Regulation of Gastrointestinal Immunity by Metabolites
Abstract
:1. Introduction
2. Intestinal Immunity
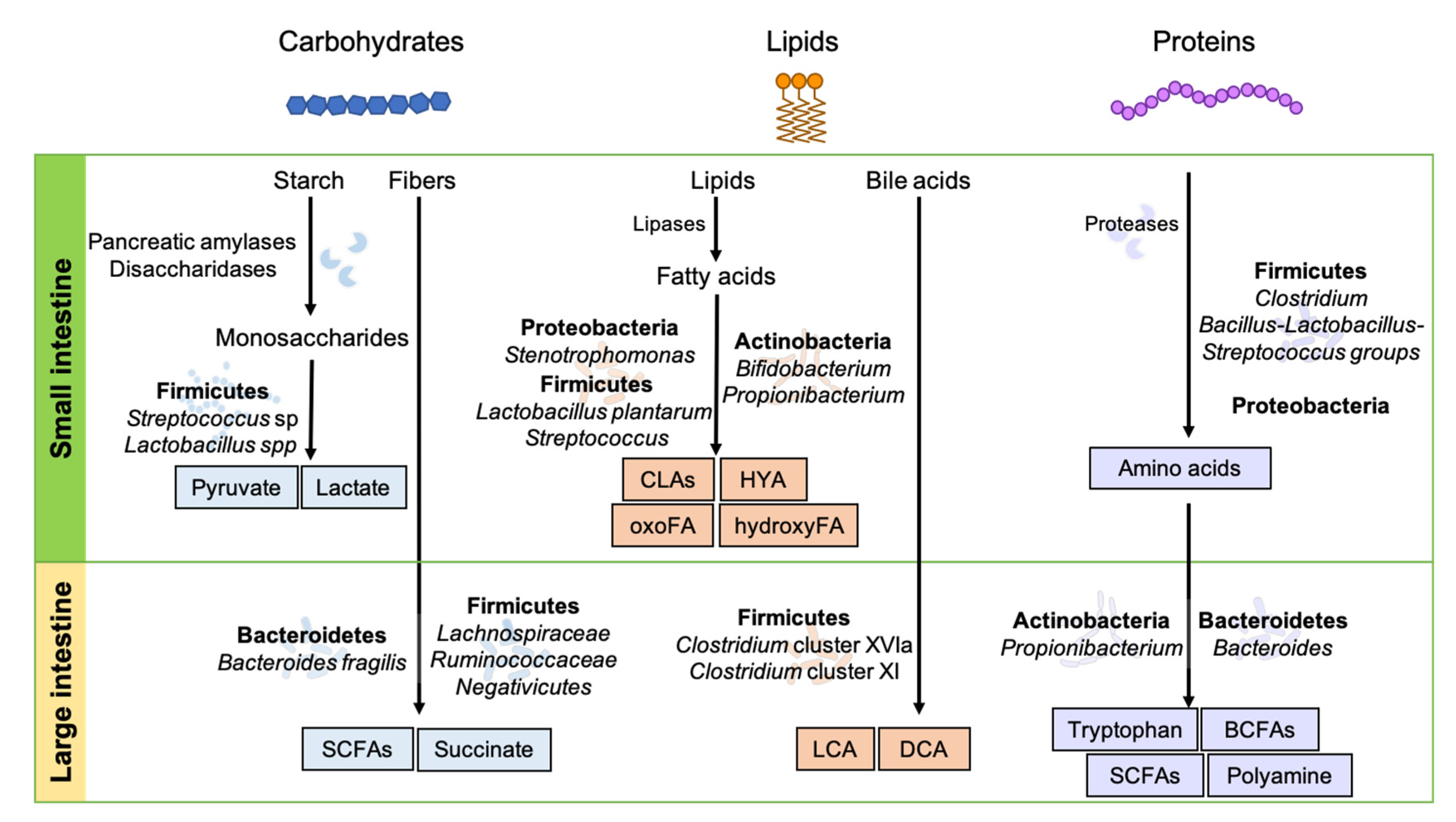
3. Dietary and Microbial Metabolites Produced in the Gastrointestinal Tract
3.1. Production of Dietary Metabolites in the Small Intestine
3.1.1. Carbohydrate Metabolites
3.1.2. Lipid and Bile Acid Metabolites
3.1.3. Protein Metabolites
3.2. Production of Dietary Metabolites in the Colon
3.2.1. Carbohydrate Metabolites
3.2.2. Lipid and Bile Acid Metabolites
3.2.3. Protein Metabolites
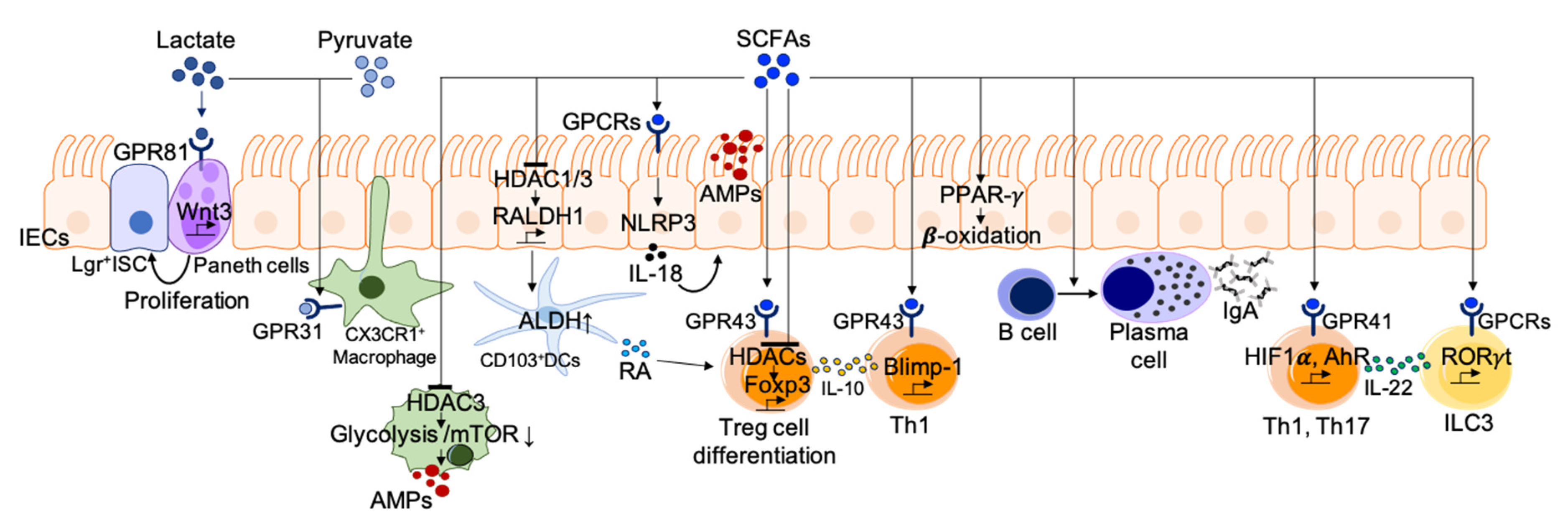
4. Mucosal Immune Regulatory Mechanism of Gut Metabolites
4.1. Carbohydrate Metabolites
- (1)
- Integration of cellular metabolism. As SCFAs are small, they can be absorbed into various cells and are then integrated into cellular metabolism. The most prominent role of SCFAs is as an energy source for colonocytes and immune cells as well [73]. It is well known that dietary fiber and SCFAs support intestinal epithelial proliferation. SCFAs are converted to acetyl-CoA for energy production through the tricarboxylic acid cycle and lipid synthesis [73]. In particular, butylate (C4), known as the primary energy source for colonocytes, activates peroxisome-proliferator-activated receptor (PPAR)-γ signaling, which maintains the energy metabolism of colonocytes toward the mitochondrial β-oxidation of fatty acids [105,106]. The PPAR-γ dependent activation of mitochondrial β-oxidation preserves epithelial hypoxia and limits host-derived nitrate availability in the lumen, thereby preventing the dysbiotic expansion of pathogenic facultative anaerobic bacteria [106]. Immune cells need a significant portion of metabolic building blocks during activation. For instance, adaptive lymphocytes utilize available nutrients to produce energy (e.g., adenosine triphosphate, ATP), undergo biogenesis of cellular components, and produce effector molecules such as antibodies and cytokines. Moreover, SCFAs integrate cellular metabolism to support functional changes in adaptive immune cells. Both glycolysis and oxidative phosphorylation for ATP production are prerequisite steps in B cell activation and plasma B cell differentiation [107]. Kim and colleagues reported that dietary fiber and SCFAs support plasma B cell differentiation through metabolic integration [35]. In fact, SCFA (C3 or SCFA mixture) administration was shown to rescue impaired