Plasma Metabolome Alterations Associated with Extrauterine Growth Restriction
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection and Processing
2.3. Untargeted Metabolomics Analysis
2.3.1. Chemicals
2.3.2. Metabolite Extraction
2.3.3. Separation and Detection
2.3.4. Untargeted Metabolomics by LC-TOF-MS
2.3.5. Untargeted Metabolomics by GC-Q-MS
2.3.6. Untargeted Metabolomics by CE-TOF-MS
2.4. Data Management
2.4.1. Metabolomics Data Processing
2.4.2. Quality Assurance Procedure
2.5. Data Pre-Treatment
2.6. Statistical Analysis
2.7. Metabolite Identification
3. Results
3.1. Clinical Characteristics of the Study Subjects
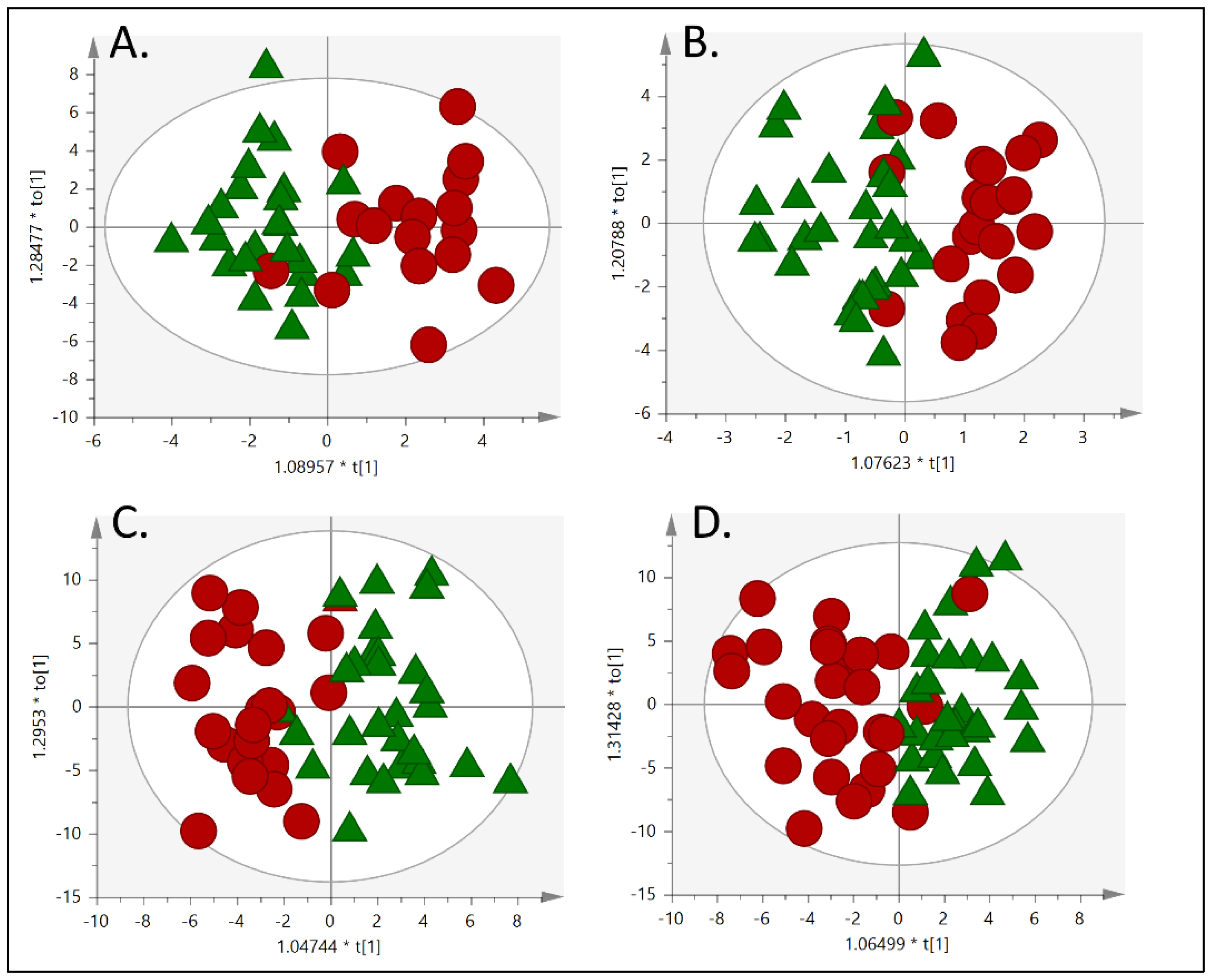
3.2. Metabolic Fingerprinting
4. Discussion
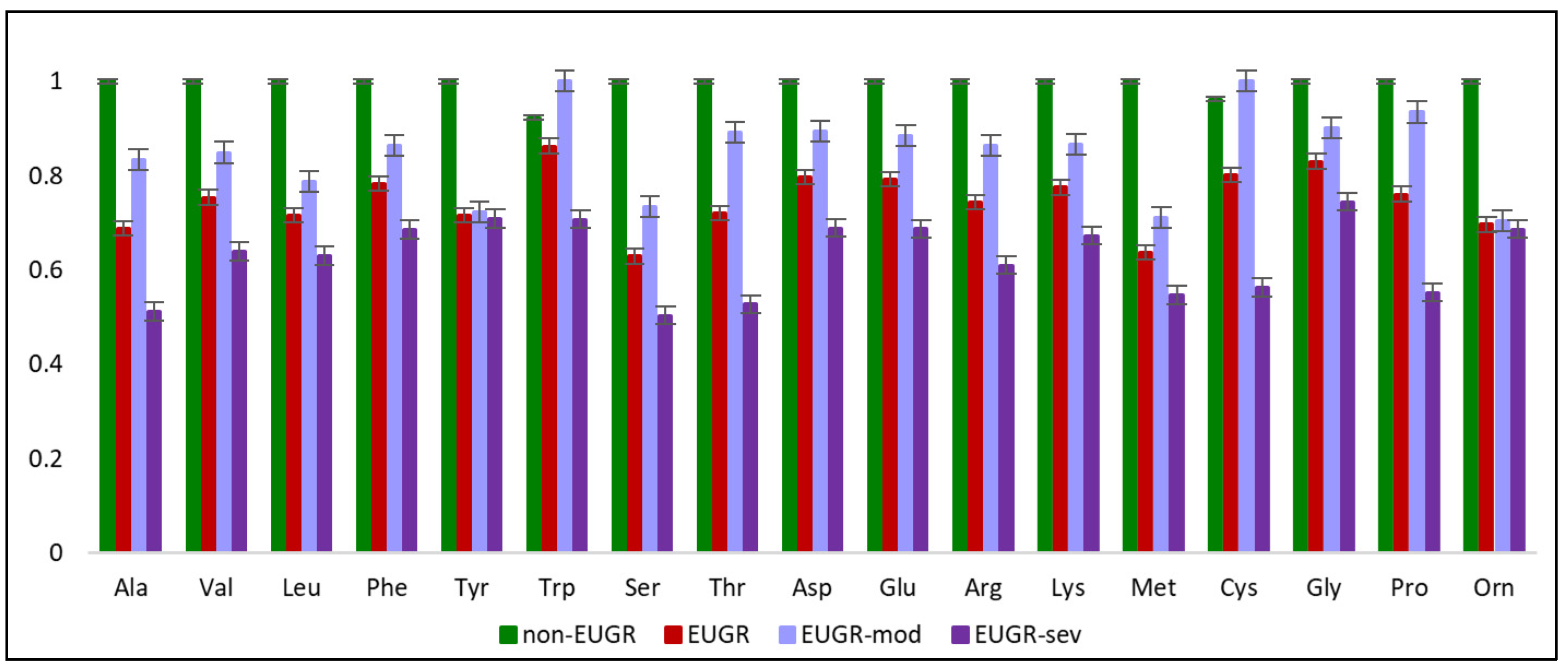
4.1. EUGR is Associated With Disrupted Amino Acid Metabolism
4.2. EUGR is Associated With Disrupted Lipid Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Izquierdo Renau, M.; Aldecoa-bilbao, V.; Balcells Esponera, C.; del Rey Hurtado de Mendoza, B.; Iriondo Sanz, M.; Iglesias Platas, I. Applying Methods for Postnatal Growth Assessment in the Clinical Setting: Evaluation in a Longitudinal Cohort of Very Preterm Infants. Nutrients 2019, 11, 2772. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, R.A.; Das, A.; Wrage, L.A.; Poindexter, B.B.; Higgins, R.D.; Stoll, B.J.; Oh, W. Early nutrition mediates the influence of severity of illness on extremely LBW infants. Pediatr. Res. 2011, 69, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, R.A. Growth in the Neonatal Intensive Care Unit Influences Neurodevelopmental and Growth Outcomes of Extremely Low Birth Weight Infants. Pediatrics 2006, 117, 1253–1261. [Google Scholar] [CrossRef]
- Neubauer, V.; Griesmaier, E.; Pehböck-Walser, N.; Pupp-Peglow, U.; Kiechl-Kohlendorfer, U. Poor postnatal head growth in very preterm infants is associated with impaired neurodevelopment outcome. Acta Paediatr. Int. J. Paediatr. 2013, 102, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Fumeaux, C.J.F.; Duerden, E.G.; Guo, T.; Foong, J.; Graz, M.B.; Hagmann, P.; Chakravarty, M.M.; Hüppi, P.S.; Beauport, L.; et al. Nutrient intake in the first two weeks of life and brain growth in preterm neonates. Pediatrics 2018, 141, e20172169. [Google Scholar] [CrossRef]
- Cormack, B.E.; Bloomfield, F.H. Increased protein intake decreases postnatal growth faltering in ELBW babies. Arch. Dis. Child. Fetal Neonatal Ed. 2013, 98, 399–404. [Google Scholar] [CrossRef]
- Andrews, E.T.; Ashton, J.J.; Pearson, F.; Beattie, R.M.; Johnson, M.J. Early postnatal growth failure in preterm infants is not inevitable. Arch Dis Child Fetal Neonatal Ed 2019, 104, F235–F241. [Google Scholar] [CrossRef]
