Identifying Interactions between Dietary Sodium, Potassium, Sodium–Potassium Ratios, and FGF5 rs16998073 Variants and Their Associated Risk for Hypertension in Korean Adults
Abstract
1. Introduction
2. Materials and Methods
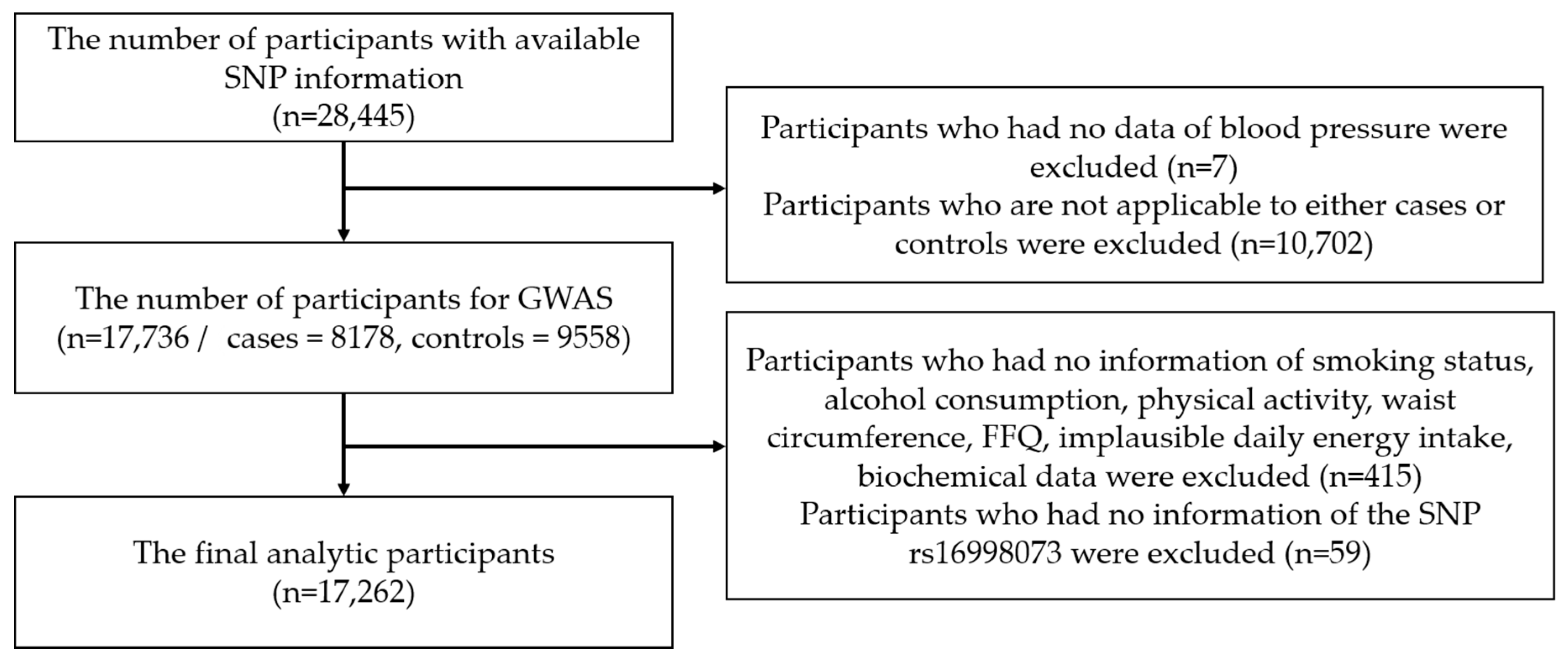
2.1. Data Source and Study Population
2.2. General Characteristics, Anthropometric, and Biochemical Variables
2.3. Dietary Assessment
2.4. Ascertainment of Hypertension
2.5. Genome-Wide Genotyping and Quality Control
2.6. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kearney, P.M.; Whelton, M.; Reynolds, K.; Muntner, P.; Whelton, P.K.; He, J. Global burden of hypertension: Analysis of worldwide data. Lancet 2005, 365, 217–223. [Google Scholar] [CrossRef]
- World Health Organization. WHO publishes definitive atlas on global heart disease and stroke epidemic. Indian J. Med. Sci. 2004, 58, 405–406. [Google Scholar]
- Korean Centers for Disease Control and Prevention. 2017 National Health Statistics. Available online: https://knhanes.cdc.go.kr/knhanes/sub01/sub01_05.do#s5_02 (accessed on 5 June 2020).