germinal center formation and IgA expression in mice fed a low-fiber diet. SCFAs promote metabolic processes such as fatty acid oxidation and mitochondrial respiration in B cells during their activation. These results suggest that SCFAs partially mediate the positive effects of dietary fiber intake on gut IgA responses by supporting metabolic processes [35].
- (2)
- Act as HDAC inhibitors. Histone acetylation is an epigenetic modification method for histone, which promotes the formation of open chromatin by adding acetyl groups to the lysine residues and activates transcription. On the contrary, HDACs remove acetyl groups and repress gene transcription. SCFAs inhibit HDACs, thereby promoting gene expression in epithelial cells, T cells, and macrophages. SCFAs promote gut barrier integrity through the induction of AMPs such as RegIIIγ, and β-defensin in epithelial cells via the activation of mammalian target of rapamycin signaling pathway (mTOR) and STAT3 [108,109]. Such functional regulation is mediated by HDAC inhibition together with AMP-activated protein kinase (AMPK) activation. Diet-derived SCFAs are also known to stimulate IECs to induce mucosal tolerogenic DCs through HDAC inhibitory action [110]. SCFAs induce an increase in RALDH1 by inhibiting HDAC1 and HDAC3 in IECs. The increased RALDH1 expression in IECs correlates with the ALDH activity of CD103+ tolerogenic DCs, along with increased numbers of intestinal Treg and high luminal IgA [110]. Among SCFA species, butyrate has strong HDAC-inhibitory activity. HDCA3 inhibition by butyrate induces the differentiation of macrophages possessing antimicrobial activity. The inhibition of metabolic programming in macrophages by glycolysis and mTOR inhibition results in the enhancement of AMPs, such as S100A8/A9/A12 and lysozymes [111]. SCFAs, as HDAC inhibitors, also affect T cell differentiation through the regulation of metabolic sensors [112]. For instance, HDAC inhibition by SCFAs enhances the acetylation of key metabolic sensors for the mTOR pathway (p70S6 kinase and phosphorylation rS6), supporting IL-10 expressing Treg differentiation. Butyrate directly induces Treg generation to provide immune tolerance in response to commensal bacteria by enhancing acetylation of the genetic locus for Foxp3 [34,113,114]. In addition to Treg, SCFAs can directly promote naïve CD4+ T-cell differentiation into Th1 or Th17 cells [112].
