and Salt Taste Responsivity in Ghrelin and GOAT Null Mice. PLoS ONE 2013, 8, e76553. [Google Scholar] [CrossRef]
- Berrichi, M.; Hichami, A.; Addou-Klouche, L.; Khan, A.S.; Khan, N. CD36 and GPR120 Methylation Associates with Orosensory Detection Thresholds for Fat and Bitter in Algerian Young Obese Children. J. Clin. Med. 2020, 9, 1956. [Google Scholar] [CrossRef] [PubMed]
- Abdoul-Azize, S.; Selvakumar, S.; Sadou, H.; Besnard, P.; Khan, N.A. Ca2+ signaling in taste bud cells and spontaneous preference for fat: Unresolved roles of CD36 and GPR120. Biochimie 2014, 96, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, B.; Hichami, A.; Khan, A.S.; Plesnik, J.; Sery, O.; Dietrich, A.; Birnbaumer, L.; Khan, N.A. Implication of TRPC3 channel in gustatory perception of dietary lipids. Acta Physiol. 2021, 231, 13554. [Google Scholar] [CrossRef]
- Yoshida, R.; Ohkuri, T.; Jyotaki, M.; Yasuo, T.; Horio, N.; Yasumatsu, K.; Sanematsu, K.; Shigemura, N.; Yamamoto, T.; Margolskee, R.; et al. Endocannabinoids selectively enhance sweet taste. Proc. Natl. Acad. Sci. USA 2010, 107, 935–939. [Google Scholar] [CrossRef]
- Niki, M.; Jyotaki, M.; Yoshida, R.; Yasumatsu, K.; Shigemura, N.; DiPatrizio, N.V.; Piomelli, D.; Ninomiya, Y. Modulation of sweet taste sensitivities by endogenous leptin and endocannabinoids in mice. J. Physiol. 2015, 593, 2527–2545. [Google Scholar] [CrossRef]
- Tarragon, E.; Moreno, J.J. Role of Endocannabinoids on Sweet Taste Perception, Food Preference, and Obesity-related Disorders. Chem. Senses 2017, 43, 3–16. [Google Scholar] [CrossRef]
- Brissard, L.; Leemput, J.; Hichami, A.; Passilly-Degrace, P.; Maquart, G.; Demizieux, L.; Degrace, P.; Khan, N.A. Orosensory detection of dietary fatty acids is altered in cb1r(−/−) mice. Nutrients 2018, 10, 1347. [Google Scholar] [CrossRef]
- Mahler, S.V.; Smith, K.S.; Berridge, K. Endocannabinoid Hedonic Hotspot for Sensory Pleasure: Anandamide in Nucleus Accumbens Shell Enhances ‘Liking’ of a Sweet Reward. Neuropsychopharmacology 2007, 32, 2267–2278. [Google Scholar] [CrossRef]
- De Luca, M.; Solinas, M.; Bimpisidis, Z.; Goldberg, S.; Di Chiara, G. Cannabinoid facilitation of behavioral and biochemical hedonic taste responses. Neuropharmacology 2012, 63, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Banerjee, R.; Thomas, B.; Zhou, P.; Qian, L.; Jia, T.; Ma, X.; Ma, Y.; Iadecola, C.; Beal, M.F.; et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 2009, 207, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, B.; Häussler, A.; Vannoni, E.; Wolfer, D.P.; Tegeder, I. Learning and memory with neuropathic pain: Impact of old age and progranulin deficiency. Front. Behav. Neurosci. 2013, 7, 174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hardt, S.; Valek, L.; Zeng-Brouwers, J.; Wilken-Schmitz, A.; Schaefer, L.; Tegeder, I. Progranulin Deficient Mice Develop Nephrogenic Diabetes Insipidus. Aging Dis. 2018, 9, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Voikar, V.; Colacicco, G.; Gruber, O.; Vannoni, E.; Lipp, H.-P.; Wolfer, D.P. Conditioned response suppression in the IntelliCage: Assessment of mouse strain differences and effects of hippocampal and striatal lesions on acquisition and retention of memory. Behav. Brain Res. 2010, 213, 304–312. [Google Scholar] [CrossRef]