- Kim, Y.-I.; Kim, S.T. National Health Insurance Statistical Yearbook; Health Insurance Review & Assessment Service, National Health Insurance Service: Wonju-si, Korea, 2019. [Google Scholar]
- Du, S.; Batis, C.; Wang, H.; Zhang, B.; Zhang, J.; Popkin, B.M. Understanding the patterns and trends of sodium intake, potassium intake, and sodium to potassium ratio and their effect on hypertension in China. Am. J. Clin. Nutr. 2014, 99, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.L.; Cogswell, M.E.; Zhao, L.; Terry, A.L.; Wang, C.Y.; Wright, J.; Coleman King, S.M.; Bowman, B.; Chen, T.C.; Merritt, R.; et al. Association Between Urinary Sodium and Potassium Excretion and Blood Pressure Among Adults in the United States: National Health and Nutrition Examination Survey, 2014. Circulation 2018, 137, 237–246. [Google Scholar] [CrossRef]
- Weaver, C.M. Potassium and health. Adv. Nutr. 2013, 4, 368s–377s. [Google Scholar] [CrossRef]
- Perez, V.; Chang, E.T. Sodium-to-potassium ratio and blood pressure, hypertension, and related factors. Adv. Nutr. 2014, 5, 712–741. [Google Scholar] [CrossRef]
- Sacks, F.M.; Svetkey, L.P.; Vollmer, W.M.; Appel, L.J.; Bray, G.A.; Harsha, D.; Obarzanek, E.; Conlin, P.R.; Miller, E.R., 3rd; Simons-Morton, D.G.; et al. Effects on blood pressure of reduced dietary sodium and the Dietary Approaches to Stop Hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N. Engl. J. Med. 2001, 344, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Qin, L.; Barnes, C.; Charisse, K.; Yi, T.; Zhang, X.; Ali, R.; Medina, P.P.; Yu, J.; Slack, F.J.; et al. FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2012, 2, 1684–1696. [Google Scholar] [CrossRef]
- Niu, W.; Zhang, Y.; Ji, K.; Gu, M.; Gao, P.; Zhu, D. Confirmation of top polymorphisms in hypertension genome wide association study among Han Chinese. Clin. Chim. Acta Int. J. Clin. Chem. 2010, 411, 1491–1495. [Google Scholar] [CrossRef]
- Xi, B.; Shen, Y.; Reilly, K.H.; Wang, X.; Mi, J. Recapitulation of four hypertension susceptibility genes (CSK, CYP17A1, MTHFR, and FGF5) in East Asians. Metabolism 2013, 62, 196–203. [Google Scholar] [CrossRef]
