Simultaneous miRNA and mRNA Transcriptome Profiling of Differentiating Equine Satellite Cells Treated with Gamma-Oryzanol and Exposed to Hydrogen Peroxide
Abstract
1. Introduction
2. Material and Methods
2.1. Muscle Samples and Cell Culture
2.2. Satellite Cells Isolation
2.3. Satellite Cells Proliferation, Differentiation, and Treatment
2.4. RNA Isolation
2.5. Microarray Analysis
2.6. Real-Time qPCR
2.7. Ontological Analysis and Target Gene Prediction
2.8. Cell Viability, Cell Damage, and Oxidative Stress
3. Results
3.1. MiRNA Expression and Gene Ontology
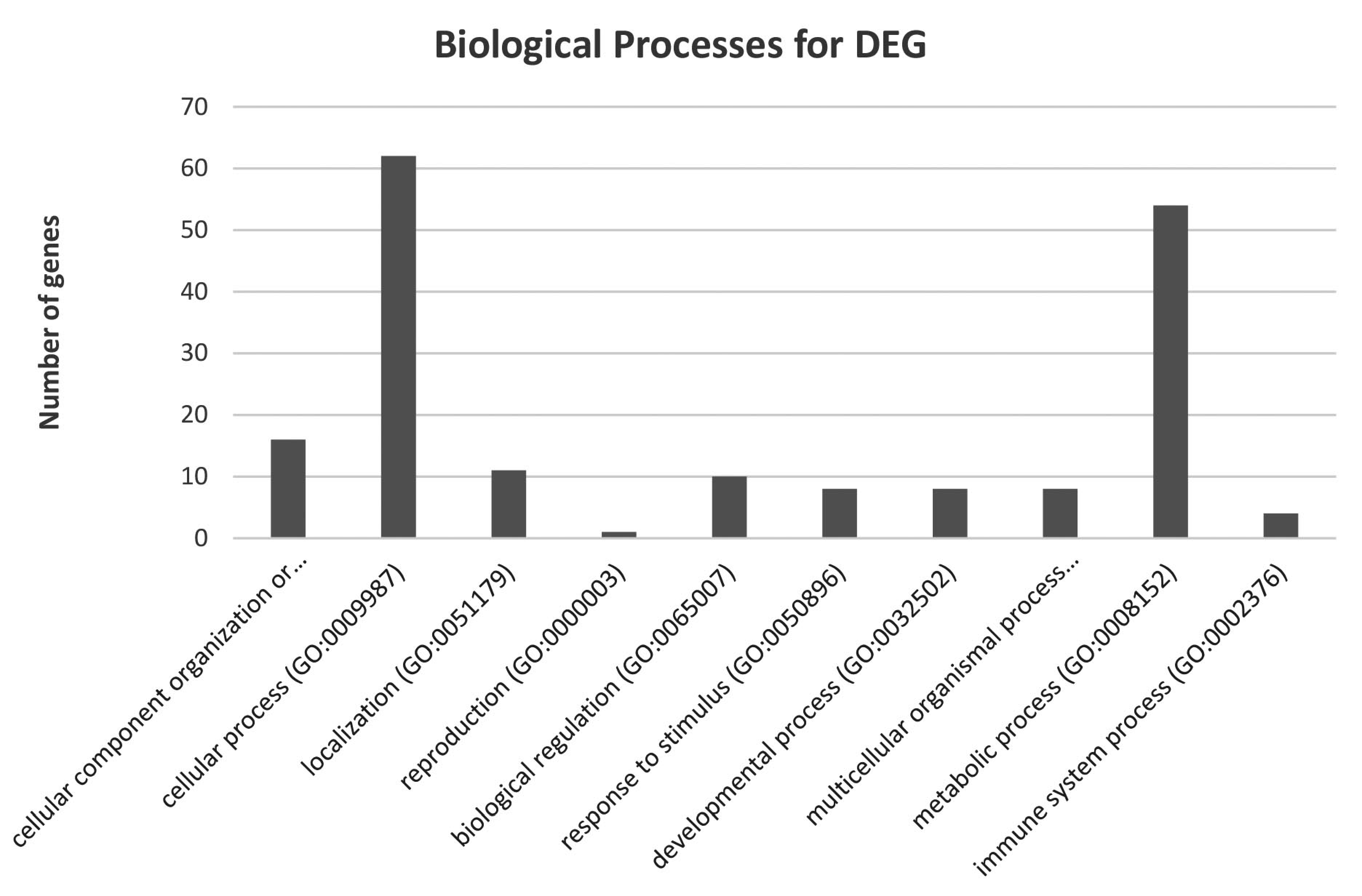
3.2. mRNA Expression and Gene Ontology
3.3. RT-qPCR Validation
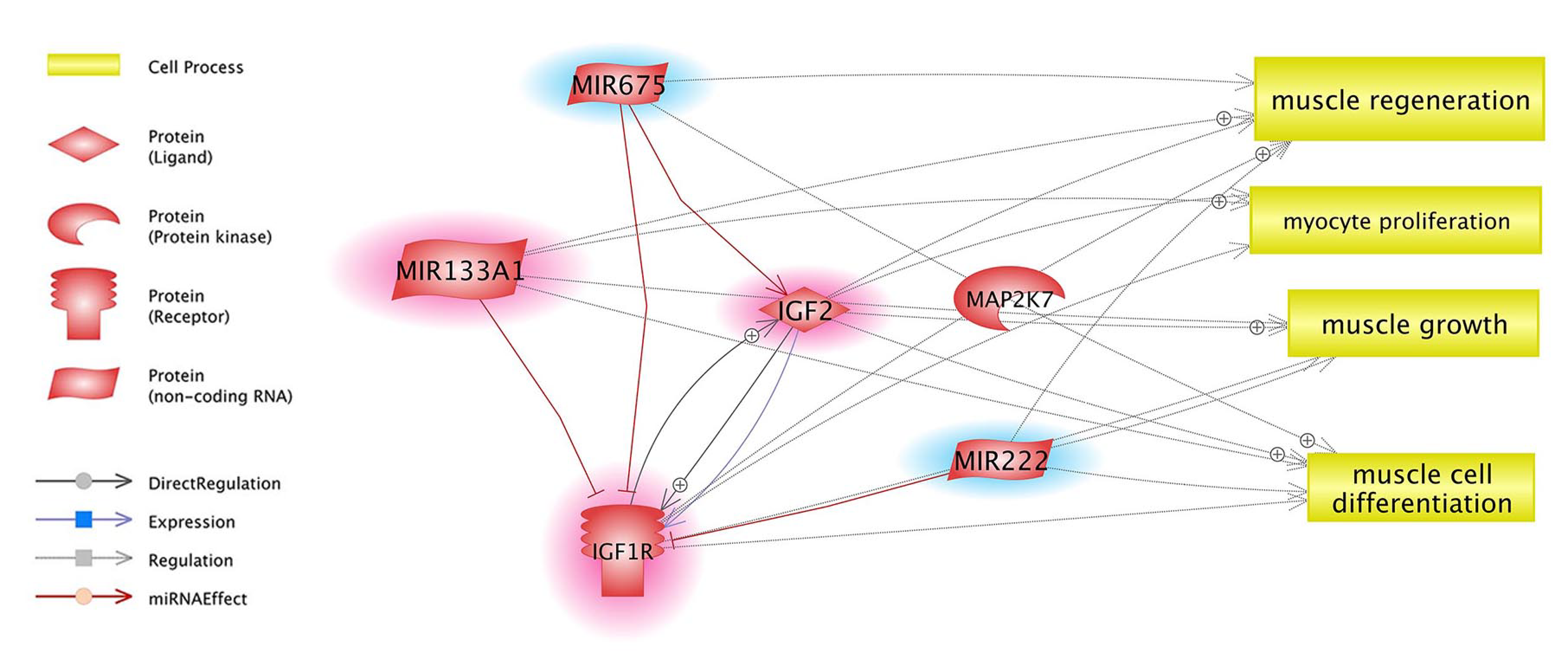
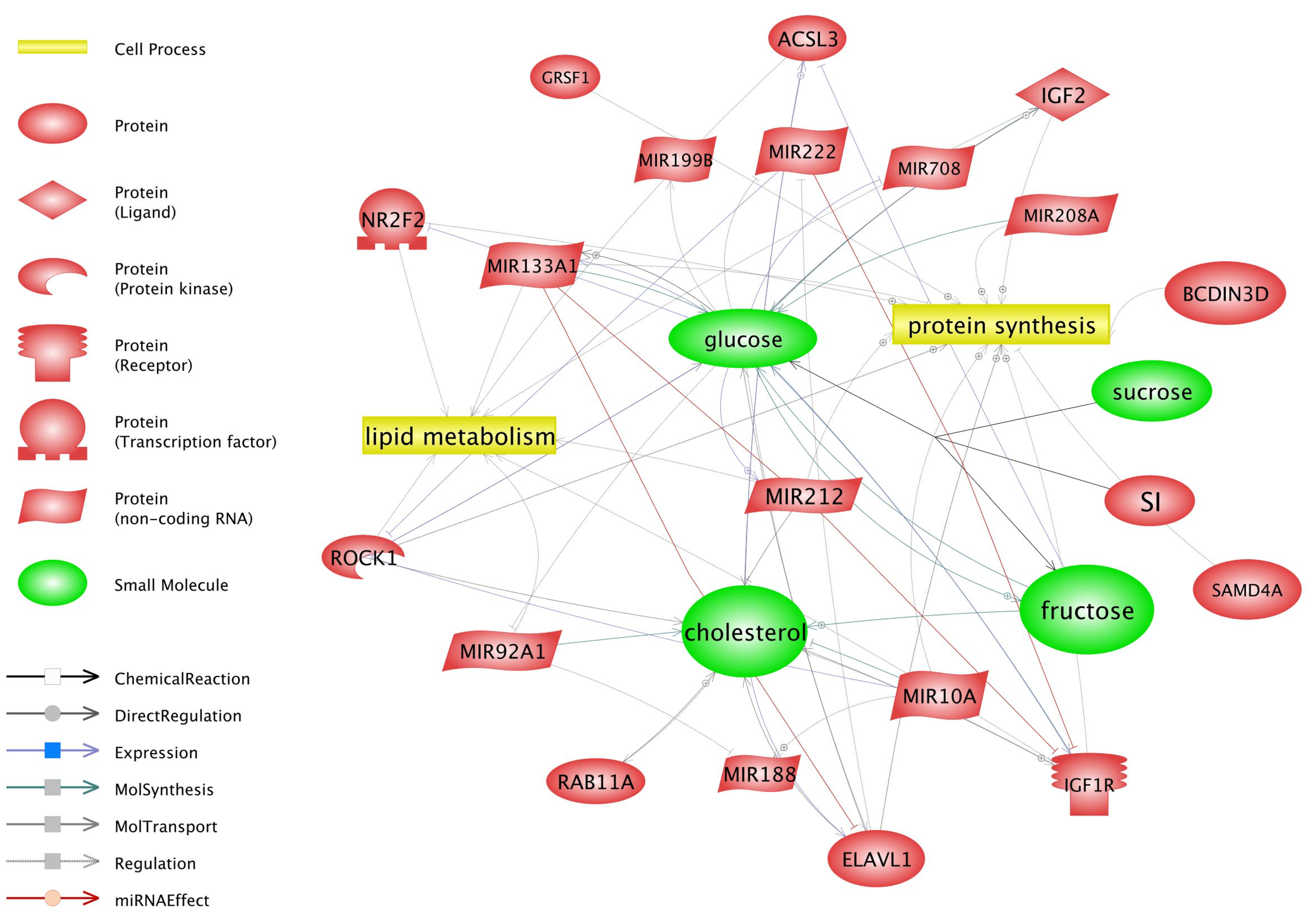
3.4. Prediction of miRNA Targets and Gene Ontology Enrichment
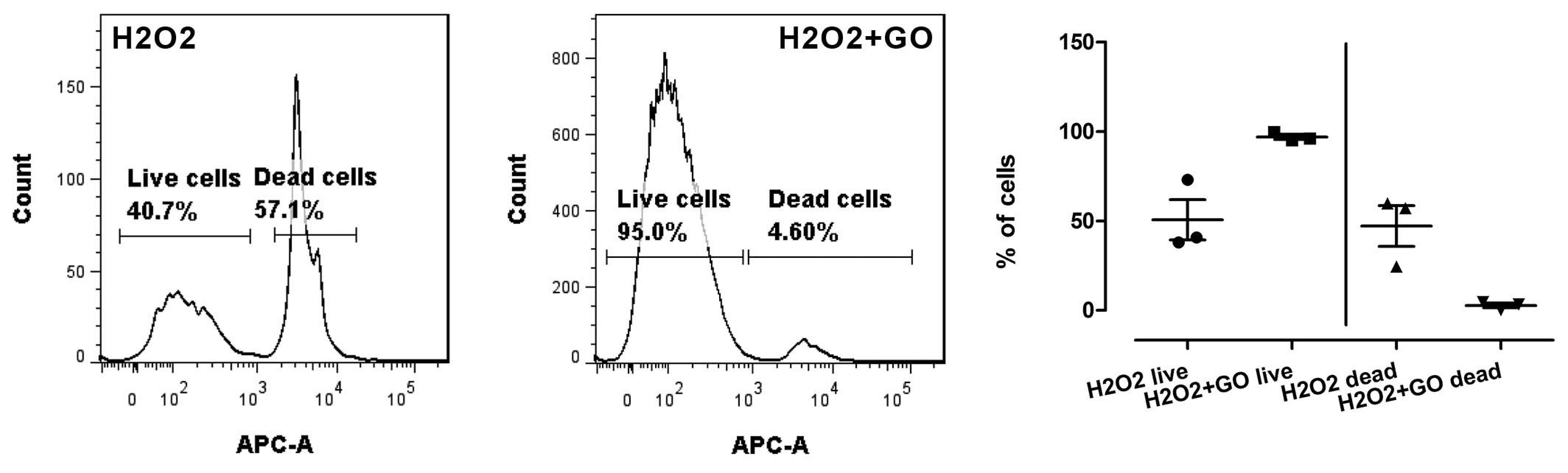
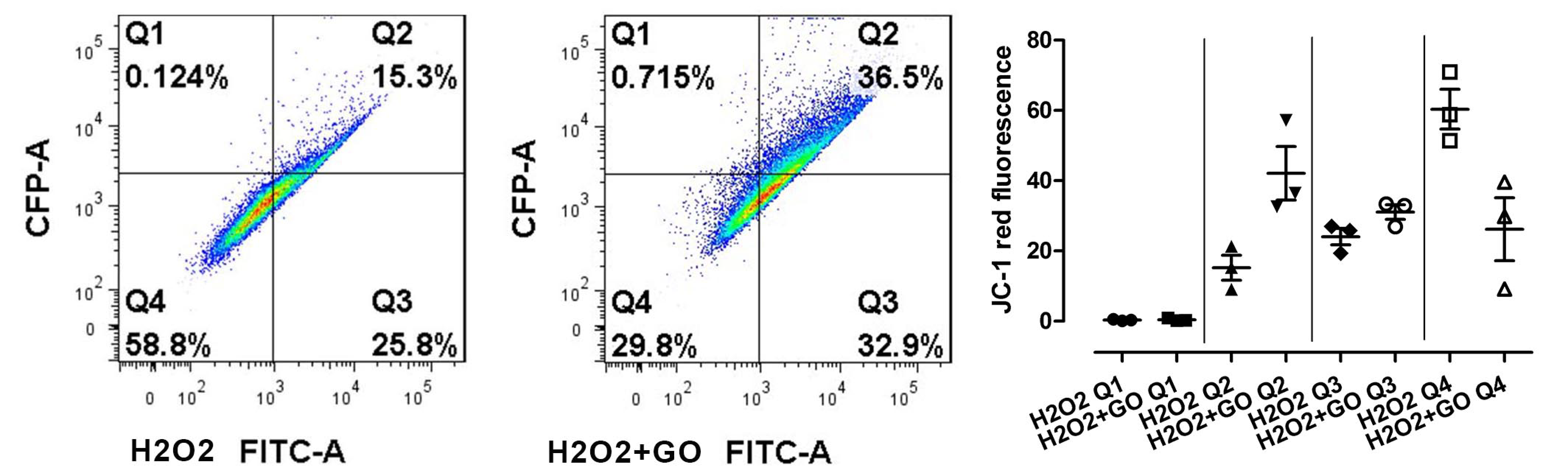
3.5. Cell Viability, Cell Damage, and Oxidative Stress
3.5.1. Cell Viability
3.5.2. Oxidative Stress and Cell Damage
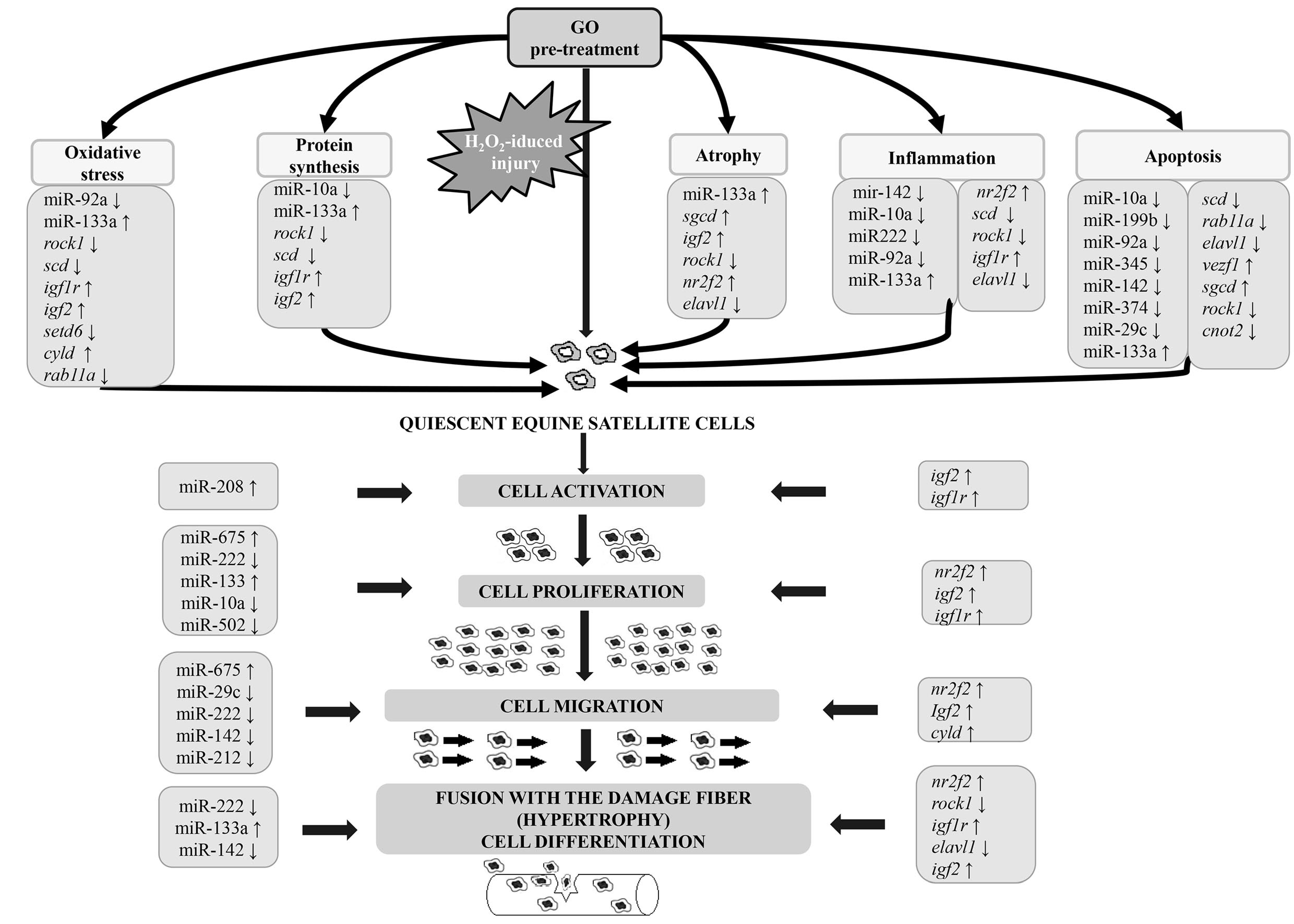
4. Discussion
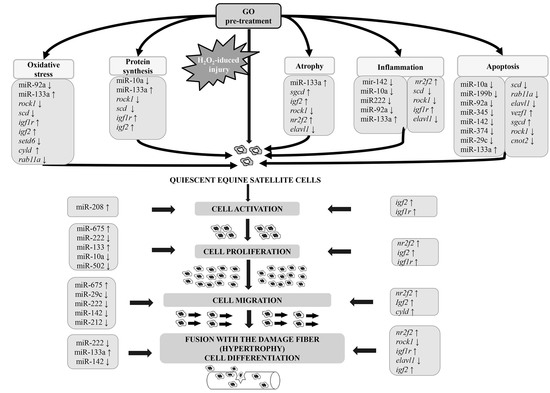
4.1. Gamma-Oryzanol Affects miRNAs and Genes Related to Muscle Cell Proliferation and Differentiation and Processes Related to Muscle Cell Injury and Regeneration
4.2. Proliferation, Differentiation, and Muscle-Related miRNA and Genes
4.3. Apoptosis, Oxidative Stress, and Immunity Related miRNAs and Genes
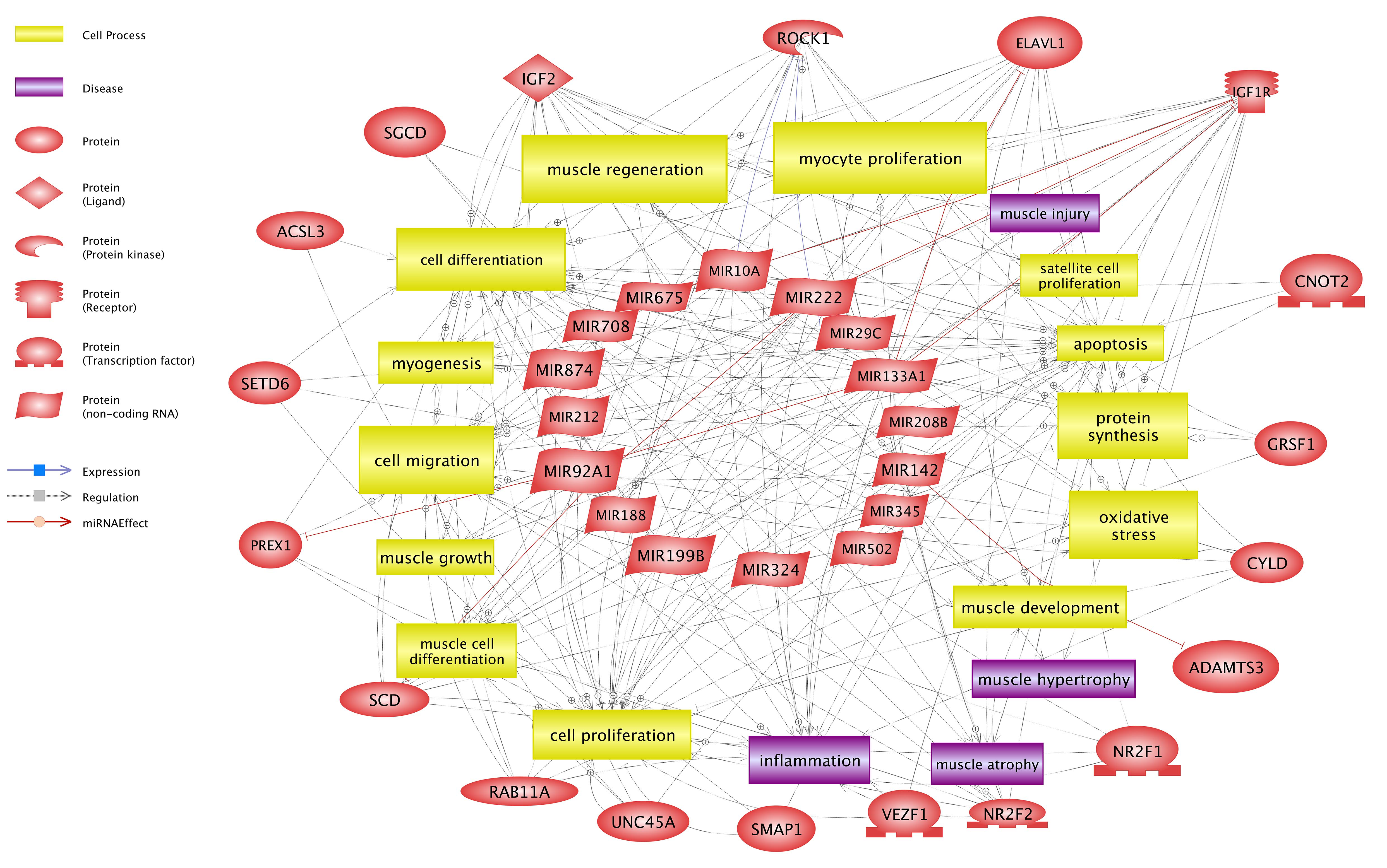
4.4. Target Genes and Their Relation with Identified miRNAs and Selected Pathways
4.5. Insulin/Insulin-Like Growth Factor, (IGF) Pathway-Mitogen Activated Protein Kinase Kinase/MAP Kinase Cascade, AMPK Signalling Pathway and Gamma-Oryzanol Related Muscle Growth-miRNAs and Genes
4.6. Gamma-Oryzanol May Affect Metabolism-Related Genes and miRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | Antibiotic |
| CTRL | control condition |
| DEG | differentially expressed genes |
| DET | differentially expressed target genes |
| DM | differentiation medium |
| DMSO | dimethylsulfoxide |
| ESC | equine satellite cells |
| FBS | fetal bovine serum |
| FEI | Federation Equestre Internationale |
| GE | gene expression |
| GM | growth medium |
| GO | gamma-oryzanol |
| HS | horse serum |
| IB | incubation buffer |
| PBS | phosphate-buffered saline |
| ROS | reactive oxygen species |
References
- Diack, M.; Saska, M. Separation of vitamin E and γ-oryzanols from rice bran by normal-phase chromatography. J. Am. Oil Chem. Soc. 1994, 71, 1211–1217. [Google Scholar] [CrossRef]
- Lerma-García, M.J.; Herrero-Martínez, J.M.; Simó-Alfonso, E.F.; Mendonça, C.R.; Ramis-Ramos, G. Composition, industrial processing and applications of rice bran γ-oryzanol. Food Chem. 2009, 115, 389–404. [Google Scholar] [CrossRef]
- Jeng, T.L.; Ho, P.T.; Shih, Y.J.; Lai, C.C.; Wu, M.T.; Sung, J.M. Comparisons of protein, lipid, phenolics, γ-oryzanol, vitamin E, and mineral contents in bran layer of sodium azide-induced red rice mutants. J. Sci. Food Agric. 2011, 91, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Cicero, A.F.G.; Gaddi, A. Rice bran oil and γ-oryzanol in the treatment of hyperlipoproteinaemias and other conditions. Phytother. Res. 2001, 15, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.A.; Nicolosi, R.J.; Woolfrey, B.; Kritchevsky, D. Rice bran oil and oryzanol reduce plasma lipid and lipoprotein cholesterol concentrations and aortic cholesterol ester accumulation to a greater extent than ferulic acid in hypercholesterolemic hamsters. J. Nutr. Biochem. 2007, 18, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Rico, C.W.; Nam, S.H.; Kang, M.Y. Effect of oryzanol and ferulic acid on the glucose metabolism of mice fed with a high-fat diet. J. Food Sci. 2011, 76, H7–H10. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Murata, T.; Fujisawa, M.; Nagasaka, R.; Ushio, H.; Bari, A.M.; Ozaki, H. Anti-inflammatory effects of phytosteryl ferulates in colitis induced by dextran sulphate sodium in mice. Br. J. Pharmacol. 2008, 154, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Hiramitsu, T.; Armstrong, D. Preventive effect of antioxidants on lipid peroxidation in the retina. Ophthalmic Res. 1991, 23, 196–203. [Google Scholar] [CrossRef]
- Patel, M.; Naik, S.N. Gamma-oryzanol from rice bran oil—A review. J. Sci. Ind. Res. 2004, 63, 569–578. [Google Scholar]
- Eslami, S.; Esa, N.M.; Marandi, S.M.; Ghasemi, G.; Eslami, S. Effects of gamma-oryzanol supplementation on anthropometric measurements & muscular strength in healthy males following chronic resistance training. Indian J. Med. Res. 2014, 139, 857. [Google Scholar]
- Wang, L.; Lin, Q.; Yang, T.; Liang, Y.; Nie, Y.; Luo, Y.; Luo, F. Oryzanol modifies high fat diet-induced obesity, liver gene expression profile, and inflammation response in mice. J. Agric. Food Chem. 2017, 65, 8374–8385. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Kim, H.W.; Jang, H.H.; Hwang, Y.J.; Choe, J.S.; Kim, J.B.; Lee, Y.H. γ-Oryzanol suppresses COX-2 expression by inhibiting reactive oxygen species-mediated Erk1/2 and Egr-1 signaling in LPS-stimulated RAW264. 7 macrophages. Biochem. Biophys. Res. Commun. 2017, 491, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Rungratanawanich, W.; Abate, G.; Serafini, M.M.; Guarienti, M.; Catanzaro, M.; Marziano, M.; Uberti, D. Characterization of the Antioxidant Effects of γ-Oryzanol: Involvement of the Nrf2 Pathway. Oxid. Med. Cell Longev. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, K.A.; Ciecierska, A.; Ostaszewski, P.; Sadkowski, T. Transcriptomic profile adaptations following exposure of equine satellite cells to nutriactive phytochemical gamma-oryzanol. Genes Nutr. 2016, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.; Tabor, G. The effects of Myofascial Release to the equine longissimus dorsi muscle on thoracic range of motion and mechanical nociceptive threshold. In Proceedings of the Horses Inside Out Conference, Cirencester, UK, 20–21 February 2016. [Google Scholar]
- Valberg, S.J. Muscle conditions affecting sport horses. Vet. Clin. N. Am. Equine Pract. 2018, 34, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Aladrović, J.; Beer Ljubić, B.; Prvanović Babić, N.; Vranković, L.; Klepo, E.; Iličić, L.; Stojević, Z. The effect of training and the Alka competition on oxidative stress and metabolic parameters in two horse breeds. Vet. Arh. 2018, 88, 295–308. [Google Scholar] [CrossRef]
- Dmitriev, P.; Saada, Y.B.; Dib, C.; Ansseau, E.; Barat, A.; Hamade, A.; Dupuy, C. DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients, Free Radic. Biol. Med. 2016, 99, 244–258. [Google Scholar] [CrossRef]
- Yuan, H.S.H.; Katyal, S.; Anderson, J.E. A mechanism for semaphorin-induced apoptosis: DNA damage of endothelial and myogenic cells in primary cultures from skeletal muscle. Oncotarget 2018, 9, 22618. [Google Scholar] [CrossRef]
- Rahimi, R.; Mirzaei, B.; Rahmani-Nia, F.; Salehi, Z. Effects of creatine monohydrate supplementation on exercise-induced apoptosis in athletes: A randomized, double-blind, and placebo-controlled study. J. Res. Med. Sci. 2015, 20, 733. [Google Scholar] [CrossRef]
- Jejurikar, S.S.; Kuzon, W.M. Satellite cell depletion in degenerative skeletal muscle. Apoptosis 2003, 8, 573–578. [Google Scholar] [CrossRef]