- Krackow, S.; Vannoni, E.; Codita, A.; Mohammed, A.H.; Cirulli, F.; Branchi, I.; Alleva, E.; Reichelt, A.; Willuweit, A.; Voikar, V.; et al. Consistent behavioral phenotype differences between inbred mouse strains in the IntelliCage. Genes, Brain Behav. 2010, 9, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Bishay, P.; Häussler, A.; Lim, H.-Y.; Oertel, B.; Galve-Roperh, I.; Ferreirós, N.; Tegeder, I. Anandamide deficiency and heightened neuropathic pain in aged mice. Neuropharmacology 2013, 71, 204–215. [Google Scholar] [CrossRef]
- Bishay, P.; Schmidt, H.; Marian, C.; Häussler, A.; Wijnvoord, N.; Ziebell, S.; Metzner, J.; Koch, M.; Myrczek, T.; Bechmann, I.; et al. R-Flurbiprofen Reduces Neuropathic Pain in Rodents by Restoring Endogenous Cannabinoids. PLoS ONE 2010, 5, e10628. [Google Scholar] [CrossRef]
- Sailler, S.; Schmitz, K.; Jäger, E.; Ferreiros, N.; Wicker, S.; Zschiebsch, K.; Pickert, G.; Geisslinger, G.; Walter, C.; Tegeder, I.; et al. Regulation of circulating endocannabinoids associated with cancer and metastases in mice and humans. Oncoscience 2014, 1, 272–282. [Google Scholar] [CrossRef]
- Schmitz, K.; de Bruin, N.; Bishay, P.; Männich, J.; Häussler, A.; Altmann, C.; Ferreirós, N.; Lotsch, J.; Ultsch, A.; Parnham, M.J.; et al. R-flurbiprofen attenuates experimental autoimmune encephalomyelitis in mice. EMBO Mol. Med. 2014, 6, 1398–1422. [Google Scholar] [CrossRef]
- Brunkhorst-Kanaan, N.; Klatt-Schreiner, K.; Hackel, J.; Schröter, K.; Trautmann, S.; Hahnefeld, L.; Wicker, S.; Reif, A.; Thomas, D.; Geisslinger, G.; et al. Targeted lipidomics reveal derangement of ceramides in major depression and bipolar disorder. Metabolism 2019, 95, 65–76. [Google Scholar] [CrossRef]
- Dramane, G.; Abdoul-Azize, S.; Hichami, A.; Vögtle, T.; Akpona, S.; Chouabe, C.; Sadou, H.; Nieswandt, B.; Besnard, P.; Khan, N.A. STIM1 regulates calcium signaling in taste bud cells and preference for fat in mice. J. Clin. Investig. 2012, 122, 2267–2282. [Google Scholar] [CrossRef] [PubMed]
- Sihag, J.; Jones, P.J.H. Oleoylethanolamide: The role of a bioactive lipid amide in modulating eating behaviour. Obes. Rev. 2017, 19, 178–197. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, I.; Cuartero, M.I.; Pradillo, J.M.; de la Parra, J.; Perez-Ruiz, A.; Corbi, A.; Ricote, M.; Hamilton, J.A.; Sobrado, M.; Vivancos, J.; et al. Rosiglitazone-induced cd36 up-regulation resolves inflammation by ppargamma and 5-lo-dependent pathways. J. Leukoc. Biol. 2014, 95, 587–598. [Google Scholar] [CrossRef]
- Lin, L.; Rideout, T.C.; Yurkova, N.; Yang, H.; Eck, P.; Jones, P.J. Fatty acid ethanolamides modulate CD36-mRNA through dietary fatty acid manipulation in Syrian Golden hamsters. Appl. Physiol. Nutr. Metab. 2013, 38, 870–878. [Google Scholar] [CrossRef]
