Diet, Perceived Intestinal Well-Being and Compositions of Fecal Microbiota and Short Chain Fatty Acids in Oat-Using Subjects with Celiac Disease or Gluten Sensitivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Study Design
2.2. Dietary Intake Using Food Diaries
2.3. Gut Symptom Diaries
2.4. Fecal Samples and DNA Extraction
2.5. 16S Library Preparation
2.6. 16S rDNA Sequencing
2.7. Short Chain Fatty Acids Assay
2.8. β-Glucuronidase and Ammoniacal Nitrogen Assays
2.9. Statistical and Data Analyses
3. Results
3.1. Dietary Intake and the Quality of Diet
3.2. Gut Symptom Diaries
3.3. Intestinal Microbiota Signatures
3.4. Microbial Metabolic Activity
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
References
- Sapone, A.; Lammers, K.M.; Casolaro, V.; Cammarota, M.; Giuliano, M.T.; De Rosa, M.; Stefanile, R.; Mazzarella, G.; Tolone, C.; Russo, M.I. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: Celiac disease and gluten sensitivity. BMC Med. 2011, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Carroccio, A.; Mansueto, P.; Iacono, G.; Soresi, M.; D’Alcamo, A.; Cavataio, F.; Brusca, I.; Florena, A.M.; Ambrosiano, G.; Seidita, A.; et al. Non-Celiac Wheat Sensitivity Diagnosed by Double-Blind Placebo-Controlled Challenge: Exploring a New Clinical Entity. Am. J. Gastroenterol. 2012, 107, 1898–1906. [Google Scholar] [CrossRef]
- Lee, A.R.; Ng, D.L.; Dave, E.; Ciaccio, E.J.; Green, P.H.R. The effect of substituting alternative grains in the diet on the nutritional profile of the gluten-free diet. J. Hum. Nutr. Diet. 2009, 22, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Vici, G.; Belli, L.; Biondi, M.; Polzonetti, V. Gluten free diet and nutrient deficiencies: A review. Clin. Nutr. 2016, 35, 1236–1241. [Google Scholar] [CrossRef]
- Melini, V.; Melini, F. Gluten-Free Diet: Gaps and Needs for a Healthier Diet. Nutrients 2019, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Peräaho, M.; Collin, P.; Kaukinen, K.; Kekkonen, L.; Miettinen, S.; Mäki, M. Oats can diversify a gluten-free diet in celiac disease and dermatitis herpetiformis. J. Am. Diet. Assoc. 2004, 104, 1148–1150. [Google Scholar] [CrossRef]
- Aaltonen, K.; Laurikka, P.; Huhtala, H.; Mäki, M.; Kaukinen, K.; Kurpp, K. The long-term con-sumption of oats in celiac disease patients is safe: A large cross-sectional study. Nutrients 2017, 9, 611. [Google Scholar] [CrossRef]
- Fritz, R.D.; Chen, Y. Oat safety for celiac disease patients: Theoretical analysis correlates adverse symptoms in clinical studies to contaminated study oats. Nutr. Res. 2018, 60, 54–67. [Google Scholar] [CrossRef]
- Pinto-Sanchez, M.I.; Calo, N.S.C.; Leffler, D.A.; Verdú, E.F.; Green, P.; Bercik, P.; Ford, A.C.; Murray, J.A.; Armstrong, D.; Semrad, C.; et al. Safety of Adding Oats to a Gluten-Free Diet for Patients With Celiac Disease: Systematic Review and Meta-analysis of Clinical and Observational Studies. Gastroenterology 2017, 153, 395–409. [Google Scholar] [CrossRef]
- Walker, W.A. Initial Intestinal Colonization in the Human Infant and Immune Homeostasis. Ann. Nutr. Metab. 2013, 63, 8–15. [Google Scholar] [CrossRef]
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Collado, M.C.; Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 2009, 102, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Marasco, G.; Di Biase, A.R.; Schiumerini, R.; Eusebi, L.H.; Iughetti, L.; Ravaioli, F.; Scaioli, E.; Colecchia, A.; Festi, D. Gut Microbiota and Celiac Disease. Dig. Dis. Sci. 2016, 61, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhang, J.-Q. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 2. [Google Scholar] [CrossRef]
- Catassi, C.; Elli, L.; Bonaz, B.; Bouma, G.; Carroccio, A.; Castillejo, G.; Cellier, C.; Cristofori, F.; De Magistris, L.; Dolinsek, J.; et al. Diagnosis of Non-Celiac Gluten Sensitivity (NCGS): The Salerno Experts’ Criteria. Nutrients 2015, 7, 4966–4977. [Google Scholar] [CrossRef]
- Leppälä, J.; Lagström, H.; Kaljonen, A.; Laitinen, K. Construction and evaluation of a self-contained index for assessment of diet quality. Scand. J. Public Health 2010, 38, 794–802. [Google Scholar] [CrossRef]
- Finnish Food Composition Database. National Institute for Health and Welfare, Nutrition Unit Fineli. Available online: https://fineli.fi/fineli/en/index? (accessed on 20 June 2018).