- Izquierdo, M.; Martínez-Monseny, A.F.; Pociello, N.; Gonzalez, P.; Del Rio, R.; Iriondo, M.; Iglesias-Platas, I. Changes in Parenteral Nutrition during the First Week of Life Influence Early but Not Late Postnatal Growth in Very Low-Birth-Weight Infants. Nutr. Clin. Pract. 2016, 31, 666–672. [Google Scholar] [CrossRef]
- Stevens, T.P.; Shields, E.; Campbell, D.; Combs, A.; Horgan, M.; La Gamma, E.F.; Xiong, K.N.; Kacica, M. Variation in Enteral Feeding Practices and Growth Outcomes among Very Premature Infants: A Report from the New York State Perinatal Quality Collaborative. Am. J. Perinatol. 2015, 33, 9–19. [Google Scholar]
- Pereira-da-silva, L.; Virella, D.; Fusch, C. Nutritional Assessment in Preterm Infants: A Practical Approach in the NICU. Nutrients 2019, 11, 1999. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Khusial, R.D.; Cioffi, C.E.; Caltharp, S.A.; Krasinskas, A.M.; Alazraki, A.; Knight-Scott, J.; Cleeton, R.; Castillo-Leon, E.; Jones, D.P.; Pierpont, B.; et al. Development of a Plasma Screening Panel for Pediatric Nonalcoholic Fatty Liver Disease Using Metabolomics. Hepatol. Commun. 2019, 3, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Zorawski, M.; Skotnicki, M.; Zarzycki, W.; García, A.; Angulo, S.; Lorenzo, M.P.; Barbas, C.; Ramos, M.P. GC–MS based Gestational Diabetes Mellitus longitudinal study: Identification of 2-and 3-hydroxybutyrate as potential prognostic biomarkers. J. Pharm. Biomed. Anal. 2017, 144, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Revello, R.; Barbas, C.; Bartha, J.L. LC - MS-based metabolomics identification of novel biomarkers of chorioamnionitis and its associated perinatal neurological damage. J. Proteome Res. 2015, 14, 1432–1444. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Martos-Moreno, G.; García, A.; Barrios, V.; Rupérez, F.J.; Chowen, J.A.; Barbas, C.; Argente, J. Insulin resistance in prepubertal obese children correlates with sex-dependent early onset metabolomic alterations. Int. J. Obes. 2016, 40, 1494–1502. [Google Scholar] [CrossRef]
- Rzehak, P.; Hellmuth, C.; Uhl, O.; Kirchberg, F.F.; Peissner, W.; Harder, U.; Grote, V.; Weber, M.; Xhonneux, A.; Langhendries, J.P.; et al. Rapid growth and childhood obesity are strongly associated with LysoPC(14:0). Ann. Nutr. Metab. 2014, 64, 294–303. [Google Scholar] [CrossRef]