- (3)
- GPCR activation. SCFAs can activate many cell types via GPCRs expressed on diverse cells including IECs, neutrophils, macrophages, DCs, B and T cells, and ILCs. Acetate is known to be a ligand of GPR43, and propionate is a ligand of GPR43 and GPR41. Butyrate induces activation through GPR41 and GPR109A [115]. SCFAs facilitate the production of inflammatory effector molecules by IECs in response to immune challenges including ethanol-induced breach, TNBS, and Citrobacter rodentium infection [116]. GPR41- and GPR43-deficient animals display abnormally low inflammatory responses in the gut [116]. SCFAs recognized by GPR43 and GRP109A on IECs activate the NLRP3 inflammasome, leading to IL-18 secretion for maintenance of the gut barrier integrity in chronic inflammation and colorectal cancer cases [115,117,118,119]. SCFAs activate antigen-specific Th2 cells to produce the immunosuppressive cytokines IL-10 [120] and IL-22 [121,122] via GPCRs. The binding of butyrate to GPR41 promotes IL-22 production in Th1 and Th17 as well as ILC3 by upregulation of hypoxia-inducible factor 1α and AhR [122]. The binding of propionate to GPR43 regulates colonic IL-22 expression in ILC3 via AKT and STAT3 signaling. GPR43-deficient ILC3s enhance the susceptibility to colonic inflammation and C. rodentium infection [121]. Neutrophils express the SCFA receptor GPR43, and its activation induces chemotaxis and functional activation, which could play roles in intestinal homeostasis. SCFAs also indirectly regulate Treg generation by inducing the expression of anti-inflammatory molecules (IL-10 and Aldh1a) on DCs and macrophages [115]. Propionate also induces an increase in GPR15 expression, which regulates the homing of Treg to the large intestine [34,123].
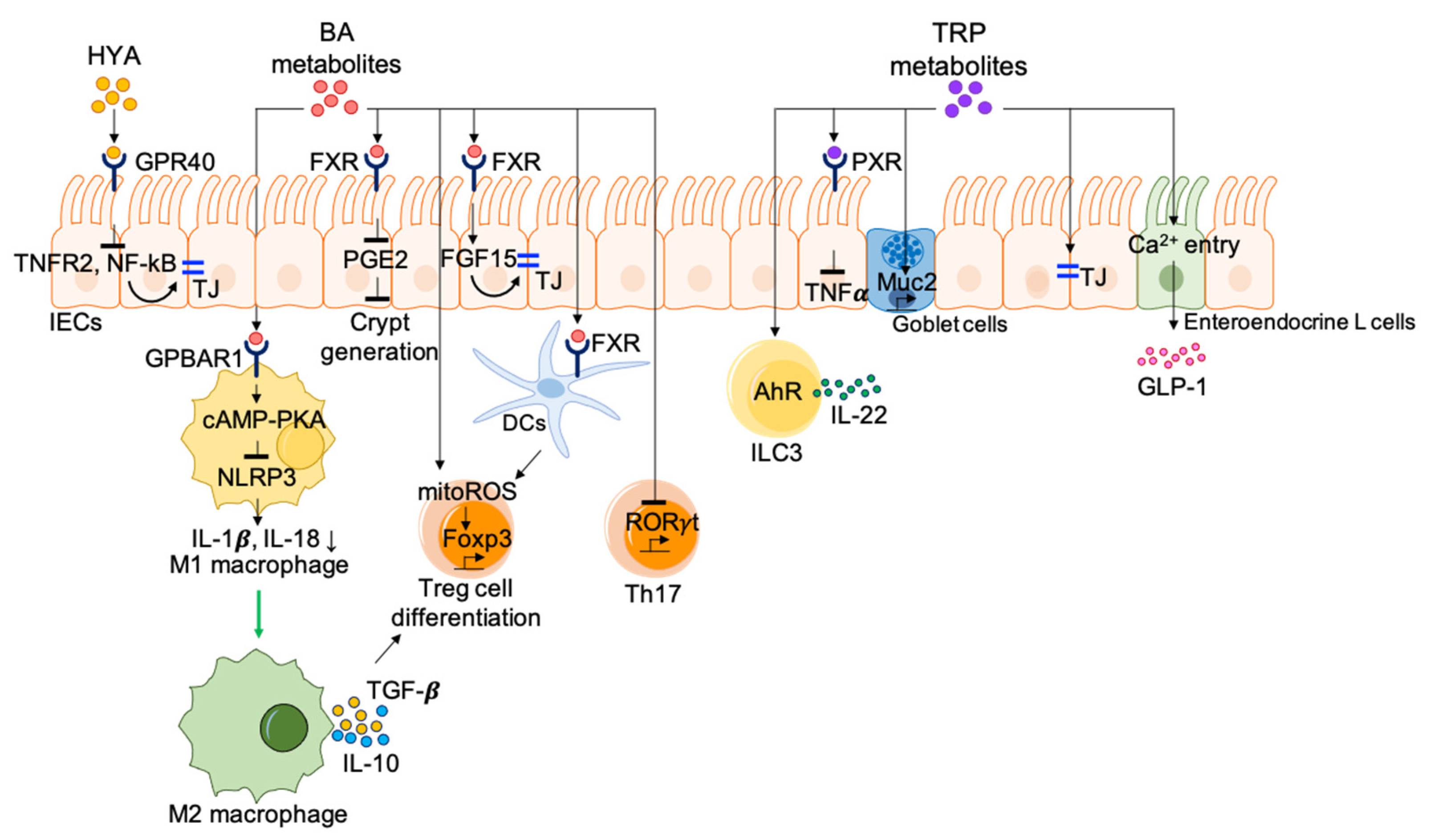
4.2. Lipid and Bile Acid Metabolites
4.3. Protein Metabolites
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H. Intestinal M cells. J. Biochem. 2016, 159, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reboldi, A.; Cyster, J.G. Peyer’s patches: Organizing B-cell responses at the intestinal frontier. Immunol. Rev. 2016, 271, 230–245. [Google Scholar] [CrossRef]
- Randall, T.D.; Mebius, R.E. The development and function of mucosal lymphoid tissues: A balancing act with micro-organisms. Mucosal Immunol. 2014, 7, 455–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, S.A.; Cerovic, V.; Thomson, C.; Brewer, J.; Mowat, A.M.; Milling, S. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol. 2016, 9, 468–478. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef] [Green Version]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [Green Version]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.-Y.; Ko, H.-J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Neish, A. Role of gut microbiota in intestinal wound healing and barrier function. Tissue Barriers 2018, 6, 1539595. [Google Scholar] [CrossRef]
- Lebedev, K.A.; Poniakina, I.D. Immunophysiology of epithelial cells and pattern-recognition receptors. Fiziol. Cheloveka 2006, 32, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Gourbeyre, P.; Berri, M.; Lippi, Y.; Meurens, F.; Vincent-Naulleau, S.; Laffitte, J.; Rogel-Gaillard, C.; Pinton, P.; Oswald, I.P. Pattern recognition receptors in the gut: Analysis of their expression along the intestinal tract and the crypt/villus axis. Physiol. Rep. 2015, 3, e12225. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; O’Leary, C.E.; von Moltke, J.; Liang, H.-E.; Ang, Q.Y.; Turnbaugh, P.J.; Radhakrishnan, S.; Pellizzon, M.; Ma, A.; Locksley, R.M. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 2018, 174, 271–284.e14. [Google Scholar] [CrossRef] [Green Version]
- Burrows, K.; Ngai, L.; Wong, F.; Won, D.; Mortha, A. ILC2 Activation by Protozoan Commensal Microbes. Int. J. Mol. Sci. 2019, 20, 4865. [Google Scholar] [CrossRef] [Green Version]
- Gause, W.C.; Rothlin, C.; Loke, P. Heterogeneity in the initiation, development and function of type 2 immunity. Nat. Rev. Immunol. 2020, 20, 603–614. [Google Scholar] [CrossRef]
