Metabolomic Associations with Serum Bone Turnover Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Study Design
2.2. Demographic and Clinical Information
2.3. Assessment of Serum P1NP and CTX
2.4. Plasma High-Resolution Metabolomics
2.5. Biostatistics and Bioinformatics
3. Results
3.1. Characteristics of the Study Population
3.2. Metabolomic Associations with P1NP
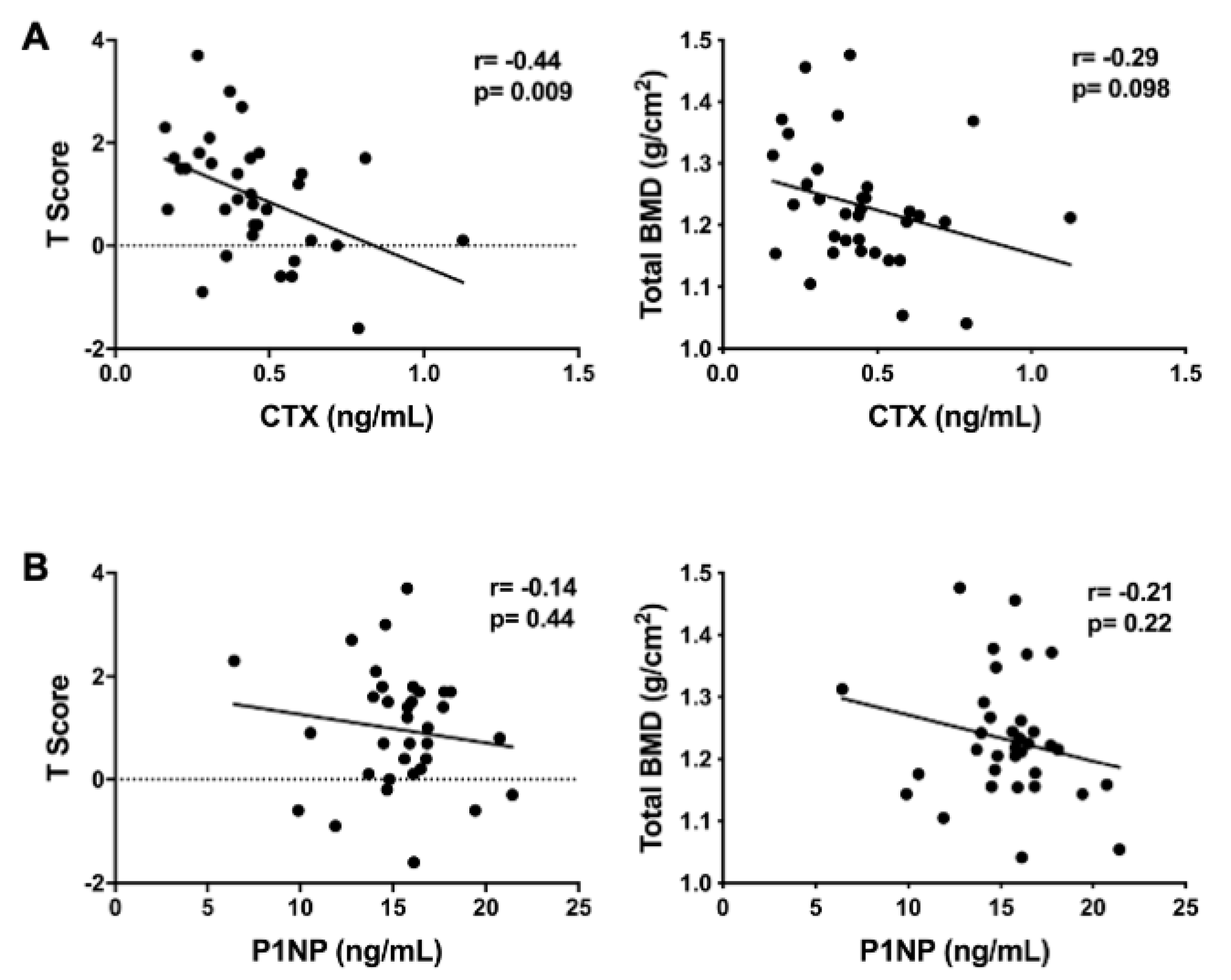
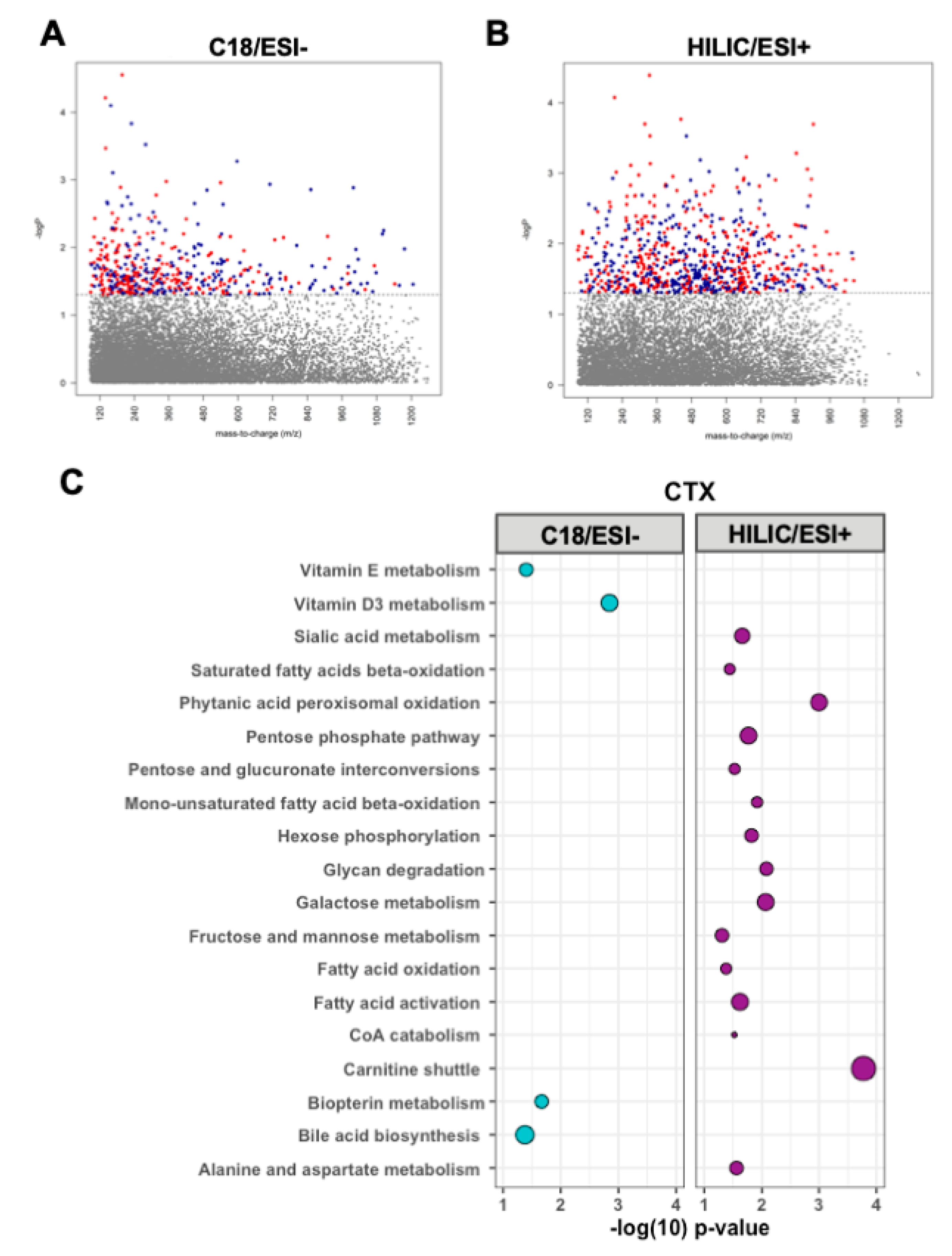
3.3. Metabolomic Associations with CTX
3.4. Targeted Analyses of Plasma Metabolites
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jilka, R.L. Biology of the basic multicellular unit and the pathophysiology of osteoporosis. Med. Pediatr. Oncol. 2003, 41, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.L.; Moreau, R. Emerging role of alpha-lipoic acid in the prevention and treatment of bone loss. Nutr. Rev. 2015, 73, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast-osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef]
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Cummings, S.R.; Melton, L.J. Epidemiology and outcomes of osteoporotic fractures. Lancet 2002, 359, 1761–1767. [Google Scholar] [CrossRef]
- Johnell, O.; Kanis, J.A. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteopor. Int. 2006, 17, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Leeming, D.J.; Alexandersen, P.; Karsdal, M.A.; Qvist, P.; Schaller, S.; Tanko, L.B. An update on biomarkers of bone turnover and their utility in biomedical research and clinical practice. Eur. J. Clin. Pharm. 2006, 62, 781–792. [Google Scholar] [CrossRef]
- Vasikaran, S.; Eastell, R.; Bruyere, O.; Foldes, A.J.; Garnero, P.; Griesmacher, A.; McClung, M.; Morris, H.A.; Silverman, S.; Trenti, T.; et al. Markers of bone turnover for the prediction of fracture risk and monitoring of osteoporosis treatment: A need for international reference standards. Osteoporos. Int. 2011, 22, 391–420. [Google Scholar] [CrossRef]
- Bone, H.G. The future of osteoporosis diagnosis and therapy. Ann. Ital. Med. Int. 1992, 7, 166S–170S. [Google Scholar]
- Garnero, P.; Ferreras, M.; Karsdal, M.A.; Nicamhlaoibh, R.; Risteli, J.; Borel, O.; Qvist, P.; Delmas, P.D.; Foged, N.T.; Delaisse, J.M. The type I collagen fragments ICTP and CTX reveal distinct enzymatic pathways of bone collagen degradation. J. Bone Miner. Res. 2003, 18, 859–867. [Google Scholar] [CrossRef]