- Newton-Cheh, C.; Johnson, T.; Gateva, V.; Tobin, M.D.; Bochud, M.; Coin, L.; Najjar, S.S.; Zhao, J.H.; Heath, S.C.; Eyheramendy, S.; et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009, 41, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Ando, M.; Nakatochi, M.; Maruyama, S.; Yasuda, Y.; Honda, H.; Kuwatsuka, Y.; Kato, S.; Kondo, T.; Iwata, M.; et al. Association of interactions between dietary salt consumption and hypertension-susceptibility genetic polymorphisms with blood pressure among Japanese male workers. Clin. Exp. Nephrol. 2017, 21, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Kwock, C.K.; Kim, K.; Kim, J.; Yang, Y.J. Interaction between Single Nucleotide Polymorphism and Urinary Sodium, Potassium, and Sodium-Potassium Ratio on the Risk of Hypertension in Korean Adults. Nutrients 2017, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.H.; Shin, C. Interaction according to urinary sodium excretion level on the association between ATP2B1 rs17249754 and incident hypertension: The Korean genome epidemiology study. Clin. Exp. Hypertens. (N. Y. 1993) 2016, 38, 352–358. [Google Scholar] [CrossRef]
- The Korean Nutrition Society. Recommended Dietary Allowances for Koreans, 7th ed.; The Korean Nutrition Society: Seoul, Korea, 2000. [Google Scholar]
- Ahn, Y.; Kwon, E.; Shim, J.E.; Park, M.K.; Joo, Y.; Kimm, K.; Park, C.; Kim, D.H. Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur. J. Clin. Nutr. 2007, 61, 1435–1441. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.; Ahn, Y.O.; Paik, H.Y.; Ahn, Y.; Tokudome, Y.; Hamajima, N.; Inoue, M.; Tajima, K. Development of a food frequency questionnaire in Koreans. Asia Pac. J. Clin. Nutr. 2003, 12, 243–250. [Google Scholar]
- Kim, M.; Kim, M.; Yoo, H.J.; Yun, R.; Lee, S.H.; Lee, J.H. Estrogen-related receptor γ gene (ESRRG) rs1890552 A>G polymorphism in a Korean population: Association with urinary prostaglandin F2α concentration and impaired fasting glucose or newly diagnosed type 2 diabetes. Diabetes Metab. 2017, 43, 385–388. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Minister of Health and Welfare; The Korean Nutrition Society. Dietary Reference Intakes for Koreans 2015; Ministry of Health and Welfare: Sejong, Korea, 2016. [Google Scholar]
- He, F.J.; Li, J.; Macgregor, G.A. Effect of longer term modest salt reduction on blood pressure: Cochrane systematic review and meta-analysis of randomised trials. BMJ 2013, 346, f1325. [Google Scholar] [CrossRef]
- Bayorh, M.A.; Socci, R.R.; Eatman, D.; Wang, M.; Thierry-Palmer, M. The role of gender in salt-induced hypertension. Clin. Exp. Hypertens. (N. Y. 1993) 2001, 23, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Fahimi, S.; Singh, G.M.; Micha, R.; Khatibzadeh, S.; Engell, R.E.; Lim, S.; Danaei, G.; Ezzati, M.; Powles, J. Global sodium consumption and death from cardiovascular causes. N. Engl. J. Med. 2014, 371, 624–634. [Google Scholar] [CrossRef] [PubMed]