- Keir, M.E.; Yi, T.; Lu, T.T.; Ghilardi, N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Zhang, H. NLRP3 Inflammasome and Inflammatory Bowel Disease. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Elinav, E.; Strowig, T.; Kau, A.L.; Henao-Mejia, J.; Thaiss, C.A.; Booth, C.J.; Peaper, D.R.; Bertin, J.; Eisenbarth, S.C.; Gordon, J.I.; et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell 2011, 145, 745–757. [Google Scholar] [CrossRef] [Green Version]
- Merad, M.; Sathe, P.; Helft, J.; Miller, J.; Mortha, A. The Dendritic Cell Lineage: Ontogeny and Function of Dendritic Cells and Their Subsets in the Steady State and the Inflamed Setting. Annu. Rev. Immunol. 2013, 31, 563–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Contreras, O.; Geem, D.; Laur, O.; Williams, I.R.; Lira, S.A.; Nusrat, A.; Parkos, C.A.; Denning, T.L. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J. Clin. Investig. 2011, 121, 4787–4795. [Google Scholar] [CrossRef] [PubMed]
- Sawai, C.M.; Serpas, L.; Neto, A.G.; Jang, G.; Rashidfarrokhi, A.; Kolbeck, R.; Sanjuan, M.A.; Reizis, B.; Sisirak, V. Plasmacytoid Dendritic Cells Are Largely Dispensable for the Pathogenesis of Experimental Inflammatory Bowel Disease. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Arimura, K.; Takagi, H.; Uto, T.; Fukaya, T.; Nakamura, T.; Choijookhuu, N.; Hishikawa, Y.; Yamashita, Y.; Sato, K. Crucial role of plasmacytoid dendritic cells in the development of acute colitis through the regulation of intestinal inflammation. Mucosal Immunol. 2017, 10, 957–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Erturk-Hasdemir, D.; Ochoa-Reparaz, J.; Reinecker, H.-C.; Kasper, D.L. Plasmacytoid Dendritic Cells Mediate Anti-inflammatory Responses to a Gut Commensal Molecule via Both Innate and Adaptive Mechanisms. Cell Host Microbe 2014, 15, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Albert-Bayo, M.; Paracuellos, I.; González-Castro, A.M.; Rodríguez-Urrutia, A.; Rodríguez-Lagunas, M.J.; Alonso-Cotoner, C.; Santos, J.; Vicario, M. Intestinal Mucosal Mast Cells: Key Modulators of Barrier Function and Homeostasis. Cells 2019, 8, 135. [Google Scholar] [CrossRef] [Green Version]
- Travers, J.; Rothenberg, M.E. Eosinophils in mucosal immune responses. Mucosal Immunol. 2015, 8, 464–475. [Google Scholar] [CrossRef]
- Shimokawa, C.; Kanaya, T.; Hachisuka, M.; Ishiwata, K.; Hisaeda, H.; Kurashima, Y.; Kiyono, H.; Yoshimoto, T.; Kaisho, T.; Ohno, H. Mast Cells Are Crucial for Induction of Group 2 Innate Lymphoid Cells and Clearance of Helminth Infections. Immunity 2017, 46, 863–874.e4. [Google Scholar] [CrossRef] [Green Version]
- Gieseck, R.L.; Wilson, M.S.; Wynn, T.A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 2018, 18, 62–76. [Google Scholar] [CrossRef]
- Le Bourhis, L.; Guerri, L.; Dusseaux, M.; Martin, E.; Soudais, C.; Lantz, O. Mucosal-associated invariant T cells: Unconventional development and function. Trends Immunol. 2011, 32, 212–218. [Google Scholar] [CrossRef]
- Olivares-Villagómez, D.; Van Kaer, L. Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends Immunol. 2018, 39, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.L.; Solomon, B.D.; Hsieh, C.-S. T-cell selection and intestinal homeostasis. Immunol. Rev. 2014, 259, 60–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wijk, F.; Cheroutre, H. Mucosal T cells in gut homeostasis and inflammation. Expert Rev. Clin. Immunol. 2010, 6, 559–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, H.N.; Moroney, J.B.; Gan, H.; Shen, T.; Im, J.L.; Li, T.; Taylor, J.R.; Zan, H.; Casali, P. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat. Commun. 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- Cani, P.; Delzenne, N. The Role of the Gut Microbiota in Energy Metabolism and Metabolic Disease. Curr. Pharm. Des. 2009, 15, 1546–1558. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metabolites: Messengers between the microbiota and the immune system. Genes Dev. 2016, 30, 1589–1597. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50, 212–224.e4. [Google Scholar] [CrossRef] [Green Version]