- Nylund, L.; Heilig, H.G.; Salminen, S.; De Vos, W.M.; Satokari, R. Semi-automated extraction of microbial DNA from feces for qPCR and phylogenetic microarray analysis. J. Microbiol. Methods 2010, 83, 231–235. [Google Scholar] [CrossRef]
- Fiorini, D.; Pacetti, D.; Gabbianelli, R.; Gabrielli, S.; Ballini, R. A salting out system for improving the efficiency of the headspace solid-phase microextraction of short and medium chain free fatty acids. J. Chromatogr. A 2015, 1409, 282–287. [Google Scholar] [CrossRef]
- Nist Chemistry Webbook. National Institute of Standards and Technology. Available online: https://webbook.nist.gov/chemistry/ (accessed on 27 September 2018).
- Koroleff, F. Direct spectrophotometric determination of ammonia in precipitation. Tellus 1966, 18, 562–565. [Google Scholar] [CrossRef]
- Shen, R.L.; Dang, X.Y.; Dong, J.L.; Hu, X.Z. Effects of oat beta-glucan and barley beta-glucan on fecal characteristics, intestinal microflora, and intestinal bacterial metabolites in rats. J. Agric. Food Chem. 2012, 60, 11301–11308. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- RDP Taxonomic Training Data Formatted for DADA2 (RDP Trainset 16/Release 11.5). Available online: https://zenodo.org/record/801828#.X0CLyjURWUl (accessed on 1 October 2018).
- Wright, E.S. DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinform. 2015, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Introduction to the Microbiome R Package Version 2.1.26. Available online: https://microbiome.github.io/microbiome/ (accessed on 1 October 2018).
- Oksanen, J.; Blanchet, F.G.; Friendly, M. Community Ecology Package. R Package Version 2.5-6. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 October 2018).
- Hallert, C.; Grant, C.; Grehn, S.; Granno, C.; Hulten, S.; Midhagen, G.; Strom, M.; Svensson, H.; Valdimason, T. Evidence of poor vitamin status in coeliac patients on a gluten-free diet for 10 years. Aliment. Pharmacol. Ther. 2002, 16, 1333–1339. [Google Scholar] [CrossRef]