- D’Elia, L.; Barba, G.; Cappuccio, F.P.; Strazzullo, P. Potassium intake, stroke, and cardiovascular disease a meta-analysis of prospective studies. J. Am. Coll. Cardiol. 2011, 57, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- He, F.J.; MacGregor, G.A. Fortnightly review: Beneficial effects of potassium. BMJ 2001, 323, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kwock, C.K.; Yang, Y.J. The Effect of the Sodium to Potassium Ratio on Hypertension Prevalence: A Propensity Score Matching Approach. Nutrients 2016, 8, 482. [Google Scholar] [CrossRef] [PubMed]
- Huggins, C.E.; O’Reilly, S.; Brinkman, M.; Hodge, A.; Giles, G.G.; English, D.R.; Nowson, C.A. Relationship of urinary sodium and sodium-to-potassium ratio to blood pressure in older adults in Australia. Med. J. Aust. 2011, 195, 128–132. [Google Scholar] [CrossRef]
- Silva-Antonialli, M.M.; Tostes, R.C.; Fernandes, L.; Fior-Chadi, D.R.; Akamine, E.H.; Carvalho, M.H.; Fortes, Z.B.; Nigro, D. A lower ratio of AT1/AT2 receptors of angiotensin II is found in female than in male spontaneously hypertensive rats. Cardiovasc. Res. 2004, 62, 587–593. [Google Scholar] [CrossRef]
- Tran, L.T.; Yuen, V.G.; McNeill, J.H. The fructose-fed rat: A review on the mechanisms of fructose-induced insulin resistance and hypertension. Mol. Cell. Biochem. 2009, 332, 145–159. [Google Scholar] [CrossRef]
- Song, D.; Arikawa, E.; Galipeau, D.M.; Yeh, J.N.; Battell, M.L.; Yuen, V.G.; McNeill, J.H. Chronic estrogen treatment modifies insulin-induced insulin resistance and hypertension in ovariectomized rats. Am. J. Hypertens. 2005, 18, 1189–1194. [Google Scholar] [CrossRef][Green Version]
- Ramirez, L.A.; Sullivan, J.C. Sex Differences in Hypertension: Where We Have Been and Where We Are Going. Am. J. Hypertens. 2018, 31, 1247–1254. [Google Scholar] [CrossRef]
- Korc, M.; Friesel, R.E. The role of fibroblast growth factors in tumor growth. Curr. Cancer Drug Targets 2009, 9, 639–651. [Google Scholar] [CrossRef]
- Szebenyi, G.; Fallon, J.F. Fibroblast growth factors as multifunctional signaling factors. Int. Rev. Cytol. 1999, 185, 45–106. [Google Scholar]