- Kamradt, M.C.; Chen, F.; Sam, S.; Cryns, V.L. The small heat shock protein αB-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J. Biol. Chem. 2002. [CrossRef] [PubMed]
- Jejurikar, S.S.; Henkelman, E.A.; Cederna, P.S.; Marcelo, C.L.; Urbanchek, M.G.; Kuzon Jr, W.M. Aging increases the susceptibility of skeletal muscle derived satellite cells to apoptosis. Exp. Gerontol. 2006, 41, 828–836. [Google Scholar] [CrossRef]
- Chodkowska, K.A.; Sadkowski, T.; Ostaszewski, P. MicroRNA function in domestic animal physiology and diseases: A promising diagnostic tool for veterinary use. Med. Veter. 2017, 73, 156–165. [Google Scholar] [CrossRef]
- Chodkowska, K.A.; Ciecierska, A.; Majchrzak, K.; Ostaszewski, P.; Sadkowski, T. Effect of β-hydroxy-β-methylbutyrate on miRNA expression in differentiating equine satellite cells exposed to hydrogen peroxide. Genes Nutr. 2018, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Mossman, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Israeli, D.; Poupiot, J.; Amor, F.; Charton, K.; Lostal, W.; Jeanson-Leh, L.; Richard, I. Circulating miRNAs are generic and versatile therapeutic monitoring biomarkers in muscular dystrophies. Sci. Rep. 2016, 6, 28097. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228. [Google Scholar] [CrossRef]
- Sadkowski, T.; Ciecierska, A.; Oprządek, J.; Balcerek, E. Breed-dependent microRNA expression in the primary culture of skeletal muscle cells subjected to myogenic differentiation. BMC Genom. 2018, 19, 109. [Google Scholar] [CrossRef]
- Cardinali, B.; Castellani, L.; Fasanaro, P.; Basso, A.; Alema, S.; Martelli, F.; Falcone, G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE 2009, 4, e7607. [Google Scholar] [CrossRef]
- Cardinali, B.; Cappella, M.; Provenzano, C.; Garcia-Manteiga, J.M.; Lazarevic, D.; Cittaro, D.; Falcone, G. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2017, 7, e2086. [Google Scholar] [CrossRef]
- Togliatto, G.; Trombetta, A.; Dentelli, P.; Cotogni, P.; Rosso, A.; Tschöp, M.H.; Brizzi, M.F. Unacylated ghrelin promotes skeletal muscle regeneration following hindlimb ischemia via SOD-2–mediated miR-221/222 expression. J. Am. Heart Assoc. 2013, 2, e000376. [Google Scholar] [CrossRef] [PubMed]
- Magenta, A.; Greco, S.; Gaetano, C.; Martelli, F. Oxidative stress and microRNAs in vascular diseases. Int. J. Mol. Sci. 2013, 14, 17319–17346. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; He, H.B.; Zhang, W.Y.; Zhang, H.X.; Bai, J.B.; Liu, H.Z.; Zhao, S.H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013, 4, e668. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Zhen, L.; Yuan, T.; Huang, J.; Deng, F.; Zhang, H.; Zhang, Y. miR-10a regulates proliferation of human cardiomyocyte progenitor cells by targeting GATA6. PLoS ONE 2014, 9, e103097. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Genom. Precis. Med. 2010, 3, 499–506. [Google Scholar] [CrossRef]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Yao, B.; Xu, W.; Chen, J.; Zhou, X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 2016, 6, 36340. [Google Scholar] [CrossRef]
- Rivas, D.A.; Lessard, S.J.; Rice, N.P.; Lustgarten, M.S.; So, K.; Goodyear, L.J.; Fielding, R.A. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014, 28, 4133–4147. [Google Scholar] [CrossRef]
- Salic, K.; De Windt, L.J. MicroRNAs as biomarkers for myocardial infarction. Curr. Atheroscler. Rep. 2012, 14, 193–200. [Google Scholar] [CrossRef]
- Kovanda, A.; Leonardis, L.; Zidar, J.; Koritnik, B.; Dolenc-Groselj, L.; Kovacic, S.R.; Rogelj, B. Differential expression of microRNAs and other small RNAs in muscle tissue of patients with ALS and healthy age-matched controls. Sci. Rep. 2018, 8, 5609. [Google Scholar] [CrossRef]