- Leal-Witt, M.J.; Ramon-Krauel, M.; Samino, S.; Llobet, M.; Cuadras, D.; Jimenez-Chillaron, J.C.; Yanes, O.; Lerin, C. Untargeted metabolomics identifies a plasma sphingolipid-related signature associated with lifestyle intervention in prepubertal children with obesity. Int. J. Obes. 2018, 42, 72–78. [Google Scholar] [CrossRef]
- Vlaardingerbroek, H.; Roelants, J.A.; Rook, D.; Dorst, K.; Schierbeek, H.; Vermes, A.; Vermeulen, M.J.; van Goudoever, J.B.; van den Akker, C.H.P. Adaptive regulation of amino acid metabolism on early parenteral lipid and high-dose amino acid administration in VLBW infants - A randomized, controlled trial. Clin. Nutr. 2014, 33, 982–990. [Google Scholar] [CrossRef]
- Stewart, C.J.; Embleton, N.D.; Marrs, E.C.L.; Smith, D.P.; Fofanova, T.; Nelson, A.; Skeath, T.; Perry, J.D.; Petrosino, J.F.; Berrington, J.E.; et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017, 5, 75. [Google Scholar] [CrossRef]
- Stewart, C.J.; Embleton, N.D.; Marrs, E.C.L.; Smith, D.P.; Nelson, A.; Abdulkadir, B.; Skeath, T.; Petrosino, J.F.; Perry, J.D.; Berrington, J.E.; et al. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 2016, 4, 67. [Google Scholar] [CrossRef]
- Stewart, C.J.; Nelson, A.; Treumann, A.; Skeath, T.; Cummings, S.P.; Embleton, N.D.; Berrington, J.E. Metabolomic and proteomic analysis of serum from preterm infants with necrotising entercolitis and late-onset sepsis. Pediatr. Res. 2016, 79, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Morniroli, D.; Dessì, A.; Giannì, M.L.; Roggero, P.; Noto, A.; Atzori, L.; Lussu, M.; Fanos, V.; Mosca, F. Is the body composition development in premature infants associated with a distinctive nuclear magnetic resonance metabolomic profiling of urine? J. Matern. Neonatal Med. 2019, 32, 2310–2318. [Google Scholar] [CrossRef] [PubMed]
- Younge, N.E.; Newgard, C.B.; Cotten, C.M.; Goldberg, R.N.; Muehlbauer, M.J.; Bain, J.R.; Stevens, R.D.; O’Connell, T.M.; Rawls, J.F.; Seed, P.C.; et al. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019, 9, 8167. [Google Scholar] [CrossRef]
- Programa de Salut Maternoinfantil, Direcció General de Salut Pública, D. de S. Corbes de referència de pes, perímetre cranial i longitud en néixer de nounats d ’ embarassos únics, de bessons i de trigèmins a Catalunya; Prous Science, S. A.: Barcelona, Spain, 2008; ISBN 9788481242539. [Google Scholar]
- de Onis, M.; Martorell, R.; Garza, C.; Lartey, A.; Members of the WHO Multicentre Growth Reference Study Group. WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr. 2006, 95, 76–85. [Google Scholar]
- Garcia, A.; Barbas, C. Gas Chromatography-Mass Spectrometry (GC-MS)-Based Metabolomics. In Metabolic Profiling. Methods in Molecular Biology; Metz, T., Ed.; Springer: Clifton, NY, USA, 2011; pp. 191–204. [Google Scholar]