- Civitelli, R.; Armamento-Villareal, R.; Napoli, N. Bone turnover markers: Understanding their value in clinical trials and clinical practice. Osteoporos. Int. 2009, 20, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Zaiss, M.M.; Jones, R.M.; Schett, G.; Pacifici, R. The gut-bone axis: How bacterial metabolites bridge the distance. J. Clin. Investig. 2019, 129, 3018–3028. [Google Scholar] [CrossRef]
- Bozec, A.; Zaiss, M.M.; Kagwiria, R.; Voll, R.; Rauh, M.; Chen, Z.; Mueller-Schmucker, S.; Kroczek, R.A.; Heinzerling, L.; Moser, M.; et al. T cell costimulation molecules CD80/86 inhibit osteoclast differentiation by inducing the IDO/tryptophan pathway. Sci. Transl. Med. 2014, 6, 235ra260. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P.; Park, Y.; Ziegler, T.R. Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012, 32, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, M.P.; Ziegler, T.R.; Jones, D.P.; Liu, K.H.; Fernandes, J.; Roberts, J.L.; Weitzmann, M.N.; Pacifici, R.; Alvarez, J.A. Plasma high-resolution metabolomics identifies linoleic acid and linked metabolic pathways associated with bone mineral density. Clin. Nutr. 2020. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Lee, K.; Soltow, Q.A.; Strobel, F.H.; Brigham, K.L.; Parker, R.E.; Wilson, M.E.; Sutliff, R.L.; Mansfield, K.G.; Wachtman, L.M.; et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012, 295, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Soltow, Q.A.; Strobel, F.H.; Mansfield, K.G.; Wachtman, L.; Park, Y.; Jones, D.P. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013, 9, S132–S143. [Google Scholar] [CrossRef]
- Liu, K.H.; Nellis, M.; Uppal, K.; Ma, C.; Tran, V.; Liang, Y.; Walker, D.I.; Jones, D.P. Reference standardization for quantification and harmonization of large-scale metabolomics. Anal. Chem. 2020. [Google Scholar] [CrossRef]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS—Adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Bonjour, J.P.; Theintz, G.; Law, F.; Slosman, D.; Rizzoli, R. Peak bone mass. Osteoporos. Int. 1994, 4 (Suppl. 1), 7–13. [Google Scholar] [CrossRef] [PubMed]