- Eckburg, P.B. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Hui, S.; Lu, W.; Cowan, A.J.; Morscher, R.J.; Lee, G.; Liu, W.; Tesz, G.J.; Birnbaum, M.J.; Rabinowitz, J.D. The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab. 2018, 27, 351–361.e3. [Google Scholar] [CrossRef] [Green Version]
- Horecker, B.L.; Mehler, A.H. Carbohydrate Metabolism. Annu. Rev. Biochem. 1955, 24, 207–274. [Google Scholar] [CrossRef]
- Rogatzki, M.J.; Ferguson, B.S.; Goodwin, M.L.; Gladden, L.B. Lactate is always the end product of glycolysis. Front. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Hove, H.; Nørgaard, H.; Brøbech Mortensen, P. Lactic acid bacteria and the human gastrointestinal tract. Eur. J. Clin. Nutr. 1999, 53, 339–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, M.C.; Small, D.M.; Bliss, C.M. Lipid Digestion and Absorption. Annu. Rev. Physiol. 1983, 45, 651–677. [Google Scholar] [CrossRef] [PubMed]
- Epstein, F.H.; Lewis, R.A.; Austen, K.F.; Soberman, R.J. Leukotrienes and Other Products of the 5-Lipoxygenase Pathway. N. Engl. J. Med. 1990, 323, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, G.; Ecker, J. The opposing effects of n−3 and n−6 fatty acids. Prog. Lipid Res. 2008, 47, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Rabot, S.; Membrez, M.; Bruneau, A.; Gérard, P.; Harach, T.; Moser, M.; Raymond, F.; Mansourian, R.; Chou, C.J. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010, 24, 4948–4959. [Google Scholar] [CrossRef] [Green Version]
- Velagapudi, V.R.; Hezaveh, R.; Reigstad, C.S.; Gopalacharyulu, P.; Yetukuri, L.; Islam, S.; Felin, J.; Perkins, R.; Borén, J.; Orešič, M.; et al. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010, 51, 1101–1112. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef]
- Kishino, S.; Takeuchi, M.; Park, S.-B.; Hirata, A.; Kitamura, N.; Kunisawa, J.; Kiyono, H.; Iwamoto, R.; Isobe, Y.; Arita, M.; et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA 2013, 110, 17808–17813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, A.; Kishino, S.; Park, S.-B.; Takeuchi, M.; Kitamura, N.; Ogawa, J. A novel unsaturated fatty acid hydratase toward C16 to C22 fatty acids from Lactobacillus acidophilus. J. Lipid Res. 2015, 56, 1340–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.Y.; Liang, N.Y.; Curtis, J.M.; Gänzle, M.G. Characterization of Linoleate 10-Hydratase of Lactobacillus plantarum and Novel Antifungal Metabolites. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.-R.; Seo, M.-J.; Shin, K.-C.; Park, J.-B.; Oh, D.-K. Comparison of Biochemical Properties of the Original and Newly Identified Oleate Hydratases from Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. In Comprehensive Physiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Wang, C.-Y.; Liu, S.; Xie, X.-N.; Tan, Z.-R. Regulation profile of the intestinal peptide transporter 1 (PepT1). Drug Des. Devel. Ther. 2017, 11, 3511–3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, R.E.; Benesh, A.E.; Mao, S.; Tabb, D.L.; Tyska, M.J. Proteomic analysis of the enterocyte brush border. Am. J. Physiol. Liver Physiol. 2011, 300, G914–G926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Z.-L. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bröer, S. Amino Acid Transport Across Mammalian Intestinal and Renal Epithelia. Physiol. Rev. 2008, 88, 249–286. [Google Scholar] [CrossRef] [PubMed]
- Stevens, B.R.; Kaunitz, J.D.; Wright, E.M. Intestinal Transport of Amino Acids and Sugars: Advances Using Membrane Vesicles. Annu. Rev. Physiol. 1984, 46, 417–433. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koepsell, H. Glucose transporters in the small intestine in health and disease. Pflügers Arch. Eur. J. Physiol. 2020, 472, 1207–1248. [Google Scholar] [CrossRef]
- Röder, P.V.; Geillinger, K.E.; Zietek, T.S.; Thorens, B.; Koepsell, H.; Daniel, H. The Role of SGLT1 and GLUT2 in Intestinal Glucose Transport and Sensing. PLoS ONE 2014, 9, e89977. [Google Scholar] [CrossRef] [PubMed]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Bhutia, Y.D.; Yang, S.; Ganapathy, V. Short-Chain Fatty Acid Transporters: Role in Colonic Homeostasis. In Comprehensive Physiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 299–314. [Google Scholar]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [Green Version]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [Green Version]