- Ucar, A.; Gupta, S.K.; Fiedler, J.; Erikci, E.; Kardasinski, M.; Batkai, S.; Remke, J. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012, 3, 1078. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Cheng, P.; Liang, M.K.; Chen, Y.S.; Lu, Q.; Wang, J.Y.; Liao, E.Y. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. Eur. J. Clin. Investig. 2015, 125, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Feng, C. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Bonnamy, B.; Barrey, E.J.; Mata, X.; Chaffaux, S.; Guerin, G. Muscular microRNA expressions in healthy and myopathic horses suffering from polysaccharide storage myopathy or recurrent exertional rhabdomyolysis. Equine Vet. J. 2010, 42, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Carraro, G.; Shrestha, A.; Rostkovius, J.; Contreras, A.; Chao, C.M.; El Agha, E.; Günther, A. miR-142-3p balances proliferation and differentiation of mesenchymal cells during lung development. Development 2014, 141, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Kaneda, R.; Ono, T.; Tohyama, S.; Hashimoto, H.; Endo, J.; Fukuda, K. miR-142-3p is essential for hematopoiesis and affects cardiac cell fate in zebrafish. Biochem. Biophys. Res. Commun. 2012, 425, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, P.; Stankevicins, L.; Ansseau, E.; Petrov, A.; Barat, A.; Dessen, P.; Belayew, A. Defective regulation of miRNAs target genes in myoblasts from facioscapulohumeral dystrophy patients. J. Biol. Chem. 2013, 288, 34989–35002. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Ding, S.L.; Long, B.; Liu, C.Y.; Li, P.F. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013, 4, e709. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Jeon, H.S.; Sun, Z.; Aubry, M.C.; Tang, H.; Park, C.H.; Park, J.Y. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin. Cancer Res. 2012, 18, 3658–3667. [Google Scholar] [CrossRef]
- Xie, X.; Tsai, S.Y.; Tsai, M.J. COUP-TFII regulates satellite cell function and muscular dystrophy. J. Clin. Investig. 2016, 126, 3929–3941. [Google Scholar] [CrossRef]
- Xie, X.; Wu, S.P.; Tsai, M.J.; Tsai, S. The role of COUP-TFII in striated muscle development and disease. Curr. Top. Dev. Biol. 2017, 125, 375–403. [Google Scholar] [CrossRef]
- Lee, H.J.; Kao, C.Y.; Lin, S.C.; Xu, M.; Xie, X.; Tsai, S.Y.; Tsai, M.J. Dysregulation of nuclear receptor COUP-TFII impairs skeletal muscle development. Sci. Rep. 2017, 7, 3136. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, A.; Cuadrado, A.; Fan, J.; Atasoy, U.; Muscat, G.E.; Munoz-Canoves, P.; Gorospe, M.; Munoz, A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 2003, 23, 4991–5004. [Google Scholar] [CrossRef] [PubMed]
- Cornelison, D.D.W.; Wilcox-Adelman, S.A.; Goetinck, P.F.; Rauvala, H.; Rapraeger, A.C.; Olwin, B.B. Essential and separable roles for Syndecan-3 and Syndecan-4 in 559 skeletal muscle development and regeneration. Genes Dev. 2004, 18, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.; Gabunia, K.; Vrakas, C.N.; Herman, A.B.; Kako, F.; Kelemen, S.E.; Autieri, M.V. Genetic deletion of IL-19 (Interleukin-19) exacerbates atherogenesis in Il19−/−× Ldlr−/− double knockout mice by dysregulation of mRNA stability protein huR (Human Antigen R). Arter. Thromb. Vasc. Biol. 2018, 38, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Morlando, M.; Mangiavacchi, A.; Fatica, A.; Bozzoni, I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 2014, 53, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Ryu, B.K.; Lee, M.G.; Han, J.; Lee, J.H.; Ha, T.K.; Lee, K.Y. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology 2008, 135, 2030–2042. [Google Scholar] [CrossRef]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef]