- Malmir, H.; Shab-Bidar, S.; Djafarian, K. Vitamin C intake in relation to bone mineral density and risk of hip fracture and osteoporosis: A systematic review and meta-analysis of observational studies. Br. J. Nutr. 2018, 119, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Gabbay, K.H.; Bohren, K.M.; Morello, R.; Bertin, T.; Liu, J.; Vogel, P. Ascorbate synthesis pathway: Dual role of ascorbate in bone homeostasis. J. Biol. Chem. 2010, 285, 19510–19520. [Google Scholar] [CrossRef]
- Choi, H.K.; Kim, G.J.; Yoo, H.S.; Song, D.H.; Chung, K.H.; Lee, K.J.; Koo, Y.T.; An, J.H. Vitamin C activates osteoblastogenesis and inhibits osteoclastogenesis via Wnt/beta-Catenin/ATF4 signaling pathways. Nutrients 2019, 11, 506. [Google Scholar] [CrossRef]
- Urban, K.; Höhling, H.J.; Lüttenberg, B.; Szuwart, T.; Plate, U. An in vitro study of osteoblast vitality influenced by the vitamins C and E. Head Face Med 2012, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Hirayama, A.; Sato, Y.; Koboyashi, T.; Katsuyama, E.; Kanagawa, H.; Fujie, A.; Morita, M.; Watanabe, R.; Tando, T.; et al. Metabolomics-based profiles predictive of low bone mass in menopausal women. Bone Rep. 2018, 9, 11–18. [Google Scholar] [CrossRef]
- Zhao, Q.; Shen, H.; Su, K.J.; Zhang, J.G.; Tian, Q.; Zhao, L.J.; Qiu, C.; Zhang, Q.; Garrett, T.J.; Liu, J.; et al. Metabolomic profiles associated with bone mineral density in US Caucasian women. Nutr. Metab. 2018, 15, 57. [Google Scholar] [CrossRef]
- You, Y.S.; Lin, C.Y.; Liang, H.J.; Lee, S.H.; Tsai, K.S.; Chiou, J.M.; Chen, Y.C.; Tsao, C.K.; Chen, J.H. Association between the metabolome and low bone mineral density in Taiwanese women determined by (1)H NMR spectroscopy. J. Bone Miner. Res. 2014, 29, 212–222. [Google Scholar] [CrossRef]
- Jennings, A.; MacGregor, A.; Spector, T.; Cassidy, A. Amino acid intakes are associated with bone mineral density and prevalence of low bone mass in women: Evidence from discordant monozygotic twins. J. Bone Miner. Res. 2016, 31, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Barbul, A. Proline precursors to sustain mammalian collagen synthesis. J. Nutr. 2008, 138, 2021S–2024S. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.E.; Choi, J.Y.; Shin, Y.O.; Park, D.S.; Kang, J.W.; Nam, D.; Choi, D.Y.; Lee, J.D. Arginine enhances osteoblastogenesis and inhibits adipogenesis through the regulation of Wnt and NFATc signaling in human mesenchymal stem cells. Int. J. Mol. Sci. 2014, 15, 13010–13029. [Google Scholar] [CrossRef]
