Exercise Training-Induced PPARβ Increases PGC-1α Protein Stability and Improves Insulin-Induced Glucose Uptake in Rodent Muscles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Cell Lines
2.3. Insulin Responsiveness
2.4. Exercise Programs
2.5. DNA Constructs and Muscle Electroporation
2.6. Western Blot Analysis
2.7. Gene Expression Analysis
2.8. Glucose Uptake
2.9. Immunofluorescence Analysis for GLUT4 Localization
2.10. Statistical Analysis
3. Results
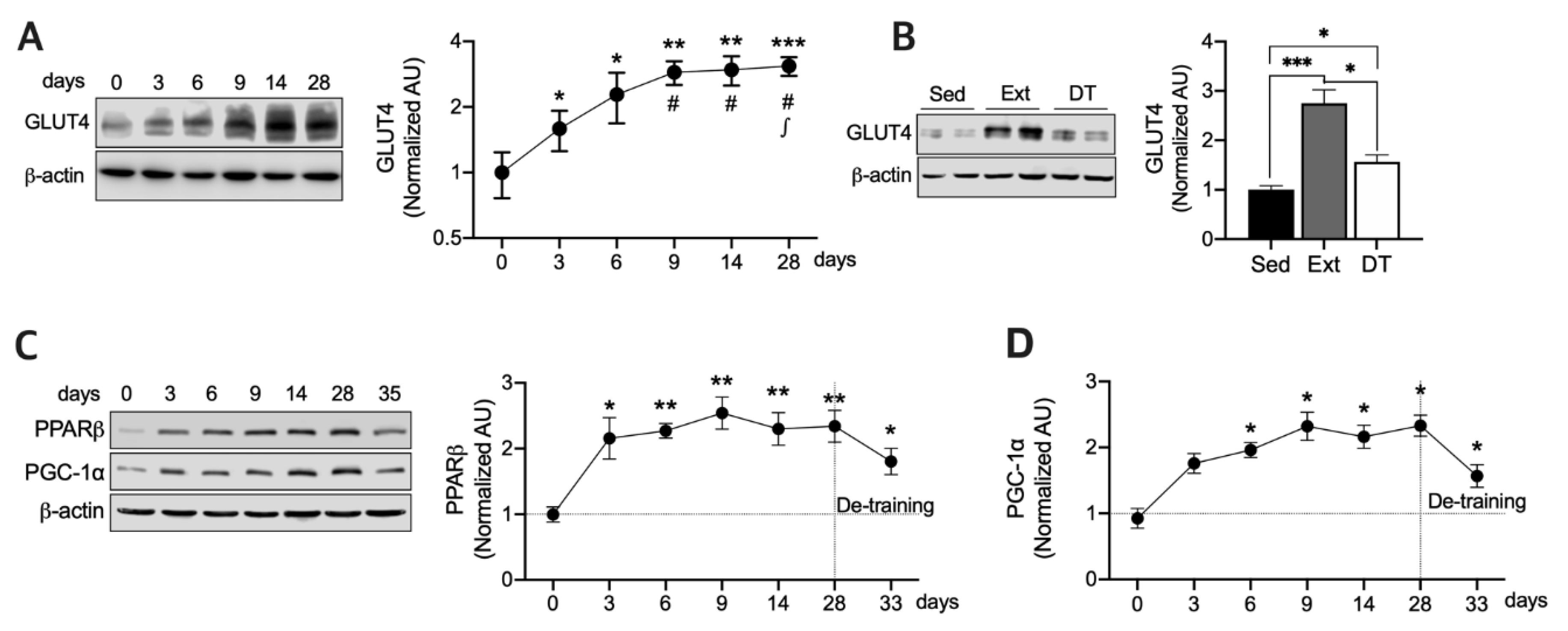
3.1. Exercise Training Upregulates GLUT4, PPARβ, and PGC-1a Protein Expression and those Half-Lives in Resting State
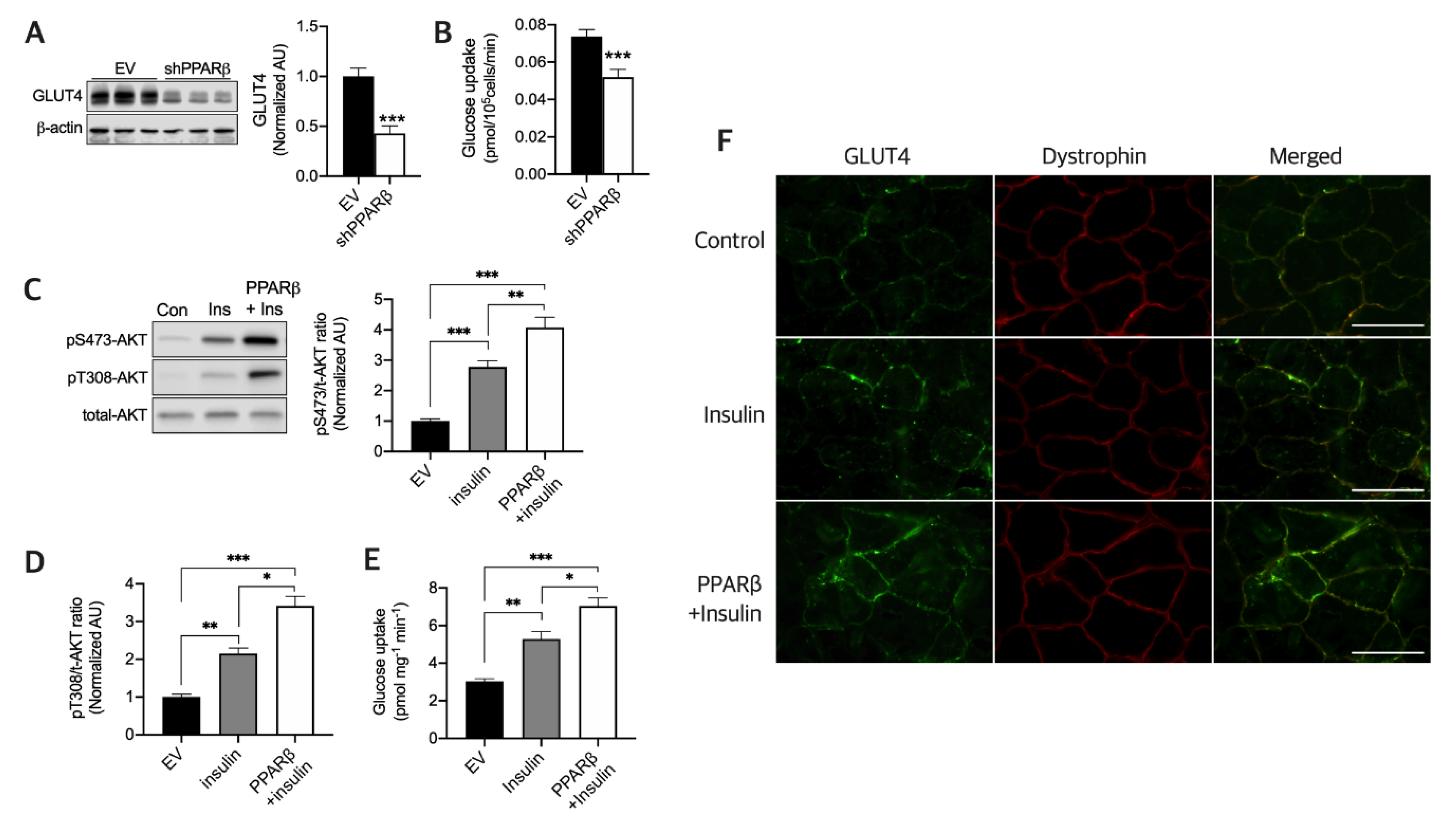
3.2. PPARβ Is Required to Maintain Normal Glucose Uptake and Maximal Insulin Sensitivity in Myotubes
3.3. A Post-Translational Cascade Induced by PPARβ Influences PGC-1α Protein Levels During and After Ext
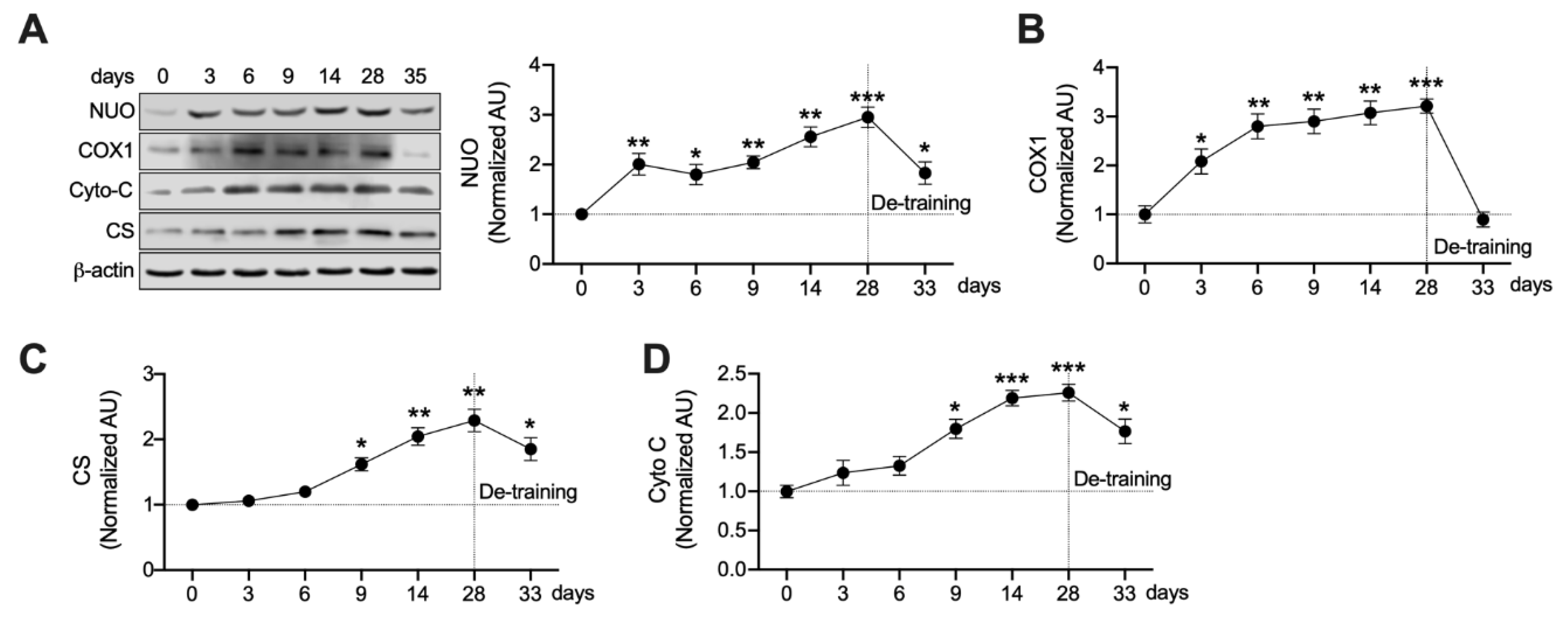
3.4. Mitochondrial Enzymes Are Differently Regulated by Ext
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Holloszy, J.O. A forty-year memoir of research on the regulation of glucose transport into muscle. Am. J. Physiol.-Endocrinol. Metab. 2003, 284, E453–E467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holloszy, J.O. Regulation by exercise of skeletal muscle content of mitochondria and GLUT4. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2008, 59, 5–18. [Google Scholar]
- Ren, J.M.; Semenkovich, C.F.; Gulve, E.A.; Gao, J.; Holloszy, J.O. Exercise induces rapid increases in GLUT4 expression, glucose transport capacity, and insulin-stimulated glycogen storage in muscle. J. Biol. Chem. 1994, 269, 14396–14401. [Google Scholar] [PubMed]
