Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. DNA Extraction and DNA Methylation Analysis
2.3. Treatment of Methylation Raw Data
2.4. Ingenuity Pathway Analysis
2.5. Differential CpG Site Selection
2.6. Blood Cell Type Composition
2.7. Statistical Analysis of Participants
3. Results
3.1. Participants’ Differences in Anthropometric and Biochemical Features, Food Consumption and Blood Cell Type Composition
3.2. Relevant Differentially Methylated CpGs Were Related to Metabolism, Inflammation, Intracellular Signals and Diabetes
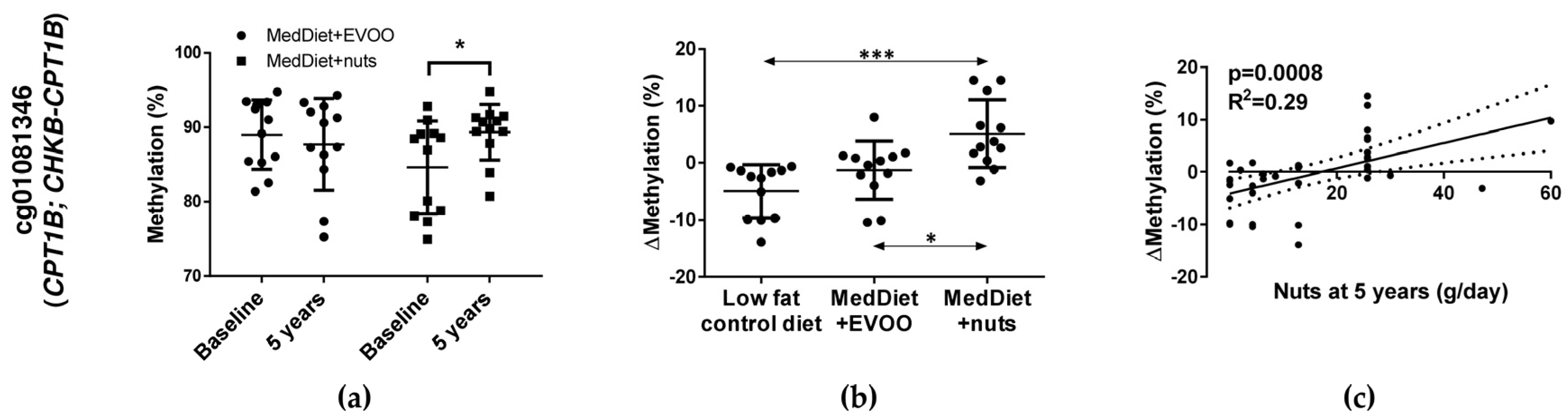
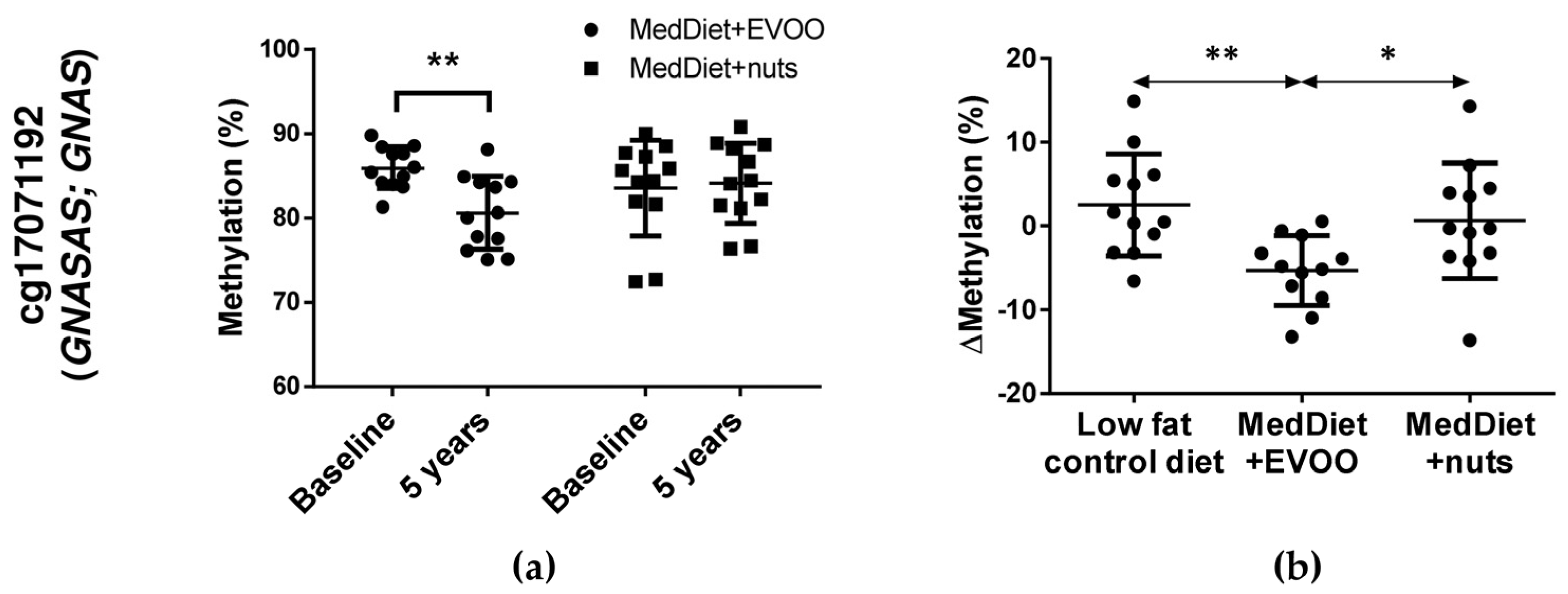
3.3. Two Differentially Methylated CpGs (cg01081346 and cg17071192) Were Selected
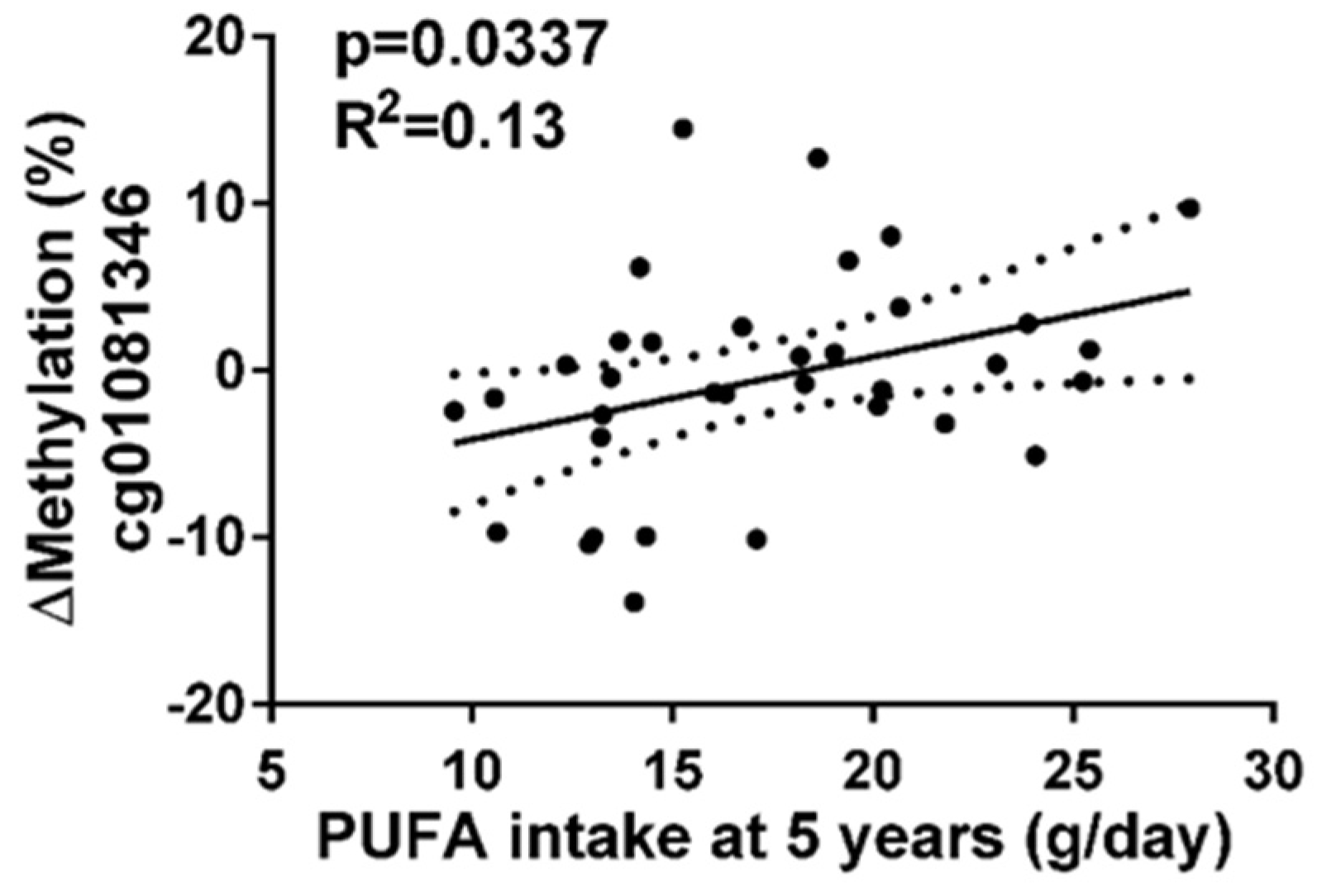
3.4. CpG cg01081346 Was Associated with PUFAs
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Milagro, F.I.; Campion, J.; Garcia-Diaz, D.F.; Goyenechea, E.; Paternain, L.; Martinez, J.A. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J. Physiol. Biochem. 2009, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Ordovás, J.M. Biomarkers: Background, classification and guidelines for applications in nutritional epidemiology. Nutrición Hospitalaria 2015, 31, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Friso, S. Epigenetics: A new bridge between nutrition and health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Goni, L.; Cuervo, M.; Milagro, F.I.; Martinez, J.A. Future perspectives of personalized weight loss interventions based on nutrigenetic, epigenetic, and metagenomic data. J. Nutr. 2016, 146, 905S–912S. [Google Scholar] [CrossRef] [PubMed]
- Barres, R.; Zierath, J.R. DNA methylation in metabolic disorders. Am. J. Clin. Nutr. 2011, 93, 897S–900S. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Hamet, P. Impact of genetic and epigenetic factors from early life to later disease. Metabolism 2008, 57 (Suppl. 2), S27–S31. [Google Scholar] [CrossRef] [PubMed]