- Gérard, P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Molinero, N.; Ruiz, L.; Sánchez, B.; Margolles, A.; Delgado, S. Intestinal Bacteria Interplay with Bile and Cholesterol Metabolism: Implications on Host Physiology. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.-B.; Guo, C.-J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Lam-Galvez, B.R.; Kirsop, J.; Wang, Z.; Levison, B.S.; Gu, X.; Copeland, M.F.; Bartlett, D.; Cody, D.B.; Dai, H.J.; et al. l-Carnitine in omnivorous diets induces an atherogenic gut microbial pathway in humans. J. Clin. Investig. 2018, 129, 373–387. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Oh, S.F.; Olszak, T.; Neves, J.F.; Avci, F.Y.; Erturk-Hasdemir, D.; Lu, X.; Zeissig, S.; Blumberg, R.S.; Kasper, D.L. Sphingolipids from a Symbiotic Microbe Regulate Homeostasis of Host Intestinal Natural Killer T Cells. Cell 2014, 156, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, E.M.; Ke, X.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 2019, 25, 668–680.e7. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K. Diet, Microbiota, and Metabolic Health: Trade-Off between Saccharolytic and Proteolytic Fermentation. Annu. Rev. Food Sci. Technol. 2018, 9, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Mariotti, F.; Huneau, J.F.; Tomé, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlane, G.T.; Cummings, J.H.; Allison, C. Protein Degradation by Human Intestinal Bacteria. Microbiology 1986, 132, 1647–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, G.; Seelig, S.; Kumar, S. Isolated adrenal cortex cells in suspension: Stimulation and inhibition of steroidogenesis by analogues of ACTH. J. Steroid Biochem. 1975, 6, 371–375. [Google Scholar] [CrossRef]
- Mortensen, P.B.; Clausen, M.R. Short-Chain Fatty Acids in the Human Colon: Relation to Gastrointestinal Health and Disease. Scand. J. Gastroenterol. 1996, 31, 132–148. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.-B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10, 117864691769193. [Google Scholar] [CrossRef] [Green Version]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [Green Version]
- Certo, M.; Tsai, C.-H.; Pucino, V.; Ho, P.-C.; Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Shanmugam, A.; Swafford, D.; Suryawanshi, A.; Bhattacharjee, P.; Hussein, M.S.; Koni, P.A.; Prasad, P.D.; Kurago, Z.B.; Thangaraju, M.; et al. GPR81, a Cell-Surface Receptor for Lactate, Regulates Intestinal Homeostasis and Protects Mice from Experimental Colitis. J. Immunol. 2018, 200, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Fukuda, S.; Hase, K.; Nishiumi, S.; Izumi, Y.; Yoshida, M.; Hagiwara, T.; Kawashima, R.; Yamazaki, M.; Oshio, T.; et al. Microbiota-derived lactate accelerates colon epithelial cell turnover in starvation-refed mice. Nat. Commun. 2013, 4, 1654. [Google Scholar] [CrossRef] [Green Version]
- Morita, N.; Umemoto, E.; Fujita, S.; Hayashi, A.; Kikuta, J.; Kimura, I.; Haneda, T.; Imai, T.; Inoue, A.; Mimuro, H.; et al. GPR31-dependent dendrite protrusion of intestinal CX3CR1+ cells by bacterial metabolites. Nature 2019, 566, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Roediger, W.E.W. Utilization of Nutrients by Isolated Epithelial Cells of the Rat Colon. Gastroenterology 1982, 83, 424–429. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Price, M.J.; Patterson, D.G.; Scharer, C.D.; Boss, J.M. Progressive Upregulation of Oxidative Metabolism Facilitates Plasmablast Differentiation to a T-Independent Antigen. Cell Rep. 2018, 23, 3152–3159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Goverse, G.; Molenaar, R.; Macia, L.; Tan, J.; Erkelens, M.N.; Konijn, T.; Knippenberg, M.; Cook, E.C.L.; Hanekamp, D.; Veldhoen, M.; et al. Diet-Derived Short Chain Fatty Acids Stimulate Intestinal Epithelial Cells to Induce Mucosal Tolerogenic Dendritic Cells. J. Immunol. 2017, 198, 2172–2181. [Google Scholar] [CrossRef] [Green Version]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406.e1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Friesen, L.; Park, J.; Kim, H.M.; Kim, C.H. Microbial metabolites, short-chain fatty acids, restrain tissue bacterial load, chronic inflammation, and associated cancer in the colon of mice. Eur. J. Immunol. 2018, 48, 1235–1247. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [Green Version]