- Mendell, J.T. MiRiad roles for the miR-17-92 cluster in development and disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, M.; Qin, G.; Weintraub, N.L.; Tang, Y. MiR-92a regulates viability and angiogenesis of endothelial cells under oxidative stress. Biochem. Biophys. Res. Commun. 2014, 446, 952–958. [Google Scholar] [CrossRef]
- Srivastava, A.; Dixit, A.B.; Banerjee, J.; Tripathi, M.; Chandra, P.S. Role of inflammation and its miRNA based regulation in epilepsy: Implications for therapy. Clin. Chim. Acta 2016, 452, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.; Lin, J. MicroRNA in inflammatory bowel disease: Translational research and clinical implication. World J. Gastroenterol. 2015, 21, 12274. [Google Scholar] [CrossRef] [PubMed]
- den Hoogen, P.V.; den Akker, F.V.; Deddens, J.C.; Sluijter, J.P.G. Heart failure in chronic myocarditis: A role for microRNAs? Curr. Genom. 2015, 16, 88–94. [Google Scholar] [CrossRef]
- Siasos, G.; Kollia, C.; Tsigkou, V.; Basdra, E.K.; Lymperi, M.; Oikonomou, E.; Papavassiliou, A.G. MicroRNAs: Novel diagnostic and prognostic biomarkers in atherosclerosis. Curr. Top. Med. Chem. 2013, 13, 1503–1517. [Google Scholar] [CrossRef]
- Smith, J.; Goldsmith, C.; Ward, A.; LeDieu, R. IGF-II ameliorates the dystrophic phenotype and coordinately down-regulates programmed cell death. Cell Death Differ. 2000, 7, 1109. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y. Essential roles of growth hormone (GH) and insulin-like growth factor-I (IGF-I) in the liver. Endocr. J. 2012, 59, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Du, Y.; Song, J. MicroRNA-340 Inhibits Epithelial-Mesenchymal Transition by Impairing ROCK-1-Dependent Wnt/β-Catenin Signaling Pathway in Epithelial Cells from Human Benign Prostatic Hyperplasia. Chin. Med. J. 2018, 131, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.C.; Bonner, E.; Lewis, D.L.; Johnson, R.L.; Stone, M.H.; Kraemer, W.J. The effects of gamma-oryzanol supplementation during resistance exercise training. Int. J. Sport Nutr. Exerc. Metab. 1997, 7, 318–329. [Google Scholar] [CrossRef]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef]
- Atherton, P.J.; Babraj, J.; Smith, K.; Singh, J.; Rennie, M.J.; Wackerhage, H. Selective activation of AMPK-PGC-1α or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 2005, 19, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Raney, M.A.; Turcotte, L.P. Evidence for the involvement of CaMKII and AMPK in Ca2+-dependent signaling pathways regulating FA uptake and oxidation in contracting rodent muscle. J. Appl. Physiol. 2008, 104, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Morales-Alamo, D.; Calbet, J.A. AMPK signaling in skeletal muscle during exercise: Role of reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2016, 98, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, L.; Song, Z.; Sheikhahmadi, A.; Wang, Y.; Buyse, J. Effects of feed deprivation on the AMPK signaling pathway in skeletal muscle of broiler chickens. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2016, 191, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Masuzaki, H.; Kozuka, C.; Okamoto, S.; Yonamine, M.; Tanaka, H.; Shimabukuro, M. Brown rice-specific γ-oryzanol as a promising prophylactic avenue to protect against diabetes mellitus and obesity in humans. J. Diabetes Investig. 2018. [Google Scholar] [CrossRef] [PubMed]