- Naz, S.; Garcia, A.; Rusak, M.; Barbas, C. Method development and validation for rat serum fingerprinting with CE-MS: Application to ventilator-induced-lung-injury study. Anal. Bioanal. Chem. 2013, 405, 4849–4858. [Google Scholar] [CrossRef]
- Villaseñor, A.; Garcia-perez, I.; García, A.; Posma, J.M.; Andreas, N.J.; Modi, N.; Holmes, E.; Barbas, C. phase extrac-tion, multiplatform analytical approach Breast milk metabolome characterization in a single phase extrac- tion, multiplatform analytical approach. Anal Chem. 2014, 86, 8245–8252. [Google Scholar] [CrossRef]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef]
- Kuligowski, J.; Sánchez-Illana, Á.; Sanjuán-Herráez, D.; Vento, M.; Quintás, G. Intra-batch effect correction in liquid chromatography-mass spectrometry using quality control samples and support vector regression (QC-SVRC). Analyst 2015, 140, 7810–7817. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Gil-de-la-Fuente, A.; Godzien, J.; Saugar, S.; Garcia-Carmona, R.; Badran, H.; Wishart, D.S.; Barbas, C.; Otero, A. CEU Mass Mediator 3.0: A Metabolite Annotation Tool. J. Proteome Res. 2019, 18, 797–802. [Google Scholar] [CrossRef]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Glenn Tarcea, V.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A BCAA Related Metabolic Signature that differentiates obese and lean humans contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C. The potential risks and benefits of insulin treatment in hyperglycaemic preterm neonates. Early Hum. Dev. 2015, 91, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Beardsall, K.; Vanhaesebrouck, S.; Frystyk, J.; Ogilvy-Stuart, A.L.; Vanhole, C.; van Weissenbruch, M.; Midgley, P.; Thio, M.; Cornette, L.; Gill, B.; et al. Relationship between insulin-like growth factor I levels, early insulin treatment, and clinical outcomes of very low birth weight infants. J. Pediatr. 2014, 164, 1038–1044.e1. [Google Scholar] [CrossRef]
- Hellström, A.; Ley, D.; Hansen-Pupp, I.; Hallberg, B.; Ramenghi, L.A.; Löfqvist, C.; Smith, L.E.H.; Hård, A.L. Role of Insulinlike Growth Factor 1 in Fetal Development and in the Early Postnatal Life of Premature Infants. Am. J. Perinatol. 2016, 33, 1067–1071. [Google Scholar] [CrossRef]
- Ottestad, I.; Ulven, S.M.; Oyri, L.K.L.; Sandvei, K.S.; Gjevestad, G.O.; Bye, A.; Sheikh, N.A.; Biong, A.S.; Andersen, L.F.; Holven, K.B. Reduced plasma concentration of branched-chain amino acids in sarcopenic older subjects: A cross-sectional study. Br. J. Nutr. 2018, 120, 445–453. [Google Scholar] [CrossRef]