- Overton, H.A.; Babbs, A.J.; Doel, S.M.; Fyfe, M.C.; Gardner, L.S.; Griffin, G.; Jackson, H.C.; Procter, M.J.; Rasamison, C.M.; Tang-Christensen, M.; et al. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006, 3, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.J.; Fu, J.; Astarita, G.; Li, X.; Gaetani, S.; Campolongo, P.; Cuomo, V.; Piomelli, D. The Lipid Messenger OEA Links Dietary Fat Intake to Satiety. Cell Metab. 2008, 8, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Oveisi, F.; Gaetani, S.; Lin, E.; Piomelli, D. Oleoylethanolamide, an endogenous PPAR-α agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology 2005, 48, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, M.; Verme, J.L.; Fu, J.; Oveisi, F.; Blázquez, C.; Piomelli, D. Oleoylethanolamide Stimulates Lipolysis by Activating the Nuclear Receptor Peroxisome Proliferator-activated Receptor α (PPAR-α). J. Biol. Chem. 2004, 279, 27849–27854. [Google Scholar] [CrossRef]
- Kim, H.-K.; Shin, M.-S.; Youn, B.-S.; Namkoong, C.; Gil, S.Y.; Kang, G.M.; Yu, J.H.; Kim, M.-S. Involvement of Progranulin in Hypothalamic Glucose Sensing and Feeding Regulation. Endocrinology 2011, 152, 4672–4682. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, B.; Li, H.; Liu, J.; Du, J.; Zang, W.; Wu, S.; Sun, H. Serum Levels of Progranulin Are Closely Associated with Microvascular Complication in Type 2 Diabetes. Dis. Markers 2015, 2015, 357279. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Ono, K.; Inoue, K.; Takagi, Y.; Kikuta, K.-I.; Nishimura, M.; Yoshida, Y.; Nakashima, Y.; Matsumae, H.; Furukawa, Y.; et al. Progranulin expression in advanced human atherosclerotic plaque. Atherosclerosis 2009, 206, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, D.; Dell’Osso, B.; Fenoglio, C.; Villa, C.; Cortini, F.; Serpente, M.; Kittel-Schneider, S.; Weigl, J.; Neuner, M.; Volkert, J.; et al. Progranulin Gene Variability and Plasma Levels in Bipolar Disorder and Schizophrenia. PLoS ONE 2012, 7, e32164. [Google Scholar] [CrossRef] [PubMed]
- Kittel-Schneider, S.; Weigl, J.; Volkert, J.; Geßner, A.; Schmidt, B.; Hempel, S.; Kiel, T.; Olmes, D.G.; Bartl, J.; Weber, H.; et al. Further evidence for plasma progranulin as a biomarker in bipolar disorder. J. Affect. Disord. 2014, 157, 87–91. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schumann, L.; Wilken-Schmitz, A.; Trautmann, S.; Vogel, A.; Schreiber, Y.; Hahnefeld, L.; Gurke, R.; Geisslinger, G.; Tegeder, I. Increased Fat Taste Preference in Progranulin-Deficient Mice. Nutrients 2021, 13, 4125. https://doi.org/10.3390/nu13114125
Schumann L, Wilken-Schmitz A, Trautmann S, Vogel A, Schreiber Y, Hahnefeld L, Gurke R, Geisslinger G, Tegeder I. Increased Fat Taste Preference in Progranulin-Deficient Mice. Nutrients. 2021; 13(11):4125. https://doi.org/10.3390/nu13114125
Chicago/Turabian StyleSchumann, Lana, Annett Wilken-Schmitz, Sandra Trautmann, Alexandra Vogel, Yannick Schreiber, Lisa Hahnefeld, Robert Gurke, Gerd Geisslinger, and Irmgard Tegeder. 2021. "Increased Fat Taste Preference in Progranulin-Deficient Mice" Nutrients 13, no. 11: 4125. https://doi.org/10.3390/nu13114125
APA StyleSchumann, L., Wilken-Schmitz, A., Trautmann, S., Vogel, A., Schreiber, Y., Hahnefeld, L., Gurke, R., Geisslinger, G., & Tegeder, I. (2021). Increased Fat Taste Preference in Progranulin-Deficient Mice. Nutrients, 13(11), 4125. https://doi.org/10.3390/nu13114125