- Rodnick, K.J.; Henriksen, E.J.; James, D.E.; Holloszy, J.O. Exercise training, glucose transporters, and glucose transport in rat skeletal muscles. Am. J. Physiol. 1992, 262, C9–C14. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.H.; Hancock, C.R.; Han, D.H.; Holloszy, J.O.; Nair, K.S.; Dasari, S. AMPK and PPARbeta positive feedback loop regulates endurance exercise training-mediated GLUT4 expression in skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2019, 316, E931–E939. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.H.; Hancock, C.R.; Terada, S.; Higashida, K.; Holloszy, J.O.; Han, D.H. PPARbeta is essential for maintaining normal levels of pgc-1alpha and mitochondria and for the increase in muscle mitochondria induced by exercise. Cell Metab. 2017, 25, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Booth, F.W.; Holloszy, J.O. Cytochrome c turnover in rat skeletal muscles. J. Biol. Chem. 1977, 252, 416–419. [Google Scholar]
- Koh, J.H.; Johnson, M.L.; Dasari, S.; LeBrasseur, N.K.; Vuckovic, I.; Henderson, G.C.; Cooper, S.A.; Manjunatha, S.; Ruegsegger, G.N.; Shulman, G.I.; et al. TFAM enhances fat oxidation and attenuates high-fat diet-induced insulin resistance in skeletal muscle. Diabetes 2019, 68, 1552–1564. [Google Scholar] [CrossRef]
- Holloszy, J.O. Regulation of mitochondrial biogenesis and GLUT4 expression by exercise. Compr. Physiol. 2011, 1, 921–940. [Google Scholar] [CrossRef]
- Kim, S.H.; Koh, J.H.; Higashida, K.; Jung, S.R.; Holloszy, J.O.; Han, D.H. PGC-1alpha mediates a rapid, exercise-induced downregulation of glycogenolysis in rat skeletal muscle. J. Physiol. 2015, 593, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Bradley, H.; Shaw, C.S.; Worthington, P.L.; Shepherd, S.O.; Cocks, M.; Wagenmakers, A.J. Quantitative immunofluorescence microscopy of subcellular GLUT4 distribution in human skeletal muscle: Effects of endurance and sprint interval training. Physiol. Rep. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Baar, K.; Wende, A.R.; Jones, T.E.; Marison, M.; Nolte, L.A.; Chen, M.; Kelly, D.P.; Holloszy, J.O. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Higashida, K.; Kim, S.H.; Higuchi, M.; Holloszy, J.O.; Han, D.H. Normal adaptations to exercise despite protection against oxidative stress. Am. J. Physiol.-Endocrinol. Metab. 2011, 301, E779–E784. [Google Scholar] [CrossRef] [Green Version]