- Yamada, M.; Kimura, Y.; Ishiyama, D.; Nishio, N.; Tanaka, T.; Ohji, S.; Otobe, Y.; Koyama, S.; Sato, A.; Suzuki, M.; et al. Plasma Amino Acid Concentrations Are Associated with Muscle Function in Older Japanese Women. J. Nutr. Heal. Aging 2018, 22, 819–823. [Google Scholar] [CrossRef]
- Johnson, M.J.; Wootton, S.A.; Leaf, A.A.; Jackson, A.A. Preterm Birth and Body Composition at Term Equivalent Age: A Systematic Review and Meta-analysis. Pediatrics 2012, 130, E640–E649. [Google Scholar] [CrossRef]
- Al-Theyab, N.A.; Donovan, T.J.; Eiby, Y.A.; Colditz, P.B.; Lingwood, B.E. Fat trajectory after birth in very preterm infants mimics healthy term infants. Pediatr. Obes. 2019, 14, 1–9. [Google Scholar] [CrossRef]
- Strømmen, K.; Haag, A.; Moltu, S.J.; Veierød, M.B.; Blakstad, E.W.; Nakstad, B.; Almaas, A.N.; Brække, K.; Rønnestad, A.E.; Daniel, H.; et al. Enhanced nutrient supply to very low birth weight infants is associated with higher blood amino acid concentrations and improved growth. Clin. Nutr. ESPEN 2017, 18, 16–22. [Google Scholar] [CrossRef]
- Scott, P.H.; Berger, H.M.; Wharton, B.A. Growth velocity and plasma amino acids in the newborn. Pediatr. Res. 1985, 19, 446–450. [Google Scholar] [CrossRef]
- Prentice, P.; Koulman, A.; Matthews, L.; Acerini, C.L.; Ong, K.K.; Dunger, D.B. Lipidomic analyses, breast- and formula-feeding, and growth in infants. J. Pediatr. 2015, 166, 276–281.e6. [Google Scholar] [CrossRef] [PubMed]
- Koulman, A.; Prentice, P.; Wong, M.C.Y.; Matthews, L.; Bond, N.J.; Eiden, M.; Griffin, J.L.; Dunger, D.B. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics 2014, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cilla, A.; Diego Quintaes, K.; Barberá, R.; Alegría, A. Phospholipids in Human Milk and Infant Formulas: Benefits and Needs for Correct Infant Nutrition. Crit. Rev. Food Sci. Nutr. 2016, 56, 1880–1892. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.B.; Færgeman, N.J. Sphingolipids: Membrane microdomains in brain development, function and neurological diseases. Open Biol. 2017, 7, pii: 170069. [Google Scholar] [CrossRef]
- Liu, H.; Radlowski, E.C.; Conrad, M.S.; Dilger, R.N.; Johnson, R.W. Early Supplementation of Phospholipids and Gangliosides Affects Brain and Cognitive Development in Neonatal Piglets Differences in cognitive development: Breast fed ( BF ) vs Formula fed ( FF ) BF children show higher cognitive development scores compa. J. Nutr. 2014, 2014,, 1903–1909. [Google Scholar] [CrossRef]