- Kohlmeier, M.; De Caterina, R.; Ferguson, L.R.; Gorman, U.; Allayee, H.; Prasad, C.; Kang, J.X.; Nicoletti, C.F.; Martinez, J.A. Guide and position of the international society of nutrigenetics/nutrigenomics on personalized nutrition: Part 2—Ethics, challenges and endeavors of precision nutrition. J. Nutrigenet. Nutrigenom. 2016, 9, 28–46. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Martinez-Gonzalez, M.A.; Corella, D.; Salas-Salvado, J.; Ruiz-Gutierrez, V.; Covas, M.I.; Fiol, M.; Gomez-Gracia, E.; Lopez-Sabater, M.C.; Vinyoles, E.; et al. Effects of a mediterranean-style diet on cardiovascular risk factors: A randomized trial. Ann. Intern. Med. 2006, 145, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Estruch, R.; Corella, D.; Fito, M.; Ros, E.; Predimed, I. Benefits of the mediterranean diet: Insights from the predimed study. Prog Cardiovasc. Dis. 2015, 58, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Mitjavila, M.T.; Fandos, M.; Salas-Salvado, J.; Covas, M.I.; Borrego, S.; Estruch, R.; Lamuela-Raventos, R.; Corella, D.; Martinez-Gonzalez, M.A.; Sanchez, J.M.; et al. The mediterranean diet improves the systemic lipid and DNA oxidative damage in metabolic syndrome individuals. A randomized, controlled, trial. Clin. Nutr. 2013, 32, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Ordovas, J.; Sorli, J.; Asensio, E.; Ortega, C.; Carrasco, P.; Portoles, O.; Coltell, O. Effect of the mediterranean diet on DNA methylation of selected genes in the predimed-valencia intervention trial. FASEB J. 2015, 29, LB242. [Google Scholar]