- Ren, Y.; Jiao, X.; Zhang, L. Expression level of fibroblast growth factor 5 (FGF5) in the peripheral blood of primary hypertension and its clinical significance. Saudi J. Biol. Sci. 2018, 25, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Volzke, H.; Fung, G.; Ittermann, T.; Yu, S.; Baumeister, S.E.; Dorr, M.; Lieb, W.; Volker, U.; Linneberg, A.; Jorgensen, T.; et al. A new, accurate predictive model for incident hypertension. J. Hypertens. 2013, 31, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, M.H. Salt sensitivity of blood pressure in humans. Hypertension 1996, 27, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Rhee, M.Y.; Yang, S.J.; Oh, S.W.; Park, Y.; Kim, C.I.; Park, H.K.; Park, S.W.; Park, C.Y. Novel genetic variations associated with salt sensitivity in the Korean population. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2011, 34, 606–611. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, H.J.; Ji, S.M.; Kim, M.E.; Jigden, B.; Lim, J.E.; Oh, B. ANTXR2 is a potential causative gene in the genome-wide association study of the blood pressure locus 4q21. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2014, 37, 811–817. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, W.; Li, J.; Yang, L.; Cai, J. ANTXR2 Knock-Out Does Not Result in the Development of Hypertension in Rats. Am. J. Hypertens. 2017, 30, 182–187. [Google Scholar] [CrossRef][Green Version]

| No. | SNP | CHR | Minor Allele | MAF | Gene | Hypertension | SBP (n = 23,344) | DBP (n = 23,344) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Cases (n = 9558); Controls (n = 8178)) | ||||||||||||
| Cases | Controls | OR (95%CI) | Add P | Beta ± se | Add P | Beta ± se | Add P | |||||
| 1 | rs16998073 | 4 | T | 0.50 | 0.47 | FGF5 | 1.29 (1.23–1.35) | 8.02 × 10−25 | 1.14 ± 0.14 | 5.46 × 10−17 | 0.73 ± 0.09 | 1.95 × 10−15 |
| 2 | rs1458038 | 4 | T | 0.36 | 0.39 | 1.26 (1.21–1.33) | 2.93 × 10−22 | 1.05 ± 0.13 | 1.41 × 10−15 | 0.68 ± 0.09 | 2.77 × 10−14 | |
| 3 | rs77768175 | 12 | G | 0.41 | 0.37 | HECTD4 | 0.77 (0.72–0.82) | 5.89 × 10−16 | −0.9 ± 0.18 | 2.79 × 10−7 | −0.75 ± 0.12 | 2.92 × 10−10 |
| 4 | rs671 | 12 | A | 0.38 | 0.33 | ALDH2 | 0.77 (0.72–0.82) | 2.08 × 10−15 | −0.89 ± 0.18 | 3.99 × 10−7 | −0.74 ± 0.12 | 3.77 × 10−10 |
| 5 | rs11066280 | 12 | A | 0.23 | 0.20 | HECTD4 | 0.78 (0.74–0.84) | 3.15 × 10−14 | −0.83 ± 0.17 | 1.15 × 10−6 | −0.72 ± 0.12 | 4.86 × 10−10 |
| 6 | rs2074356 | 12 | A | 0.30 | 0.32 | HECTD4 | 0.78 (0.73–0.83) | 2.86 × 10−13 | −0.89 ± 0.18 | 1.14 × 10−6 | −0.72 ± 0.12 | 6.12 × 10−9 |
| 7 | rs36034102 | 4 | T | 0.32 | 0.35 | FGF5 | 1.22(1.15–1.29) | 3.20 × 10−12 | 0.87 ± 0.16 | 3.80 × 10−8 | 0.47 ± 0.11 | 9.21 × 10−6 |