- Belfort, M.; Gillman, M.; Buka, S.; McCormick, M. Preterm infant linear growth and adiposity gain: Tradeoffs for later weight status, and IQ. J. Pediatr. 2013, 163, 1–15. [Google Scholar] [CrossRef]
- Roggero, P.; Giannì, M.L.; Amato, O.; Orsi, A.; Piemontese, P.; Cosma, B.; Morlacchi, L.; Mosca, F. Postnatal growth failure in preterm infants: Recovery of growth and body composition after term. Early Hum. Dev. 2008, 84, 555–559. [Google Scholar] [CrossRef]
- McCoin, C.S.; Knotts, T.A.; Adams, S.H. Acylcarnitines-old actors auditioning for new roles in metabolic physiology. Nat. Rev. Endocrinol. 2015, 11, 617–625. [Google Scholar] [CrossRef]
- Luo, L.; Aubrecht, J.; Li, D.; Warner, R.L.; Johnson, K.J.; Kenny, J.; Colangelo, J.L. Assessment of serum bile acid profiles as biomarkers of liver injury and liver disease in humans. PLoS ONE 2018, 13, e0193824. [Google Scholar] [CrossRef]
- Satrom, K.; Gourley, G. Cholestasis in Preterm Infants. Clin. Perinatol. 2016, 43, 355–373. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [PubMed]

| Normally Grown (n = 29) | EUGR (n = 22) | p-Value | ||
|---|---|---|---|---|
| At birth | Gestational age (weeks) | 29.8 (1.8) | 29.4 (1.9) | 0.390 |
| Birthweight (g) Z-score | 1402 (294) 0.42 (0.68) | 1223 (235) –0.34 (0.74) | 0.023 0.027 | |
| Length at birth (cm) Z-score | 39.2 (3.0) 0.20 (0.80) | 37.9 (2.6) –0.17 (0.76) | 0.091 0.109 | |
| Head circumference at birth (cm) Z-score | 26.9 (1.9) –0.09 (0.57) | 25.9 (2.0) –0.55 (0.74) | 0.084 0.023 | |
| At discharge | Postmenstrual age (weeks) | 36.8 (2.1) | 37.8 (2.2) | 0.094 |
| Weight at discharge (g) Z-score Fall in weight z-score from birth | 2399 (353) –0.83 (0.34) –1.25 (0.61) | 2216 (444) –1.91 (0.43) –1.88 (0.61) | 0.108 <0.0001 0.001 | |
| Length at discharge (cm) Z-score Fall in length z-score from birth | 45.5 (1.9) –0.78 (0.69) –0.97 (0.90) | 45.3 (2.5) –38 (0.95) –1.14 (0.96) | 0.796 0.012 0.535 | |
| Head circumference at discharge (cm) Z-score Fall in HC z-score from birth | 32.2 (1.6) –0.52 (0.58) –0.42 (0.67) | 32.0 (1.4) –1.66 (1.08) –1.07 (1.19) | 0.424 <0.0001 0.033 | |
| Nutrition | Parenteral nutrition (days) | 10.1 (6.9) | 12.3 (7.8) | 0.293 |
| Age at first full enteral feeds (days) | 10.6 (6.7) | 11.8 (4.4) | 0.461 | |
| Average parenteral nutrition 1st week Energy (kcal/kg/day) Protein (g/kg/day) Lipids (g/kg/day) Protein/energy ratio (g/100kcal) | 61.3 (12.2) 2.4 (0.7) 1.7 (0.7) 3.5 (0.5) | 71.1 (11.1) 2.8 (0.5) 2.1 (0.5) 3.6 (0.3) | 0.014 0.030 0.018 0.100 | |
| Average enteral nutrition 1st week Volume (ml/kg/day) Calculated energy (kcal/kg/day) Calculated protein (g/kg/day) | 33.3 (21.2) 24.3 (16.0) 0.6 (0.4) | 21.3 (14.4) 15.2 (10.2) 0.3 (0.2) | 0.021 0.017 0.019 | |