- Arpón, A.; Riezu-Boj, J.I.; Milagro, F.I.; Marti, A.; Razquin, C.; Martinez-Gonzalez, M.A.; Corella, D.; Estruch, R.; Casas, R.; Fito, M.; et al. Adherence to mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J. Physiol. Biochem. 2017, 73, 445–455. [Google Scholar] [CrossRef]
- Rodriguez-Miguel, C.; Moral, R.; Escrich, R.; Vela, E.; Solanas, M.; Escrich, E. The role of dietary extra virgin olive oil and corn oil on the alteration of epigenetic patterns in the rat DMBA-induced breast cancer model. PLoS ONE 2015, 10, e0138980. [Google Scholar] [CrossRef] [PubMed]
- Poulose, S.; Bielinski, D.; Crott, J.; Roe, A.; Thangthaeng, N.; Shukitt-Hale, B. Effects of aging and walnut-rich diet on DNA methylation and expression of immediate-early genes in critical brain regions. FASEB J. 2015, 29, 749.7. [Google Scholar]
- Estruch, R.; Ros, E.; Salas-Salvado, J.; Covas, M.I.; Corella, D.; Aros, F.; Gomez-Gracia, E.; Ruiz-Gutierrez, V.; Fiol, M.; Lapetra, J.; et al. Primary prevention of cardiovascular disease with a mediterranean diet. N. Engl. J. Med. 2013, 368, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, M.A.; Corella, D.; Salas-Salvado, J.; Ros, E.; Covas, M.I.; Fiol, M.; Warnberg, J.; Aros, F.; Ruiz-Gutierrez, V.; Lamuela-Raventos, R.M.; et al. Cohort profile: Design and methods of the predimed study. Int. J. Epidemiol. 2012, 41, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Ballart, J.D.; Pinol, J.L.; Zazpe, I.; Corella, D.; Carrasco, P.; Toledo, E.; Perez-Bauer, M.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Martin-Moreno, J.M. Relative validity of a semi-quantitative food-frequency questionnaire in an elderly mediterranean population of spain. Br. J. Nutr. 2010, 103, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Touleimat, N.; Tost, J. Complete pipeline for infinium(®) human methylation 450k beadchip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 2012, 4, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. ISBN 978-0-387-29362-2. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene expression omnibus: Ncbi gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Volkmar, M.; Dedeurwaerder, S.; Cunha, D.A.; Ndlovu, M.N.; Defrance, M.; Deplus, R.; Calonne, E.; Volkmar, U.; Igoillo-Esteve, M.; Naamane, N.; et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012, 31, 1405–1426. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C.; Hoile, S.P.; Lillycrop, K.A. Epigenetics: Are there implications for personalised nutrition? Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Casas, R.; Sacanella, E.; Urpi-Sarda, M.; Corella, D.; Castaner, O.; Lamuela-Raventos, R.M.; Salas-Salvado, J.; Martinez-Gonzalez, M.A.; Ros, E.; Estruch, R. Long-term immunomodulatory effects of a mediterranean diet in adults at high risk of cardiovascular disease in the prevencion con dieta mediterranea (predimed) randomized controlled trial. J. Nutr. 2016, 146, 1684–1693. [Google Scholar] [CrossRef] [PubMed]
- Haynie, K.R.; Vandanmagsar, B.; Wicks, S.E.; Zhang, J.; Mynatt, R.L. Inhibition of carnitine palymitoyltransferase1b induces cardiac hypertrophy and mortality in mice. Diabetes Obes. Metab. 2014, 16, 757–760. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Kim, T.; Long, Q.; Liu, J.; Wang, P.; Zhou, Y.; Ding, Y.; Prasain, J.; Wood, P.A.; Yang, Q. Carnitine palmitoyltransferase-1b deficiency aggravates pressure overload-induced cardiac hypertrophy caused by lipotoxicity. Circulation 2012, 126, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.S.; Salehzadeh, F.; Fritz, T.; Zierath, J.R.; Krook, A.; Osler, M.E. Mitochondrial regulators of fatty acid