- Thompson, T.; Dennis, M.; Higgins, L.A.; Lee, A.R.; Sharrett, M.K. Gluten-free diet survey: Are Americans with coeliac disease consuming recommended amounts of fibre, iron, calcium and grain foods? J. Hum. Nutr. Diet. 2005, 18, 163–169. [Google Scholar] [CrossRef]
- De Souza, M.C.P.; Deschênes, M.-E.; Laurencelle, S.; Godet, P.; Roy, C.C.; Djilali-Saiah, I. Pure Oats as Part of the Canadian Gluten-Free Diet in Celiac Disease: The Need to Revisit the Issue. Can. J. Gastroenterol. Hepatol. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Hernando, A.; Mujico, J.R.; Mena, M.C.; Lombardía, M.; Méndez, E. Measurement of wheat gluten and barley hordeins in contaminated oats from Europe, the United States and Canada by Sandwich R5 ELISA. Eur. J. Gastroenterol. Hepatol. 2008, 20, 545–554. [Google Scholar] [CrossRef]
- Potter, M.D.E.; Jones, M.P.; Walker, M.M.; Koloski, N.A.; Keely, S.; Holtmann, G.; Talley, A. Incidence and prevalence of self-reported non-coeliac wheat sensitivity and gluten avoidance in Australia. Med. J. Aust. 2020, 212, 126–131. [Google Scholar] [CrossRef]
- Tovoli, F.; Granito, A.; Negrini, G.; Guidetti, E.; Faggiano, C.; Bolondi, L. Long term effects of gluten-free diet in non-celiac wheat sensitivity. Clin. Nutr. 2019, 38, 357–363. [Google Scholar] [CrossRef]
- Skodje, G.I.; Sarna, V.K.; Minelle, I.H.; Rolfsen, K.L.; Muir, J.G.; Gibson, P.R.; Veierød, M.B.; Henriksen, C.; Lundin, K.E. Fructan, Rather Than Gluten, Induces Symptoms in Patients With Self-Reported Non-Celiac Gluten Sensitivity. Gastroenterology 2018, 154, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Reig-Otero, Y.; Mañes, J.; Manyes, L. Amylase–Trypsin Inhibitors in Wheat and Other Cereals as Potential Activators of the Effects of Nonceliac Gluten Sensitivity. J. Med. Food 2018, 21, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Dale, H.F.; Biesiekierski, J.R.; Lied, G.A. Non-coeliac