- Francisqueti, F.V.; Minatel, I.O.; Ferron, A.J.T.; Bazan, S.G.Z.; Silva, V.D.S.; Garcia, J.L.; Corrêa, C.R. Effect of gamma-oryzanol as therapeutic agent to prevent cardiorenal metabolic syndrome in animals submitted to high sugar-fat diet. Nutrients 2017, 9, 1299. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, J.; Wang, C.; Li, K.; Liu, B.; Du, Y.; Guo, F. MiR-212-5p suppresses lipid accumulation by targeting FAS and SCD1. J. Mol. Endocrinol. 2017, 59, 205–217. [Google Scholar] [CrossRef]
- Miao, H.; Zeng, H.; Gong, H. MicroRNA-212 promotes lipid accumulation and attenuates cholesterol efflux in THP-1 human macrophages by targeting SIRT1. Gene 2018, 643, 55–60. [Google Scholar] [CrossRef]
- Jentzsch, C.; Leierseder, S.; Loyer, X.; Flohrschütz, I.; Sassi, Y.; Hartmann, D.; Engelhardt, S.A. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J. Mol. Cell. Cardiol. 2012, 52, 13–20. [Google Scholar] [CrossRef]
- Niculescu, L.S.; Simionescu, N.; Fuior, E.V.; Stancu, C.S.; Carnuta, M.G.; Dulceanu, M.D.; Sima, A.V. Inhibition of miR-486 and miR-92a decreases liver and plasma cholesterol levels by modulating lipid-related genes in hyperlipidemic hamsters. Mol. Biol. Rep. 2018, 1–13. [Google Scholar] [CrossRef]
- Setyowati Karolina, D.; Sepramaniam, S.; Tan, H.Z.; Armugam, A.; Jeyaseelan, K. MiR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013, 10, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Bernardo, B.C.; McMullen, J.R.; Ritchie, R.H. Diabetic cardiomyopathy: Mechanisms and new treatment strategies targeting antioxidant signaling pathways. Pharmacol. Ther. 2014, 142, 375–415. [Google Scholar] [CrossRef] [PubMed]
- Crawford, P.A.; Schaffer, J.E. Metabolic stress in the myocardium: Adaptations of gene expression. J. Mol. Cell. Cardiol. 2013, 55, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Comas, J.; Moreno-Asso, A.; Moreno-Vedia, J.; Martín, M.; Castaño, C.; Marzà-Florensa, A.; Gasa, R. Stress-induced MicroRNA-708 impairs β-cell function and growth. Diabetes 2017, 66, 3029–3040. [Google Scholar] [CrossRef] [PubMed]


| (A) | |||||
| No. | Primer name | Target sequence | Amplification time and temperature | ||
| 1 | miR-133a | UUUGGUCCCCUUCAACCAGCUG | 10 s/95 °C and 60 s/60 °C | ||
| 2 | miR-345 | GCUGACUCCUAGUCCAGUGCUC | 10 s/95 °C and 60 s/60 °C | ||
| 3 | miR-675 | UGGUGCGGAGAGGGCCCACAGUG | 10 s/95 °C and 60 s/60 °C | ||
| (B) | |||||
| No. | Primer name | Forward sequence | Reverse Sequence | Annealing time and temp. | Product length |
| 1 | nr2f2 | CCACCTCCTGCAGAACAAAAG | GGGTCTTGGTAAAGGCTCATT | 15 s/60 °C | 326 |
| 2 | gtpbp1 | CTCCAGACCACCAACAACTC | CCCCTGGGACTTCACCTTAT | 15 s/62 °C | 261 |
| 3 | nr2f2 | TCCTCCTCAGTCATAGAGCAAT | CTCTGTTTCACTCCCCTTTCTT | 15 s/60 °C | 340 |
| 4 | dtd1 | AGAGTTGGAGGAGAGCAGAT | TGGAGGGTAAACTGGCTGAT | 15 s/60 °C | 209 |
| 5 | btf3 | GCTCGCAGAAAGAAGAAGGT | GGTGCTTTTCCATCCACAGA | 15 s/60 °C | 329 |
| 6 | cs | CGAGGCTACAGTATCCCTGA | CTTTCACTGTTGAGGGCTGT | 15 s/60 °C | 257 |
| 7 | gapdh | GTTTGTGATGGGCGTGAACC | GTCTTCTGGGTGGCAGTGAT | 15 s/60 °C | 198 |
| No. | miRNA Name | FC (GO vs. ctrl) | Log FC (GO vs. ctrl) | Regulation (GO vs. ctrl) | miRbase Accession No |
|---|---|---|---|---|---|
| 1 | eca-miR-188-5p | 2.18 | 1.13 | up | MIMAT0013198 |
| 2 | eca-miR-212 | 1.68 | 0.75 | up | MIMAT0013030 |
| 3 | eca-miR-685_v14.0 | 1.61 | 0.68 | up | MIMAT0012904 |
| 4 | eca-miR-133a | 1.33 | 0.41 | up | MIMAT0012997 |
| 5 | eca-miR-208b | 1.30 | 0.38 | up | MIMAT0012900 |
| 6 | eca-miR-675 | −46.67 | −5.54 | down | MIMAT0013053 |
| 7 | eca-miR-10a | −43.24 | −5.43 | down | MIMAT0013019 |
| 8 | eca-miR-502-5p | −41.35 | −5.37 | down | MIMAT0013225 |
| 9 | eca-miR-708 | −40.90 | −5.35 | down | MIMAT0012993 |
| 10 | eca-miR-345-5p | −33.19 | −5.05 | down | MIMAT0013138 |
| 11 | eca-miR-142-3p | −28.34 | −4.82 | down | MIMAT0013023 |