metabolism reflect metabolic dysfunction in type 2 diabetes mellitus. Metabolism 2012, 61, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Cress, A.P.; Fraker, P.J.; Bieber, L.L. Carnitine and acylcarnitine levels of human peripheral blood lymphocytes and mononuclear phagocytes. Biochim. Biophys. Acta 1989, 992, 135–139. [Google Scholar] [CrossRef]
- Weinstein, L.S.; Xie, T.; Zhang, Q.H.; Chen, M. Studies of the regulation and function of the Gsα gene Gnas using gene targeting technology. Pharmacol. Ther. 2007, 115, 271–291. [Google Scholar] [CrossRef] [PubMed]
- Tobi, E.W.; Heijmans, B.T.; Kremer, D.; Putter, H.; Delemarre-van de Waal, H.A.; Finken, M.J.; Wit, J.M.; Slagboom, P.E. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics 2011, 6, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C.; Lillycrop, K.A. Bridging the gap between epigenetics research and nutritional public health interventions. Genome Med. 2010, 2, 80. [Google Scholar] [CrossRef] [PubMed]
- Crujeiras, A.B.; Diaz-Lagares, A.; Sandoval, J.; Milagro, F.I.; Navas-Carretero, S.; Carreira, M.C.; Gomez, A.; Hervas, D.; Monteiro, M.P.; Casanueva, F.F.; et al. DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: A genome-wide analysis from non-obese and obese patients. Sci. Rep. 2017, 7, 41903. [Google Scholar] [CrossRef] [PubMed]
- Covas, M.I. Olive oil and the cardiovascular system. Pharmacol. Res. 2007, 55, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Solà-Alberich, R.; Godàs-Bonfill, G.; Salas-Salvadó, J. Nutritional Value-Tree Nuts and Health: Effects of Tree Nuts on Cholesterol and Cardiovascular Diseases; Northern Nut Growers Association (NNGA): Notre Dame, IN, USA, 2011; Available online: http://www.nutgrowing.org/tree_nuts_and_health.htm (accessed on 9 November 2016).