- Zaki, M.H.; Boyd, K.L.; Vogel, P.; Kastan, M.B.; Lamkanfi, M.; Kanneganti, T.-D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010, 32, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Ma, C.; et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef] [Green Version]
- Chun, E.; Lavoie, S.; Fonseca-Pereira, D.; Bae, S.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Gallini Comeau, C.A.; Glickman, J.N.; Fuller, M.H.; et al. Metabolite-Sensing Receptor Ffar2 Regulates Colonic Group 3 Innate Lymphoid Cells and Gut Immunity. Immunity 2019, 51, 871–884.e6. [Google Scholar] [CrossRef]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 4457. [Google Scholar] [CrossRef]
- Kim, S.V.; Xiang, W.V.; Kwak, C.; Yang, Y.; Lin, X.W.; Ota, M.; Sarpel, U.; Rifkin, D.B.; Xu, R.; Littman, D.R. GPR15-Mediated Homing Controls Immune Homeostasis in the Large Intestine Mucosa. Science 2013, 340, 1456–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungaro, F.; Rubbino, F.; Danese, S.; D’Alessio, S. Actors and Factors in the Resolution of Intestinal Inflammation: Lipid Mediators as a New Approach to Therapy in Inflammatory Bowel Diseases. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Oh, D.Y.; Talukdar, S.; Bae, E.J.; Imamura, T.; Morinaga, H.; Fan, W.; Li, P.; Lu, W.J.; Watkins, S.M.; Olefsky, J.M. GPR120 Is an Omega-3 Fatty Acid Receptor Mediating Potent Anti-inflammatory and Insulin-Sensitizing Effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Bassaganya-Riera, J.; Reynolds, K.; Martino-Catt, S.; Cui, Y.; Hennighausen, L.; Gonzalez, F.; Rohrer, J.; Benninghoff, A.U.; Hontecillas, R. Activation of PPAR γ and δ by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology 2004, 127, 777–791. [Google Scholar] [CrossRef]
- Peyrin-Biroulet, L.; Beisner, J.; Wang, G.; Nuding, S.; Oommen, S.T.; Kelly, D.; Parmentier-Decrucq, E.; Dessein, R.; Merour, E.; Chavatte, P.; et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl. Acad. Sci. USA 2010, 107, 8772–8777. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, J.; Mizukure, T.; Park, S.-B.; Kishino, S.; Kimura, I.; Hirano, K.; Bergamo, P.; Rossi, M.; Suzuki, T.; Arita, M.; et al. A Gut Microbial Metabolite of Linoleic Acid, 10-Hydroxy-cis-12-octadecenoic Acid, Ameliorates Intestinal Epithelial Barrier Impairment Partially via GPR40-MEK-ERK Pathway. J. Biol. Chem. 2015, 290, 2902–2918. [Google Scholar] [CrossRef] [Green Version]
- Staudinger, J.L.; Goodwin, B.; Jones, S.A.; Hawkins-Brown, D.; MacKenzie, K.I.; LaTour, A.; Liu, Y.; Klaassen, C.D.; Brown, K.K.; Reinhard, J.; et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 3369–3374. [Google Scholar] [CrossRef] [Green Version]
- Makishima, M. Vitamin D Receptor as an Intestinal Bile Acid Sensor. Science 2002, 296, 1313–1316. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Lo, J.-L.; Huang, L.; Zhao, A.; Metzger, E.; Adams, A.; Meinke, P.T.; Wright, S.D.; Cui, J. Lithocholic Acid Decreases Expression of Bile Salt Export Pump through Farnesoid X Receptor Antagonist Activity. J. Biol. Chem. 2002, 277, 31441–31447. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Shah, Y.M.; Ma, X.; Morimura, K.; Kim, I.; Gonzalez, F.J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-κB target gene expression. Am. J. Physiol. Liver Physiol. 2007, 292, G1114–G1122. [Google Scholar] [CrossRef] [PubMed]