- Fini, M.; Torricelli, P.; Giavaresi, G.; Carpi, A.; Nicolini, A.; Giardino, R. Effect of L-lysine and L-arginine on primary osteoblast cultures from normal and osteopenic rats. Biomed. Pharm. 2001, 55, 213–220. [Google Scholar] [CrossRef]
- Torricelli, P.; Fini, M.; Giavaresi, G.; Giardino, R.; Gnudi, S.; Nicolini, A.; Carpi, A. L-arginine and L-lysine stimulation on cultured human osteoblasts. Biomed. Pharm. 2002, 56, 492–497. [Google Scholar] [CrossRef]
- Lee, W.C.; Guntur, A.R.; Long, F.; Rosen, C.J. Energy metabolism of the osteoblast: Implications for osteoporosis. Endocr. Rev. 2017, 38, 255–266. [Google Scholar] [CrossRef]
- Cohn, D.V.; Forscher, B.K. Aerobic metabolism of glucose by bone. J. Biol. Chem. 1962, 237, 615–618. [Google Scholar]
- Peck, W.A.; Birge, S.J., Jr.; Fedak, S.A. Bone cells: Biochemical and biological studies after enzymatic isolation. Science 1964, 146, 1476–1477. [Google Scholar] [CrossRef]
- Wei, J.; Shimazu, J.; Makinistoglu, M.P.; Maurizi, A.; Kajimura, D.; Zong, H.; Takarada, T.; Lezaki, T.; Pessin, J.E.; Hinoi, E.; et al. Glucose uptake and runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell 2015, 161, 1576–1591. [Google Scholar] [CrossRef]
- Zoch, M.L.; Abou, D.S.; Clemens, T.L.; Thorek, D.L.; Riddle, R.C. In vivo radiometric analysis of glucose uptake and distribution in mouse bone. Bone Res. 2016, 4, 16004. [Google Scholar] [CrossRef]
- Flanagan, B.; Nichols, G., Jr. Metabolic Studies of bone in vitro. v. glucose metabolism and collagen biosynthesis. J. Biol. Chem. 1964, 239, 1261–1265. [Google Scholar]
- Dodds, R.A.; Gowen, M.; Bradbeer, J.N. Microcytophotometric analysis of human osteoclast metabolism: Lack of activity in certain oxidative pathways indicates inability to sustain biosynthesis during resorption. J. Histochem. Cytochem. 1994, 42, 599–606. [Google Scholar] [CrossRef]
- Ishii, K.A.; Fumoto, T.; Iwai, K.; Takeshita, S.; Ito, M.; Shimohata, N.; Aburatani, H.; Taketani, S.; Lelliott, C.J.; Vidal-Puig, A.; et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat. Med. 2009, 15, 259–266. [Google Scholar] [CrossRef]
- Wei, W.; Wang, X.; Yang, M.; Smith, L.C.; Dechow, P.C.; Sonoda, J.; Evans, R.M.; Wan, Y. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010, 11, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Faccio, R.; Novack, D.V. Alternative NF-kappaB regulates RANKL-induced osteoclast differentiation and mitochondrial biogenesis via independent mechanisms. J. Bone Miner. Res. 2015, 30, 2287–2299. [Google Scholar] [CrossRef]