| 8 | rs12229654 | 12 | G | 0.23 | 0.26 | 0.79 (0.74–0.84) | 4.25 × 10−25 | −0.9 ± 0.18 | 1.02 × 10−6 | −0.7 ± 0.12 | 1.52 × 10−8 | |
| 9 | rs11066453 | 12 | G | 0.24 | 0.26 | 0.79 (0.74–0.85) | 6.66 × 10−11 | −0.41 ± 0.19 | 3.41 × 10−2 | −0.42 ± 0.13 | 1.33 × 10−3 | |
| 10 | rs2072134 | 12 | A | 0.24 | 0.26 | OAS3 | 0.78 (0.73–0.84) | 1.278 × 10−10 | −0.49 ± 0.2 | 1.55 × 10−2 | −0.46 ± 0.14 | 7.10 × 10−4 |
| 11 | rs17249754 | 12 | A | 0.24 | 0.27 | ATP2B1 | 0.87 (0.83–0.91) | 4.40 × 10−9 | −0.72 ± 0.13 | 4.69 × 10−8 | −0.39 ± 0.09 | 1.12 × 10−5 |
| 12 | rs12413409 | 10 | A | 0.36 | 0.39 | CNNM2 | 0.85 (0.81–0.9) | 4.63 × 10−9 | −0.61 ± 0.15 | 3.73 × 10−5 | −0.33 ± 0.1 | 7.66 × 10−4 |
| 13 | rs16849273 | 2 | G | 0.36 | 0.39 | 0.87 (0.83–0.91) | 4.84 × 10−9 | −0.65 ± 0.13 | 9.37 × 10−7 | −0.36 ± 0.09 | 8.19 × 10−5 | |
| 14 | rs12231049 | 12 | G | 0.37 | 0.39 | MYL2 | 0.83 (0.78–0.89) | 7.76 × 10−9 | −0.62 ± 0.17 | 2.60 × 10−4 | −0.46 ± 0.11 | 6.10 × 10−5 |
| 15 | rs732998 | 10 | C | 0.16 | 0.18 | NT5C2 | 0.85 (0.81–0.9) | 8.48 × 10−9 | −0.59 ± 0.15 | 5.85 × 10−5 | −0.32 ± 0.1 | 1.24 × 10−3 |
| 16 | rs11191548 | 10 | C | 0.16 | 0.18 | CNNM2 | 0.86 (0.81–0.9) | 1.04 × 10−8 | −0.59 ± 0.15 | 6.14 × 10−5 | −0.33 ± 0.1 | 7.54 × 10−4 |
| 17 | rs2681492 | 12 | C | 0.13 | 0.15 | ATP2B1 | 0.87 (0.83–0.91) | 1.54 × 10−8 | −0.68 ± 0.13 | 2.48 × 10−7 | −0.38 ± 0.09 | 2.47 × 10−5 |
| 18 | rs3782889 | 12 | G | 0.14 | 0.17 | MYL2 | 0.84 (0.79–0.89) | 2.03 × 10−8 | −0.58 ± 0.17 | 6.59 × 10−5 | −0.43 ± 0.11 | 1.46 × 10−4 |
| 19 | rs167479 | 19 | T | 0.13 | 0.16 | RGL3 | 0.88 (0.84–0.92) | 2.37 × 10−8 | −0.51 ± 0.13 | 6.57 × 10−5 | −0.35 ± 0.09 | 5.39 × 10−5 |
| 20 | rs112735431 | 17 | A | 0.15 | 0.17 | RNF213, LOC100294362 | 1.91 (1.52–2.41) | 3.02 × 10−8 | 3.4 ± 0.65 | 1.85 × 10−7 | 1.54 ± 0.44 | 4.89 × 10−4 |
| 21 | rs1799998 | 8 | G | 0.16 | 0.18 | CYP11B2 | 0.87 (0.83–0.91) | 3.16 × 10−8 | −0.22 ± 0.14 | 1.04 × 10−1 | −0.08 ± 0.09 | 3.66 × 10−1 |
| 22 | rs11191580 | 10 | C | 0.11 | 0.13 | NT5C2 | 0.86 (0.82–0.91) | 3.39 × 10−8 | −0.57 ± 0.15 | 1.31 × 10−4 | −0.31 ± 0.1 | 1.73 × 10−3 |
| 23 | rs7136259 | 12 | T | 0.10 | 0.12 | ATP2B1 | 0.88 (0.84–0.92) | 4.89 × 10−8 | −0.63 ± 0.13 | 1.65 × 10−6 | −0.36 ± 0.09 | 4.52 × 10−5 |
| Men | Women | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HTN (n = 3575) | No HTN (n = 2280) | p Value | HTN (n = 4355) | No HTN (n = 7052) | p Value | ||||||