gluten sensitivity and the spectrum of gluten-related disorders: An updated overview. Nutr. Res. Rev. 2018, 32, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Viitasalo, L.; Kurppa, K.; Ashorn, M.; Saavalainen, P.; Huhtala, H.; Ashorn, S.; Mäki, M.; Ilus, T.; Kaukinen, K.; Iltanen, S. Microbial Biomarkers in Patients with Nonresponsive Celiac Disease. Dig. Dis. Sci. 2018, 63, 3434–3441. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Laurikka, P.; Lindfors, K.; Collin, P.; Salmi, T.; Lähdeaho, M.L.; Saavalainen, P.; Mäki, M.; Matto, J.; Kurppa, K. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 2014, 109, 1933–1941. [Google Scholar] [CrossRef]
- Collado, M.C.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 2008, 62, 264–269. [Google Scholar] [CrossRef]
- Holm, K.; Maki, M.; Vuolteenaho, N.; Mustalahti, K.; Ashorn, M.; Ruuska, T.; Kaukinen, K. Oats in the treatment of childhood coeliac disease: A 2-year controlled trial and a long-term clinical follow-up study. Aliment. Pharmacol. Ther. 2006, 23, 1463–1472. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites andcolorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Pylkas, A.M.; Juneja, L.R.; Slavin, J.L. Comparison of Different Fibers for In Vitro Production of Short Chain Fatty Acids by Intestinal Microflora. J. Med. Food 2005, 8, 113–116. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- McOrist, A.L.; Miller, R.B.; Bird, A.R.; Keogh, J.B.; Noakes, M.; Topping, D.L.; Conlon, M.A. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in re-sistant starch. J. Nutr. 2011, 141, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Villalba, R.; Gimenez-Bastida, J.A.; Garcia-Conesa, M.T.; Tomas-Barberan, F.A.; Espin, J.C.; Larrosa, M. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 2012, 35, 1906–1913. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; Ruas-Madiedo, P.; Suárez, A.; Mayo, B. Interindividual Differences in Microbial Counts and Biochemical-Associated Variables in the Feces of Healthy Spanish Adults. Dig. Dis. Sci. 2006, 51, 737–743. [Google Scholar] [CrossRef]