| 12 | eca-miR-324-3p | −1.75 | −0.81 | down | MIMAT0013034 |
| 13 | eca-miR-199b-5p | −1.65 | −0.72 | down | MIMAT0013780 |
| 14 | eca-miR-92a | −1.47 | −0.56 | down | MIMAT0013089 |
| 15 | eca-miR-222 | −1.45 | −0.53 | down | MIMAT0013204 |
| 16 | eca-miR-874 | −1.42 | −0.50 | down | MIMAT0013069 |
| 17 | eca-miR-29c | −1.31 | −0.39 | down | MIMAT0012964 |
| No. | GO vs. Control | Fold Change | p-Value |
|---|---|---|---|
| 1 | nr2f2 | 11.78 | 0.003 |
| 2 | btf3 | −6.37 | 0.005 |
| 3 | dtd1 | −6.32 | 0.007 |
| 4 | gtpbp1 | 120.89 | 0.008 |
| 5 | otud4 | 2.78 | 0.020 |
| 6 | miR-133a | 1.71 | 0.023 |
| 7 | miR-345 | −5.33 | 0.001 |
| 8 | miR-675 | 1.39 | 0.038 |
| Pathway | Genes | p-Value | FDR |
|---|---|---|---|
| AMP-activated protein kinase (AMPK) signaling pathway | igf1r, scd, elavl1 | 1.00 × 10−2 | 9.50 |
| Proteoglycan in cancer | igf1r, rock1, igf2 | 2.80 × 10−2 | 24.0 |
| Endocytosis | igf1r, smap1, rab11a | 4.50 × 10−2 | 36.0 |
| Fatty acid metabolism Peroxisome proliferator-activated receptor | scd, acsl3 | 6.30 × 10−2 | 47.0 |
| (PPAR) signaling pathway | scd, acsl3 | 8.70 × 10−2 | 58.0 |
| No. | Target Gene Symbol | Gene Description | Biological Processes | Fold Change (Microarray) | Related Identified miRNAs |
|---|---|---|---|---|---|
| 1. | acsl3 | Acyl-CoA Synthetase Long-Chain Family Member 3 | Lipid biosynthesis and fatty acid degradation; Anabolic role in energy metabolism; Cell proliferation; Cell differentiation | −1.13 | miR-142↓ miR-222↓ |
| 2. | adamts3 | ADAM metallopeptidase with thrombospondin type 1 motif | Proteolysis; Protein processing; Inflammation | 1.18 | miR-142↓ |
| 3. | bcdin3d | BCDIN3 domain containing RNA Methyltransferase | Methylation; miRNA metabolic process | −1.13 | miR-29↓ miR-502↓ |
| 4. | cnot2 | CCR4-NOT transcription complex, subunit 2 | Inflammation; Cytoplasmic deadenylation; DNA damage response; Regulation of stem cell population maintenance | −1.12 | miR-212↑ miR-222↓ |
| 5. | cyld | Cylindromatosis (Turban Tumor Syndrome) | Cell survival, proliferation, and differentiation; Innate immunity; Negative regulation of nf-kappa B import into nucleus | 1.35 | miR-133↑ miR-212↑ |
| 6. | elavl1 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 | Embryonic cell differentiation; Cytokine-induced cachexia; Cell growth and proliferation; Regulation of translation; Regulation of stem cell population maintenance | −1.23 | miR-133a↑ miR-208↑ miR-222↓ miR-212↑ |
| 7. | grsf1 | G-rich RNA sequence binding factor 1 | RNA processing; Cell proliferation; Protein synthesis: Apoptosis | −1.23 | miR-212↑ miR-199↓ miR-208↑ |
| 8. | igf1r | Insulin like growth factor 1 receptor | Apoptosis; Cell growth, differentiation and survival control; Immune response; Regulation of MAPK cascade; Oxidative stress | 1.28 | miR-133↑ miR-502↓ miR-675↓ miR-222↓ |
| 9. | igf2 | Insulin-like growth factor 2 | Growth factor activity; Striated muscle cell differentiation; Positive regulation of glycogen Biosynthetic process; Satellite cell proliferation | 1.52 | miR-133↑ |
| 10. | mcmbp | Minichromosome Maintenance Complex Binding Protein | A key regulator of pre-replication complex | −1.21 | miR-212↑ miR-142↓ |
| 11. | nr2f2 | Nuclear receptor subfamily 2, group F, member 2 | Muscle organ development: Cell migration; Cell proliferation; Inflammation | 1.11 | miR-212↑ |
| 12. | rab11a | RAB11A A member RAS oncogene family | Metabolic process; Inflammation; Marker for myopathies | −1.17 | miR-142↓ |
| 13. | rbpms | RNA Binding Protein with Multiple Splicing | Response to oxidative stress; Heart, and gastrointestinal smooth Muscle development | −1.24 | miR-199↓ |