- Wende, A.R.; Huss, J.M.; Schaeffer, P.J.; Giguere, V.; Kelly, D.P. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: A mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell. Biol. 2005, 25, 10684–10694. [Google Scholar] [CrossRef] [Green Version]
- Trausch-Azar, J.; Leone, T.C.; Kelly, D.P.; Schwartz, A.L. Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{Akimoto, #5} via the N-terminal pathway. J. Biol. Chem. 2010, 285, 40192–40200. [Google Scholar] [CrossRef] [Green Version]
- Perry, C.G.; Lally, J.; Holloway, G.P.; Heigenhauser, G.J.; Bonen, A.; Spriet, L.L. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J. Physiol. 2010, 588, 4795–4810. [Google Scholar] [CrossRef]
- Hancock, C.R.; Han, D.H.; Higashida, K.; Kim, S.H.; Holloszy, J.O. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011, 25, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.X.; Zhang, C.L.; Yu, R.T.; Cho, H.K.; Nelson, M.C.; Bayuga-Ocampo, C.R.; Ham, J.; Kang, H.; Evans, R.M. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004, 2, e294. [Google Scholar] [CrossRef] [Green Version]
- Schuler, M.; Ali, F.; Chambon, C.; Duteil, D.; Bornert, J.M.; Tardivel, A.; Desvergne, B.; Wahli, W.; Chambon, P.; Metzger, D. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006, 4, 407–414. [Google Scholar] [CrossRef]
- Akimoto, T.; Pohnert, S.C.; Li, P.; Zhang, M.; Gumbs, C.; Rosenberg, P.B.; Williams, R.S.; Yan, Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 2005, 280, 19587–19593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, J.H.; Kim, H.S.; Ahn, N.Y.; Kim, K.J. PPARδ activation or overexpression attenuates PGC-1α protein degradation. Exerc. Sci. 2015, 24, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Marcus, R.L.; Addison, O.; LaStayo, P.C.; Hungerford, R.; Wende, A.R.; Hoffman, J.M.; Abel, E.D.; McClain, D.A. Regional muscle glucose uptake remains elevated one week after cessation of resistance training independent of altered insulin sensitivity response in older adults with type 2 diabetes. J. Endocrinol. Investig. 2013, 36, 111–117. [Google Scholar] [CrossRef]
- Ishiki, M.; Klip, A. Minireview: Recent developments in the regulation of glucose transporter-4 traffic: New signals, locations, and partners. Endocrinology 2005, 146, 5071–5078. [Google Scholar] [CrossRef]
- Haga, Y.; Ishii, K.; Suzuki, T. N-Glycosylation Is Critical for the Stability and Intracellular Trafficking of Glucose Transporter GLUT4. J. Biol. Chem. 2011, 286, 31320–31327. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Qi, Y.; Im, W. Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Sci. Rep. 2015, 5, 8926. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-S.; Holloszy, J.O.; Kim, K.; Koh, J.-H. Exercise Training-Induced PPARβ Increases PGC-1α Protein Stability and Improves Insulin-Induced Glucose Uptake in Rodent Muscles. Nutrients 2020, 12, 652. https://doi.org/10.3390/nu12030652
Park J-S, Holloszy JO, Kim K, Koh J-H. Exercise Training-Induced PPARβ Increases PGC-1α Protein Stability and Improves Insulin-Induced Glucose Uptake in Rodent Muscles. Nutrients. 2020; 12(3):652. https://doi.org/10.3390/nu12030652
Chicago/Turabian StylePark, Ju-Sik, John O. Holloszy, Kijin Kim, and Jin-Ho Koh. 2020. "Exercise Training-Induced PPARβ Increases PGC-1α Protein Stability and Improves Insulin-Induced Glucose Uptake in Rodent Muscles" Nutrients 12, no. 3: 652. https://doi.org/10.3390/nu12030652
APA StylePark, J.-S., Holloszy, J. O., Kim, K., & Koh, J.-H. (2020). Exercise Training-Induced PPARβ Increases PGC-1α Protein Stability and Improves Insulin-Induced Glucose Uptake in Rodent Muscles. Nutrients, 12(3), 652. https://doi.org/10.3390/nu12030652