- Renga, B.; Mencarelli, A.; Cipriani, S.; D’Amore, C.; Carino, A.; Bruno, A.; Francisci, D.; Zampella, A.; Distrutti, E.; Fiorucci, S. The Bile Acid Sensor FXR Is Required for Immune-Regulatory Activities of TLR-9 in Intestinal Inflammation. PLoS ONE 2013, 8, e54472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biagioli, M.; Carino, A.; Cipriani, S.; Francisci, D.; Marchianò, S.; Scarpelli, P.; Sorcini, D.; Zampella, A.; Fiorucci, S. The Bile Acid Receptor GPBAR1 Regulates the M1/M2 Phenotype of Intestinal Macrophages and Activation of GPBAR1 Rescues Mice from Murine Colitis. J. Immunol. 2017, 199, 718–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cipriani, S.; Mencarelli, A.; Chini, M.G.; Distrutti, E.; Renga, B.; Bifulco, G.; Baldelli, F.; Donini, A.; Fiorucci, S. The Bile Acid Receptor GPBAR-1 (TGR5) Modulates Integrity of Intestinal Barrier and Immune Response to Experimental Colitis. PLoS ONE 2011, 6, e25637. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef] [Green Version]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The Bile Acid Receptor FXR Is a Modulator of Intestinal Innate Immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Jain, U.; Lai, C.-W.; Xiong, S.; Goodwin, V.M.; Lu, Q.; Muegge, B.D.; Christophi, G.P.; VanDussen, K.L.; Cummings, B.P.; Young, E.; et al. Temporal Regulation of the Bacterial Metabolite Deoxycholate during Colonic Repair Is Critical for Crypt Regeneration. Cell Host Microbe 2018, 24, 353–363.e5. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blachier, F.; Boutry, C.; Bos, C.; Tomé, D. Metabolism and functions of l-glutamate in the epithelial cells of the small and large intestines. Am. J. Clin. Nutr. 2009, 90, 814S–821S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, D.; Hou, Y.; Wang, L.; Ouyang, W.; Long, M.; Zhao, D.; Ding, B.; Liu, Y.; Wu, G. l-Glutamine enhances enterocyte growth via activation of the mTOR signaling pathway independently of AMPK. Amino Acids 2015, 47, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Gould, R.L.; Pazdro, R. Impact of Supplementary Amino Acids, Micronutrients, and Overall Diet on Glutathione Homeostasis. Nutrients 2019, 11, 1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlodarska, M.; Luo, C.; Kolde, R.; D’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chimerel, C.; Emery, E.; Summers, D.K.; Keyser, U.; Gribble, F.M.; Reimann, F. Bacterial Metabolite Indole Modulates Incretin Secretion from Intestinal Enteroendocrine L Cells. Cell Rep. 2014, 9, 1202–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matteoli, G.; Mazzini, E.; Iliev, I.D.; Mileti, E.; Fallarino, F.; Puccetti, P.; Chieppa, M.; Rescigno, M. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 2010, 59, 595–604. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Bessede, A.; Gargaro, M.; Pallotta, M.T.; Matino, D.; Servillo, G.; Brunacci, C.; Bicciato, S.; Mazza, E.M.C.; Macchiarulo, A.; Vacca, C.; et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 2014, 511, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Lanis, J.M.; Alexeev, E.E.; Curtis, V.F.; Kitzenberg, D.A.; Kao, D.J.; Battista, K.D.; Gerich, M.E.; Glover, L.E.; Kominsky, D.J.; Colgan, S.P. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017, 10, 1133–1144. [Google Scholar] [CrossRef]
- Alvarado, D.M.; Chen, B.; Iticovici, M.; Thaker, A.I.; Dai, N.; VanDussen, K.L.; Shaikh, N.; Lim, C.K.; Guillemin, G.J.; Tarr, P.I.; et al. Epithelial Indoleamine 2,3-Dioxygenase 1 Modulates Aryl Hydrocarbon Receptor and Notch Signaling to Increase Differentiation of Secretory Cells and Alter Mucus-Associated Microbiota. Gastroenterology 2019, 157, 1093–1108.e11. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, B.-H.; Kim, M.; Yun, C.-H. Regulation of Gastrointestinal Immunity by Metabolites. Nutrients 2021, 13, 167. https://doi.org/10.3390/nu13010167
Gu B-H, Kim M, Yun C-H. Regulation of Gastrointestinal Immunity by Metabolites. Nutrients. 2021; 13(1):167. https://doi.org/10.3390/nu13010167
Chicago/Turabian StyleGu, Bon-Hee, Myunghoo Kim, and Cheol-Heui Yun. 2021. "Regulation of Gastrointestinal Immunity by Metabolites" Nutrients 13, no. 1: 167. https://doi.org/10.3390/nu13010167
APA StyleGu, B.-H., Kim, M., & Yun, C.-H. (2021). Regulation of Gastrointestinal Immunity by Metabolites. Nutrients, 13(1), 167. https://doi.org/10.3390/nu13010167