- Di Cagno, R.; De Angelis, M.; De Pasquale, I.; Ndagijimana, M.; Vernocchi, P.; Ricciuti, P.; Gagliardi, F.; Laghi, L.; Crecchio, C.; Guerzoni, M.E.; et al. Duodenal and faecal microbiota of celiac children: Molecular, phenotype and metabolome characterization. BMC Microbiol. 2011, 11, 219. [Google Scholar] [CrossRef]
- Pellock, S.J.; Redinbo, M.R. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J. Boil. Chem. 2017, 292, 8569–8576. [Google Scholar] [CrossRef]
- Freeman, H.J. Effects of differing purified cellulose, pectin, and hemicellulose fiber diets on fecal enzymes in 1,2-dimethylhydrazine-induced rat colon carcinogenesis. Cancer Res. 1986, 46, 5529–5532. [Google Scholar]
- McIntosh, G.; Noakes, M.; Royle, P.J.; Foster, P.R. Whole-grain rye and wheat foods and markers of bowel health in overweight middle-aged men. Am. J. Clin. Nutr. 2003, 77, 967–974. [Google Scholar] [CrossRef]
- Brahma, S.; Martinez, I.; Walter, J.; Clarke, J.; Gonzalez, T.; Menon, R.; Rose, D.J. Impact of die-tary pattern of the fecal donor on in vitro fermentation properties of whole grains and brans. J. Funct. Foods 2017, 29, 281–289. [Google Scholar] [CrossRef]
- Lupton, J.R.; Marchant, L.J. Independent effects of fiber and protein on colonic luminal am-monia concentration. J. Nutr. 1989, 119, 235–241. [Google Scholar] [CrossRef]
- Nemoto, H.; Kataoka, K.; Ishikawa, H.; Ikata, K.; Arimochi, H.; Iwasaki, T.; Ohnishi, Y.; Kuwahara, T.; Yasutomo, K. Reduced Diversity and Imbalance of Fecal Microbiota in Patients with Ulcerative Colitis. Dig. Dis. Sci. 2012, 57, 2955–2964. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, K.; Ohashi, Y.; Kawasumi, K.; Terada, A.; Fujisawa, T. Effect of apple intake on fecal microbiota and metabolites in humans. Anaerobe 2010, 16, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.M.; Topping, D.L.; Christophersen, C.T.; Bird, A.R.; Lange, K.; Saunders, I.; Cobiac, L. Butyrate esterified to starch is released in the human gastrointestinal tract. Am. J. Clin. Nutr. 2011, 94, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Slavin, J.; Feirtag, J. Chicory inulin does not increase stool weight or speed up intestinal transit time in healthy male subjects. Food Funct. 2011, 2, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Tiihonen, K.; Ouwehand, A.C.; Rautonen, N. Effect of overweight on gastrointestinal micro-biology and immunology: Correlation with blood biomarkers. Br. J. Nutr. 2010, 103, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Kaukinen, K.; Collin, P.; Huhtala, H.; Mäki, M. Long-Term Consumption of Oats in Adult Celiac Disease Patients. Nutrients 2013, 5, 4380–4389. [Google Scholar] [CrossRef]
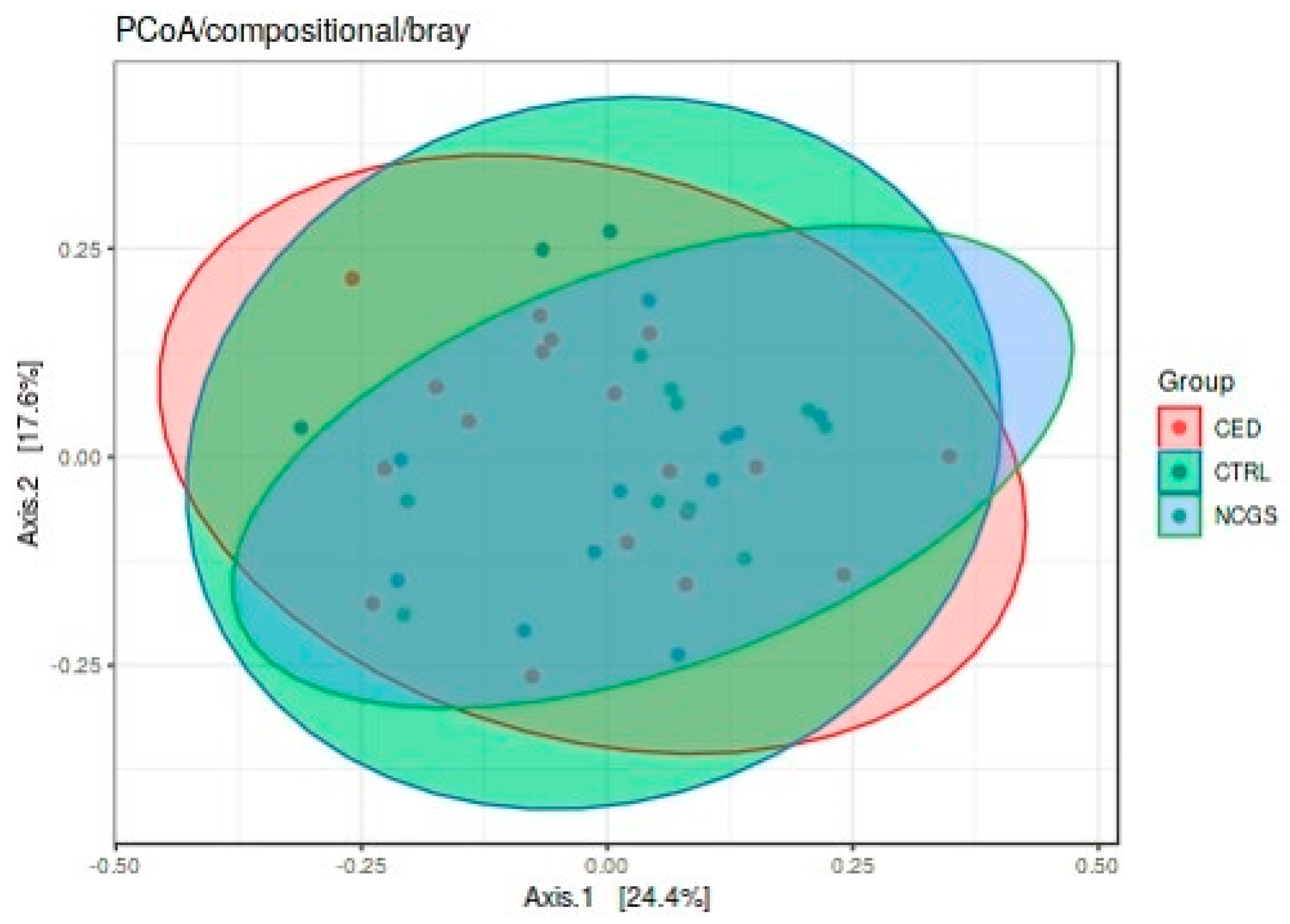

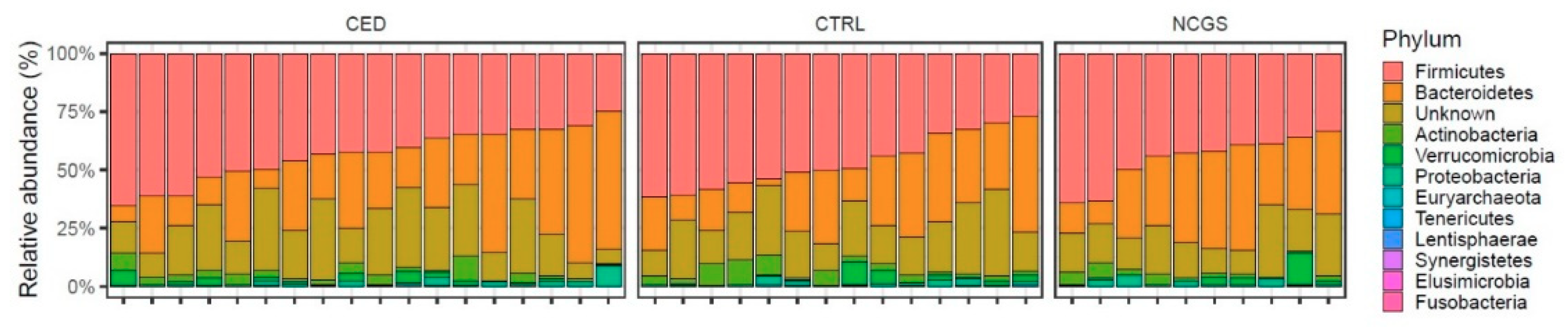
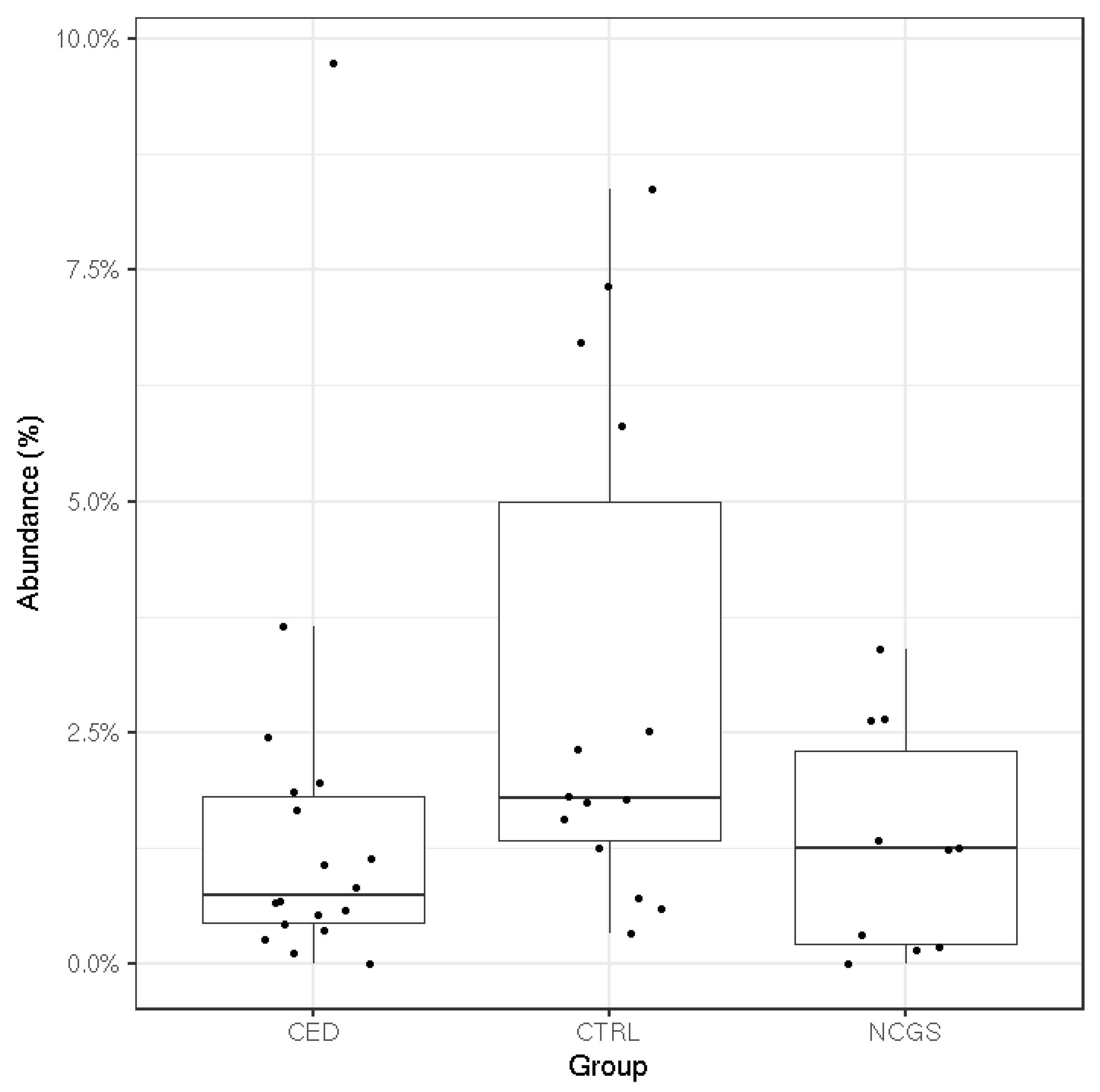
| GROUP | CeD (n = 19) | NCGS (n = 10) | CTRL (n = 14) | p-Value |
|---|---|---|---|---|
| Subjects (n) | 19 | 10 | 14 | n.s. |
| Male/Female 1 | 4/15 | 1/9 | 6/9 | n.s. |
| Age (year) 2 | 51 (24, 65) a | 34 (22, 61) b | 34 (24, 63) b | 0.020 |
| BMI (kg/m2) | 24.6 (3.2) | 23.0 (2.6) | 24.4 (2.6) | n.s. |
| Proteins (E %) | 17.1 (3.6) | 16.5 (3.4) | 15.8 (3.1) | n.s. |
| Carbohydrates (E %) | 41.9 (4.9) a,b | 40.3 (6.1) a | 45.8 (4.8) b | 0.045 |
| Fat (E %) | 36.4 (5.7) a,b | 41.0 (6.2) a | 34.7 (4.6) b | 0.025 |
| Dietary fiber (g) | 25.5 (9.1) | 27.6 (7.7) | 26.0 (7.4) | n.s. |
| Saccharose (g) | 46.2 (19.3) | 40.8 (12.3) | 52.5 (23.0) | n.s. |
| Diet Quality Index | 10.9 (1.7) | 10.2 (2.2) | 10.3 (1.5) | n.s. |
| CeD (n = 19) | NCGS (n = 10) | CTRL (n = 14) | ||||
|---|---|---|---|---|---|---|