| Global nutrition 1st week (PN + enteral) Energy (kcal/kg/day) Protein (g/kg/day) | 85.6 (8.9) 3.0 (0.5) | 86.3 (8.5) 3.2 (0.3) | 0.776 0.163 | |
| Average parenteral nutrition 2nd week Energy (kcal/kg/day) Protein (g/kg/day) Lipids (g/kg/day) Protein/energy ratio (g/100kcal) | 20.3 (30.4) 0.8 (1.2) 0.5 (0.9) 2.9 (0.5) | 31.4 (30.0) 1.1 (1.1) 0.7 (0.9) 2.9 (0.5) | 0.203 0.445 0.326 0.678 | |
| Average enteral nutrition 2nd week Volume (mL/kg/day) Calculated energy (kcal/kg/day) Calculated protein (g/kg/day) | 123.0 (47.4) 100.2 (41.6) 2.7 (1.3) | 97.9 (42.7) 77.6 (37.1) 2.1 (1.2) | 0.056 0.050 0.083 | |
| Global nutrition 2nd week (PN + enteral) Energy (kcal/kg/day) Protein (g/kg/day) | 120.6 (14.8) 3.6 (0.7) | 109.0 (16.0) 3.2 (0.8) | 0.010 0.069 | |
| Feeding at discharge Own Mother´s Milk Mixed feeding Formula | 20 (69.0) 4 (13.8) 5 (17.2) | 17 (77.3) 5 (22.7) 0 (0.0) | – * – – | |
| Exclusive own´s mother milk at discharge | 20 (69.0) | 17 (77.3) | 0.510 | |
| Nutritional intake at discharge Milk volume (ml/kg/day) Calculated protein intake Per kilogram (g/kg/day) Total (g/day) Calculated energy intake (kcal/kg/day) | 170 (16) 2.3 (1.0) 5.5 (2.2) 127 (18) | 163 (23) 1.9 (0.9) 4.3 (2.0) 124 (17) | 0.222 0.143 0.049 0.520 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudzik, D.; Iglesias Platas, I.; Izquierdo Renau, M.; Balcells Esponera, C.; del Rey Hurtado de Mendoza, B.; Lerin, C.; Ramón-Krauel, M.; Barbas, C. Plasma Metabolome Alterations Associated with Extrauterine Growth Restriction. Nutrients 2020, 12, 1188. https://doi.org/10.3390/nu12041188
Dudzik D, Iglesias Platas I, Izquierdo Renau M, Balcells Esponera C, del Rey Hurtado de Mendoza B, Lerin C, Ramón-Krauel M, Barbas C. Plasma Metabolome Alterations Associated with Extrauterine Growth Restriction. Nutrients. 2020; 12(4):1188. https://doi.org/10.3390/nu12041188
Chicago/Turabian StyleDudzik, Danuta, Isabel Iglesias Platas, Montserrat Izquierdo Renau, Carla Balcells Esponera, Beatriz del Rey Hurtado de Mendoza, Carles Lerin, Marta Ramón-Krauel, and Coral Barbas. 2020. "Plasma Metabolome Alterations Associated with Extrauterine Growth Restriction" Nutrients 12, no. 4: 1188. https://doi.org/10.3390/nu12041188
APA StyleDudzik, D., Iglesias Platas, I., Izquierdo Renau, M., Balcells Esponera, C., del Rey Hurtado de Mendoza, B., Lerin, C., Ramón-Krauel, M., & Barbas, C. (2020). Plasma Metabolome Alterations Associated with Extrauterine Growth Restriction. Nutrients, 12(4), 1188. https://doi.org/10.3390/nu12041188