- Zhou, J.; Ye, S.; Fujiwara, T.; Manolagas, S.C.; Zhao, H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J. Biol. Chem. 2013, 288, 30064–30074. [Google Scholar] [CrossRef] [PubMed]
- Lemma, S.; Sboarina, M.; Porporato, P.E.; Zini, N.; Sonveaux, P.; Di Pompo, G.; Baldini, N.; Avnet, S. Energy metabolism in osteoclast formation and activity. Int. J. Biochem. Cell Biol. 2016, 79, 168–180. [Google Scholar] [CrossRef]
- Hartley, A.; Paternoster, L.; Evans, D.M.; Fraser, W.D.; Tang, J.; Lawlor, D.A.; Tobias, J.H.; Gregson, C.L. Metabolomics analysis in adults with high bone mass identifies a relationship between bone resorption and circulating citrate which replicates in the general population. Clin. Endo 2020, 92, 29–37. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Deng, W.; Zhang, Z.; Deng, Z.; Hu, Y.; Xia, W.; Xu, L. Chinese bone turnover marker study: Reference ranges for C-terminal telopeptide of type I collagen and procollagen I N-terminal peptide by age and gender. PLoS ONE 2014, 9, e103841. [Google Scholar] [CrossRef]
- Roodman, G.D.; Ibbotson, K.J.; MacDonald, B.R.; Kuehl, T.J.; Mundy, G.R. 1,25-Dihydroxyvitamin D3 causes formation of multinucleated cells with several osteoclast characteristics in cultures of primate marrow. Proc. Natl. Acad. Sci. USA 1985, 82, 8213–8217. [Google Scholar] [CrossRef]
- Raisz, L.G.; Trummel, C.L.; Holick, M.F.; DeLuca, H.F. 1,25-dihydroxycholecalciferol: A potent stimulator of bone resorption in tissue culture. Science 1972, 175, 768–769. [Google Scholar] [CrossRef]
- Carlsson, A. Tracer experiments on the effect of vitamin D on the skeletal metabolism of calcium and phosphorus. Acta Physiol. Scand. 1952, 26, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Wolf, R.L.; Cauley, J.A.; Pettinger, M.; Jackson, R.; Lacroix, A.; Leboff, M.S.; Lewis, C.E.; Nevitt, M.C.; Simon, J.A.; Stone, K.L.; et al. Lack of a relation between vitamin and mineral antioxidants and bone mineral density: Results from the Women’s Health Initiative. Am. J. Clin. Nutr. 2005, 82, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Iwasaki, M.; Ochi, H.; Fukuda, T.; Ma, C.; Miyamoto, T.; Takitani, K.; Negishi-Koga, T.; Sunamura, S.; Kodama, T.; et