| Age (years) | 57.55 ± 7.89 | 53.66 ± 8.09 | <0.0001 | Age (years) | 57.35 ± 6.91 | 50.48 ± 7.08 | <0.0001 | ||||
| Smoking Status | <0.0001 | Smoking Status | <0.0001 | ||||||||
| Current | 796 | 22.30% | 768 | 33.70% | Current | 56 | 1.30% | 165 | 2.30% | ||
| Past | 1688 | 47.20% | 863 | 37.90% | Past | 48 | 1.00% | 73 | 1.00% | ||
| None | 1091 | 30.50% | 649 | 28.50% | None | 4251 | 97.60% | 6814 | 96.60% | ||
| Physical Activity | <0.0001 | Physical Activity | 0.916 | ||||||||
| No | 1323 | 37.00% | 962 | 42.80% | No | 2052 | 47.10% | 3322 | 47.10% | ||
| Yes | 2252 | 63.00% | 1318 | 57.80% | Yes | 2303 | 52.90% | 3730 | 52.90% | ||
| Alcohol Consumption | <0.0001 | Alcohol Consumption | <0.0001 | ||||||||
| Current | 2596 | 72.60% | 1541 | 67.60% | Current | 1036 | 23.80% | 2281 | 32.30% | ||
| Past | 304 | 8.50% | 173 | 7.60% | Past | 95 | 2.20% | 106 | 1.50% | ||
| Never | 675 | 18.90% | 566 | 24.80% | Never | 3224 | 74.00% | 4665 | 66.20% | ||
| BMI (kg/m2) | 25.22 ± 2.72 | 23.47 ± 2.54 | <0.0001 | BMI (kg/m2) | 24.96 ± 3.04 | 22.74 ± 2.58 | <0.0001 | ||||
| Waist Circumference | 87.83 ± 7.33 | 83.24 ± 7.20 | <0.0001 | Waist Circumference | 81.85 ± 8.05 | 75.64 ± 7.47 | <0.0001 | ||||
| Na Intake (mg/day) | 2667.27 ± 1468.11 | 2624.73 ± 1490.0.4 | 0.169 | Na Intake (mg/day) | 2347.85 ± 1328.80 | 2384.80 ± 1314.68 | 0.058 | ||||
| K Intake (mg/day) | 2244.55 ± 945.03 | 2248.00 ± 962.94 | 0.858 | K Intake (mg/day) | 2168.86 ± 971.31 | 2262.50 ± 1006.48 | <0.0001 | ||||
| Na–K Ratio | 1.18 ± 0.39 | 1.16 ± 0.38 | 0.042 | Na–K Ratio | 1.08 ± 0.39 | 1.06 ± 0.36 | 0.004 | ||||
| Triglyceride (mg/dL) | 160.64 ± 111.76 | 128.03 ± 88.36 | <0.0001 | Triglyceride (mg/dL) | 134.82 ± 87.20 | 100.17 ± 62.48 | <0.0001 | ||||
| HDL Cholesterol (mg/dL) | 48.13 ± 11.42 | 49.24 ± 11.84 | <0.0001 | HDL Cholesterol (mg/dL) | 52.80 ± 12.42 | 56.20 ± 12.77 | <0.0001 | ||||
| Total Cholesterol (mg/dL) | 189.85 ± 34.95 | 189.61 ± 33.99 | 0.670 | Total Cholesterol (mg/dL) | 201.02 ± 36.25 | 195.54 ± 34.67 | <0.0001 | ||||
| Men | Women | ||||||
|---|---|---|---|---|---|---|---|
| Na intake | Na intake | ||||||
| <2000 mg | ≥2000 mg | p-interaction | <2000 mg | ≥2000 mg | p-interaction | ||
| rs16998073 | <0.0001 | rs16998073 | <0.0001 | ||||
| AA (wild type) | 1 (ref.) | 1.11 (0.92–1.34) | AA (wild type) | 1 (ref.) | 0.90 (0.78–1.04) | ||
| AT | 1.51 (1.24–1.85) | 1.62 (1.35–1.95) | AT | 1.22 (1.05–1.40) | 1.25 (1.09–1.43) | ||