| Concentration | % of Total SCFA | Concentration | % of Total SCFA | Concentration | % of Total SCFA | |
| Fecal acetic acid (µg) | 2144 (1228) | 63 a,b | 2149 (1205) | 59 a | 2789 (1473) | 71 b |
| Fecal propionic acid (µg) | 806 (607) | 23 | 948 (451) | 28 | 698 (521) | 19 |
| Fecal butyric acid (µg) | 337 (128) | 14 | 456 (258) | 13 | 424 (327) | 10 |
| Total SCFA (µg) | 3287 (1786) | 3553 (1680) | 3912 (2072) | |||
| Fecal ammonia (µmol) | 18.0 (6.5) | 18.5 (4.8) | 15.7 (7.2) | |||
| Fecal β-glucuronidase (U) | 30.0 (15.0) | 25.9 (15.0) | 29.9 (18.0) | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nylund, L.; Hakkola, S.; Lahti, L.; Salminen, S.; Kalliomäki, M.; Yang, B.; Linderborg, K.M. Diet, Perceived Intestinal Well-Being and Compositions of Fecal Microbiota and Short Chain Fatty Acids in Oat-Using Subjects with Celiac Disease or Gluten Sensitivity. Nutrients 2020, 12, 2570. https://doi.org/10.3390/nu12092570
Nylund L, Hakkola S, Lahti L, Salminen S, Kalliomäki M, Yang B, Linderborg KM. Diet, Perceived Intestinal Well-Being and Compositions of Fecal Microbiota and Short Chain Fatty Acids in Oat-Using Subjects with Celiac Disease or Gluten Sensitivity. Nutrients. 2020; 12(9):2570. https://doi.org/10.3390/nu12092570
Chicago/Turabian StyleNylund, Lotta, Salla Hakkola, Leo Lahti, Seppo Salminen, Marko Kalliomäki, Baoru Yang, and Kaisa M. Linderborg. 2020. "Diet, Perceived Intestinal Well-Being and Compositions of Fecal Microbiota and Short Chain Fatty Acids in Oat-Using Subjects with Celiac Disease or Gluten Sensitivity" Nutrients 12, no. 9: 2570. https://doi.org/10.3390/nu12092570
APA StyleNylund, L., Hakkola, S., Lahti, L., Salminen, S., Kalliomäki, M., Yang, B., & Linderborg, K. M. (2020). Diet, Perceived Intestinal Well-Being and Compositions of Fecal Microbiota and Short Chain Fatty Acids in Oat-Using Subjects with Celiac Disease or Gluten Sensitivity. Nutrients, 12(9), 2570. https://doi.org/10.3390/nu12092570