al. Vitamin E decreases bone mass by stimulating osteoclast fusion. Nat. Med. 2012, 18, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gaspa, S.; Guanabens, N.; Jurado, S.; Dubreuil, M.; Combalia, A.; Peris, P.; Monegal, A.; Pares, A. Bile acids and bilirubin effects on osteoblastic gene profile. Implications in the pathogenesis of osteoporosis in liver diseases. Gene 2020, 725, 144167. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; An, J.H.; Park, H.; Yang, J.Y.; Choi, H.J.; Kim, S.W.; Park, Y.J.; Kim, S.Y.; Yim, M.; Baek, W.Y.; et al. Positive regulation of osteogenesis by bile acid through FXR. J. Bone Miner. Res. 2013, 28, 2109–2121. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Bessede, A.; Gargaro, M.; Pallotta, M.T.; Matino, D.; Servillo, G.; Brunacci, C.; Bicciato, S.; Mazza, E.M.; Macchiarulo, A.; Vacca, C.; et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 2014, 511, 184–190. [Google Scholar] [CrossRef]
- Schepper, J.D.; Collins, F.L.; Rios-Arce, N.D.; Raehtz, S.; Schaefer, L.; Gardinier, J.D.; Britton, R.A.; Parameswaran, N.; McCabe, L.R. Probiotic lactobacillus reuteri prevents postantibiotic bone loss by reducing intestinal dysbiosis and preventing barrier disruption. J. Bone Miner. Res. 2019, 34, 681–698. [Google Scholar] [CrossRef]
- Collins, F.L.; Rios-Arce, N.D.; Schepper, J.D.; Parameswaran, N.; McCabe, L.R. The potential of probiotics as a therapy for osteoporosis. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Collins, F.L.; Irwin, R.; Bierhalter, H.; Schepper, J.; Britton, R.A.; Parameswaran, N.; McCabe, L.R. Lactobacillus reuteri 6475 increases bone density in intact females only under an inflammatory setting. PLoS ONE 2016, 11, e0153180. [Google Scholar] [CrossRef] [PubMed]
- Britton, R.A.; Irwin, R.; Quach, D.; Schaefer, L.; Zhang, J.; Lee, T.; Parameswaran, N.; McCabe, L.R. Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J. Cell Physiol. 2014, 229, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Chassaing, B.; Tyagi, A.M.; Vaccaro, C.; Luo, T.; Adams, J.; Darby, T.M.; Weitzmann, M.N.; Mulle, J.G.; Gewirtz, A.T.; et al. Sex steroid deficiency–associated bone loss is microbiota dependent and prevented by probiotics. J. Clin. Investig. 2016, 126, 2049–2063. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.M.; Yu, M.; Darby, T.M.; Vaccaro, C.; Li, J.-Y.; Owens, J.A.; Hsu, E.; Adams, J.; Weitzmann, M.N.; Jones, R.M.; et al. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity 2018, 49, 1116–1131.e1117. [Google Scholar] [CrossRef]