| TT | 2.04 (1.48–2.83) | 2.41 (1.84–3.15) | TT | 1.67 (1.35–2.07) | 1.33 (1.09–1.62) | ||
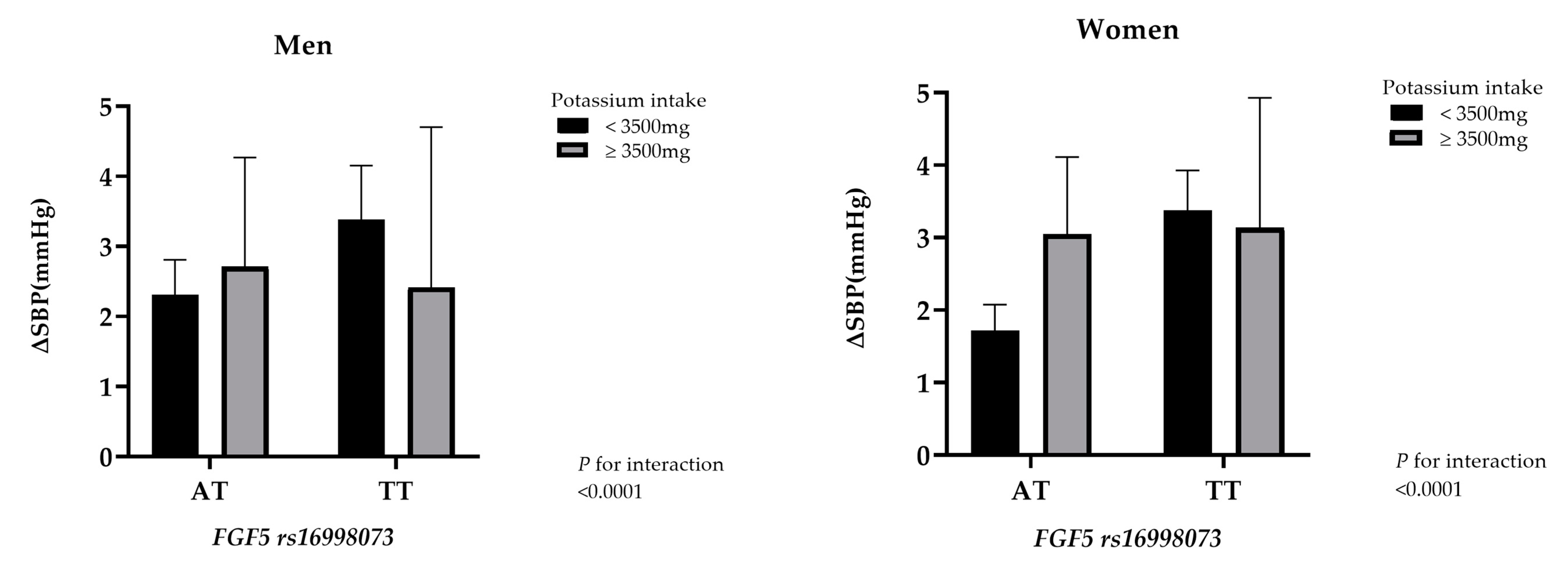
| K intake | K intake | ||||||
| <3500 mg | ≥3500 mg | p-interaction | <3500 mg | ≥3500 mg | p-interaction | ||
| rs16998073 | <0.0001 | rs16998073 | <0.0001 | ||||
| AA (wild type) | 1 (ref.) | 0.99 (0.75–1.34) | AA (wild type) | 1 (ref.) | 0.75 (0.58–0.95) | ||
| AT | 1.48 (1.30–1.68) | 1.48 (1.09–2.01) | AT | 1.27 (1.15–1.41) | 1.32 (1.05–1.65) | ||
| TT | 2.15 (1.74–2.64) | 1.85 (0.98–3.48) | TT | 1.55 (1.34–1.80) | 1.19 (0.77–1.84) | ||
| Men | Women | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Na–K Ratio | Na–K ratio | ||||||||
| Tertile 1 | Tertile 2 | Tertile 3 | p-Interaction | Tertile 1 | Tertile 2 | Tertile 3 | p-Interaction | ||
| rs16998073 | <0.0001 | rs16998073 | <0.0001 | ||||||
| AA (wild type) | 1 (ref.) | 1.17 (0.94–1.46) | 1.34 (1.08–1.67) | AA (wild type) | 1 (ref.) | 1.03 (0.87–1.22) | 1.02 (0.56–1.20) | ||
| AT | 1.55 (1.25–1.91) | 1.86 (1.80–2.31) | 1.80 (1.45–2.24) | AT | 1.34 (1.14–1.58) | 1.24 (1.05–1.46) | 1.42 (1.21–1.68) | ||
| TT | 2.26 (1.61–3.18) | 3.03 (2.14–4.29) | 2.12 (1.55–3.12) | TT | 1.82 (1.43–2.33) | 1.56 (1.22–1.99) | 1.40 (1.09–1.79) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.; Jin, H.-S.; Kim, S.-S.; Shin, D. Identifying Interactions between Dietary Sodium, Potassium, Sodium–Potassium Ratios, and FGF5 rs16998073 Variants and Their Associated Risk for Hypertension in Korean Adults. Nutrients 2020, 12, 2121. https://doi.org/10.3390/nu12072121
Jeong H, Jin H-S, Kim S-S, Shin D. Identifying Interactions between Dietary Sodium, Potassium, Sodium–Potassium Ratios, and FGF5 rs16998073 Variants and Their Associated Risk for Hypertension in Korean Adults. Nutrients. 2020; 12(7):2121. https://doi.org/10.3390/nu12072121
Chicago/Turabian StyleJeong, Hyeyun, Hyun-Seok Jin, Sung-Soo Kim, and Dayeon Shin. 2020. "Identifying Interactions between Dietary Sodium, Potassium, Sodium–Potassium Ratios, and FGF5 rs16998073 Variants and Their Associated Risk for Hypertension in Korean Adults" Nutrients 12, no. 7: 2121. https://doi.org/10.3390/nu12072121
APA StyleJeong, H., Jin, H.-S., Kim, S.-S., & Shin, D. (2020). Identifying Interactions between Dietary Sodium, Potassium, Sodium–Potassium Ratios, and FGF5 rs16998073 Variants and Their Associated Risk for Hypertension in Korean Adults. Nutrients, 12(7), 2121. https://doi.org/10.3390/nu12072121