| 14. | rock1 | Rho-associated, coiled-coil containing protein kinase 1 | Regulation of smooth muscle contraction; Regulation of actin filament-based process; Apoptotic process; Loss of muscle protein | −1.21 | miR-212↑ |
| 15. | samd4a | Sterile Alpha Motif Domain Containing 4A | Cell junction; Modulates the activities of the mechanistic target of rapamycin complex 1; Regulates muscle/fat volume; Highly muscle-specific gene; (potential marker in dmd) | 1.23 | miR-222↓ |
| 16. | scd | Stearoyl-CoA Desaturase | Fatty acid biosynthesis; Biosynthesis of membrane phospholipids, cholesterol esters, and triglycerides; Cholesterol esterification; Muscle metabolism | −1.54 | miR-199↓ |
| 17. | serf2 | Small EDRK-Rich Factor 2 | Marker of spinal and muscular atrophy; Muscle cachexia, muscle loss; Cell proliferation; Oxidative stress; Apoptosis | 1.12 | miR-199↓ |
| 18. | setd6 | SET Domain Containing 6 | Protein binding; Cell differentiation; Apoptosis | 1.21 | miR-199↓ miR-133↑ |
| 19. | sgcd | Sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) | Muscle organ/cell development; Cardiac muscle tissue development; Muscle dystrophy; Cell apoptosis | 1.44 | miR-142↓ |
| 20. | smap1 | Small ArfGAP 1 | Endocytosis; Cell differentiation; Apoptosis; Cell migration; Cell proliferation | −1.10 | miR-10a↓ |
| 21. | vezf1 | Vascular Endothelial Zinc Finger 1 | Normal and abnormal cellular Proliferation and differentiation; DNA methylation | 1.34 | miR-222↓ miR-142↓ |
| 22. | zswim6 | Zinc Finger SWIM-Type Containing 6 | Metal ion binding; Dysostosis | 1.20 | miR-208↑ |
| No. | Process | Number of Genes | Genes |
|---|---|---|---|
| 1 | Nucleobase-containing compound metabolic process (GO:0006139) | 21 | grsf1, atp6v0b, dis3, mcmbp, supt5h, nap1l1, mfn1, rab11a, rbpms, cwc22, snrpb2, samd4a, dtd1, ppil4, arhgap27, bcdin3d, strbp, ring1, nme2, sart1, dhx29 |
| 2 | Carbohydrates metabolic process (GO:0005975) | 3 | sord, reep3, cs |
| 3 | Cellular amino acid metabolic process (GO: 0006520) | 4 | psph, cs, fahd2a, apip |
| 4 | Lipid metabolic process (GO:0006629) | 3 | nr2f2, acsl3, hadhb |
| 5 | Protein metabolic process (GO:0019538) | 20 | setd6, rwdd1, icmt, pigb, ilkap, mrps17, psmb4, gphn, spcs1, fkbp1a, dtd1, psmd14, ppil4, cyld, cnot2, adamts3, rpl29, pigg, eif3m, scara3 |
| 6 | Tricarboxylic acid cycle (GO:0006099) | 1 | cs |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chodkowska, K.A.; Ciecierska, A.; Majchrzak, K.; Ostaszewski, P.; Sadkowski, T. Simultaneous miRNA and mRNA Transcriptome Profiling of Differentiating Equine Satellite Cells Treated with Gamma-Oryzanol and Exposed to Hydrogen Peroxide. Nutrients 2018, 10, 1871. https://doi.org/10.3390/nu10121871
Chodkowska KA, Ciecierska A, Majchrzak K, Ostaszewski P, Sadkowski T. Simultaneous miRNA and mRNA Transcriptome Profiling of Differentiating Equine Satellite Cells Treated with Gamma-Oryzanol and Exposed to Hydrogen Peroxide. Nutrients. 2018; 10(12):1871. https://doi.org/10.3390/nu10121871
Chicago/Turabian StyleChodkowska, Karolina A., Anna Ciecierska, Kinga Majchrzak, Piotr Ostaszewski, and Tomasz Sadkowski. 2018. "Simultaneous miRNA and mRNA Transcriptome Profiling of Differentiating Equine Satellite Cells Treated with Gamma-Oryzanol and Exposed to Hydrogen Peroxide" Nutrients 10, no. 12: 1871. https://doi.org/10.3390/nu10121871
APA StyleChodkowska, K. A., Ciecierska, A., Majchrzak, K., Ostaszewski, P., & Sadkowski, T. (2018). Simultaneous miRNA and mRNA Transcriptome Profiling of Differentiating Equine Satellite Cells Treated with Gamma-Oryzanol and Exposed to Hydrogen Peroxide. Nutrients, 10(12), 1871. https://doi.org/10.3390/nu10121871