- Weaver, C.M. Diet, gut microbiome, and bone health. Curr. Osteoporos. Rep. 2015, 13, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Quach, D.; Britton, R.A. Gut microbiota and bone health. Adv. Exp. Med. Biol. 2017, 1033, 47–58. [Google Scholar] [CrossRef]
- Cheng, S.; Qi, X.; Ma, M.; Zhang, L.; Cheng, B.; Liang, C.; Liu, L.; Li, P.; Kafle, O.P.; Wen, Y.; et al. Assessing the relationship between gut microbiota and bone mineral density. Front. Genet. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Henry, Y.M.; Eastell, R. Ethnic and gender differences in bone mineral density and bone turnover in young adults: Effect of bone size. Osteopor. Int. 2000, 11, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Wilkin, L.D.; Jackson, M.C.; Sims, T.D.; Haddock, B.L. Racial/Ethnic Differences in Bone Mineral Density of Young Adults. Int. J. Exerc. Sci. 2010, 3, 197. [Google Scholar] [PubMed]

| Characteristic | n (%) or Mean ± SD | Range |
|---|---|---|
| Female n (%) | 19 (56) | - |
| Race n (%) | ||
| Caucasian | 20 (59) | - |
| Asian | 11 (32) | - |
| African American | 3 (9) | - |
| Age (years) | 27.8 ± 4.9 | 18–36 |
| BMI | 24.7 ± 4.0 | 18.6–36.5 |
| Body Fat (%) | ||
| Females | 31.1 ± 7.3 | 17.0–48.1 |
| Males | 24.1 ± 8.6 | 9.8–40.3 |
| Total BMD (gm/cm2) | 1.23 ± 0.10 | 1.04–1.48 |
| Whole-Body BMD T-score | 0.97 ± 1.2 | −1.6–3.7 |
| Multivitamin supplement | 15 (44) | - |
| Calcium supplement | 1 (3) | - |
| Vitamin D supplement | 2 (6) | - |
| P1NP (ng/mL) | 15.4 ± 2.9 | 6.43–21.42 |
| CTX (ng/mL) | 0.45 ± 0.20 | 0.16–1.13 |
| Plasma Metabolite | m/z | Time (s) | β Estimate ± SE | p-Value |
|---|---|---|---|---|
| Associations with P1NP | ||||
| Tryptophan | 205.0971 | 36 | −0.03 ± 0.02 | 0.07 |
| 3-Hydroxykynurenine | 225.0823 | 33 | −0.02 ± 0.05 | 0.76 |
| L-Kynurenine | 209.092 | 40 | −0.05 ± 0.03 | 0.06 |
| Indole | 118.0652 | 38 | −0.03 ± 0.02 | 0.11 |
| Indole-3-Acetaldehyde | 160.0798 | 90 | 0.04 ± 0.02 | 0.01 |
| 5-Hydroxyindoleacetate | 192.0655 | 38 | –0.08 ± 0.05 | 0.09 |
| Indole-3-Ethanol | 162.0955 | 57 | 0.08 ± 0.05 | 0.14 |
| 3-Methyl-2-Oxindole | 148.0732 | 248 | 0.16 ± 0.08 | 0.04 |
| Indole-3-Acetic Acid | 176.0705 | 24 | –0.1 ± 0.07 | 0.17 |
| Methyl Indole-3-Acetate | 190.0845 | 153 | –0.14 ± 0.11 | 0.20 |
| Phenylacetic Acid | 137.0551 | 68 | 0.03 ± 0.04 | 0.40 |
| Linoleic Acid (FA 18:2) | 281.2475 | 199 | 0.01 ± 0.08 | 0.90 |
| HPODE | 313.2373 | 259 | –0.01 ± 0.05 | 0.80 |
| HODE/EpOME | 297.2211 | 221 | –0.03 ± 0.04 | 0.50 |
| Associations with CTX | ||||
| Tryptophan | 205.0971 | 36 | 0.02 ± 0.24 | 0.93 |
| 3-Hydroxykynurenine | 225.0823 | 33 | –0.56 ± 0.7 | 0.43 |
| L-Kynurenine | 209.092 | 40 | 0.37 ± 0.38 | 0.33 |
| Indole | 118.0652 | 38 | 0.22 ± 0.24 | 0.36 |
| Indole-3-Acetaldehyde | 160.0798 | 90 | 0.36 ± 0.25 | 0.16 |
| 5-Hydroxyindoleacetate | 192.0655 | 38 | 0.47 ± 0.66 | 0.48 |
| Indole-3-Ethanol | 162.0955 | 57 | 1.47 ± 0.68 | 0.04 |
| 3-Methyl-2-Oxindole | 148.0732 | 248 | 1.48 ± 0.45 | 0.58 |
| Indole-3-Acetic Acid | 176.0705 | 24 | 0.0001 ± 0.99 | 1.00 |
| Methyl Indole-3-Acetate | 190.0845 | 153 | 0.65 ± 1.16 | 0.33 |
| Phenylacetic Acid | 137.0551 | 68 | 1.02 ± 0.48 | 0.04 |
| Linoleic Acid (FA 18:2) | 281.2475 | 199 | 1.88 ± 1.04 | 0.08 |
| HPODE | 313.2373 | 259 | 0.64 ± 0.64 | 0.33 |
| HODE/EpOME | 297.2211 | 221 | 0.64 ± 0.6 | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellissimo, M.P.; Roberts, J.L.; Jones, D.P.; Liu, K.H.; Taibl, K.R.; Uppal, K.; Weitzmann, M.N.; Pacifici, R.; Drissi, H.; Ziegler, T.R.; et al. Metabolomic Associations with Serum Bone Turnover Markers. Nutrients 2020, 12, 3161. https://doi.org/10.3390/nu12103161
Bellissimo MP, Roberts JL, Jones DP, Liu KH, Taibl KR, Uppal K, Weitzmann MN, Pacifici R, Drissi H, Ziegler TR, et al. Metabolomic Associations with Serum Bone Turnover Markers. Nutrients. 2020; 12(10):3161. https://doi.org/10.3390/nu12103161
Chicago/Turabian StyleBellissimo, Moriah P., Joseph L. Roberts, Dean P. Jones, Ken H. Liu, Kaitlin R. Taibl, Karan Uppal, M. Neale Weitzmann, Roberto Pacifici, Hicham Drissi, Thomas R. Ziegler, and et al. 2020. "Metabolomic Associations with Serum Bone Turnover Markers" Nutrients 12, no. 10: 3161. https://doi.org/10.3390/nu12103161
APA StyleBellissimo, M. P., Roberts, J. L., Jones, D. P., Liu, K. H., Taibl, K. R., Uppal, K., Weitzmann, M. N., Pacifici, R., Drissi, H., Ziegler, T. R., & Alvarez, J. A. (2020). Metabolomic Associations with Serum Bone Turnover Markers. Nutrients, 12(10), 3161. https://doi.org/10.3390/nu12103161