- Voisin, S.; Almen, M.S.; Moschonis, G.; Chrousos, G.P.; Manios, Y.; Schioth, H.B. Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of greek preadolescents. Eur. J. Hum. Genet. 2015, 23, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, V.; Covas, M.I.; Munoz-Aguayo, D.; Khymenets, O.; de la Torre, R.; Saez, G.; Tormos Mdel, C.; Toledo, E.; Marti, A.; Ruiz-Gutierrez, V.; et al. In vivo nutrigenomic effects of virgin olive oil polyphenols within the frame of the mediterranean diet: A randomized controlled trial. FASEB J. 2010, 24, 2546–2557. [Google Scholar] [CrossRef] [PubMed]
- De la Puerta, R.; Ruiz Gutierrez, V.; Hoult, J.R. Inhibition of leukocyte 5-lipoxygenase by phenolics from virgin olive oil. Biochem. Pharma. 1999, 57, 445–449. [Google Scholar] [CrossRef]
- Visioli, F.; Galli, C. Biological properties of olive oil phytochemicals. Crit. Rev. Food Sci. Nutr. 2002, 42, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Falconi, A.; Di Germanio, C.; Micioni Di Bonaventura, M.V.; Costa, A.; Caramuta, S.; Del Carlo, M.; Compagnone, D.; Dainese, E.; Cifani, C.; et al. Extravirgin olive oil up-regulates CB1 tumor suppressor gene in human colon cancer cells and in rat colon via epigenetic mechanisms. J. Nutr. Biochem. 2015, 26, 250–258. [Google Scholar] [CrossRef] [PubMed]
| Low-Fat Control Diet (n = 12) | p2 | MedDiet + EVOO (n = 12) | p2 | MedDiet + nuts (n = 12) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 5 Years | Baseline | 5 Years | Baseline | 5 Years | p2 | p 03 | p 53 | p Δ3 | |||
| Female, n (%) | 6 (50) | 6 (50) | 6 (50) | ns | ||||||||
| Age(years) 1 | 64.3 (3.9) | NA | 63.5 (1.7) | NA | 63.2 (2.1) | NA | ns | |||||
| Weight(kg) 1 | 73.0 (10.9) | 73.1 (9.6) | ns | 73.3 (9.4) | 74.6 (10.7) | ns | 71.6 (7.5) | 73.2 (7.6) | ns | ns | ns | ns |
| Waist circumference(cm) 1 | 92.4 (9.6) | 92.4 (9.5) | ns | 90.1 (7.1) | 93.8 (6.4) | ns | 93.5 (6.3) | 94.3 (6.7) | ns | ns | ns | ns |
| BMI(kg/m2) 1 | 27.3 (3.2) | 27.4 (2.9) | ns | 27.9 (1.5) | 28.3 (1.8) | ns | 28.1 (1.5) | 28.8 (1.8) | ns | ns | ns | ns |
| Glycemia(mg/dL) 1 | 106.0 (27.6) | 102.2 (18.0) | ns | 129.5 (67.1) | 120.8 (52.7) | ns | 131.5 (57.4) | 127.0 (34.9) | ns | ns | ns | ns |
| HDL cholesterol(mg/dL) 1 | 52.7 (14.5) | 51.5 (13.3) | ns | 55.2 (8.3) | 55.1 (10.4) | ns | 60.9 (9.1) | 64.6 (12.0) | ns | ns | 0.022 | ns |
| LDL cholesterol(mg/dL) 1 | 132.0 (15.5) | 112.3 (27.1) | ns | 122.0 (30.5) | 113.0 (26.3) | ns | 119.1 (23.2) | 123.1 (35.2) | ns | ns | ns | ns |
| Total cholesterol(mg/dL) 1 | 209.8 (21.0) | 188.6 (27.7) | ns | 203.4 (28.7) | 191.6 (29.1) | ns | 199.2 (24.3) | 210.4 (41.2) | ns | ns | ns | ns |
| Triglycerides(mg/dL) 1 | 126.1 (49.6) | 124.1 (40.7) | ns | 131.2 (78.2) | 117.6 (47.9) | ns | 95.7 (24.6) | 113.4 (40.0) | ns | ns | ns | ns |
| Systolic arterialpressure (mmHg) 1 | 148.9 (17.2) | 150.1 (22.4) | ns | 158.5 (19.1) | 157.6 (20.5) | ns | 142.9 (16.0) | 147.7 (21.6) | ns | ns | ns | ns |
| Diastolic arterialpressure (mmHg) 1 | 85.7 (8.6) | 85.8 (11.4) | ns | 88.1 (8.4) | 83.9 (10.0) | ns | 87.9 (7.2) | 87.2 (14.0) | ns | ns | ns | ns |
| Diabetes, n (%) | 1 (8) | 0 (0) | ns | 4 (33) | 0 (0) | 0.046 | 6 (50) | 0 (0) | 0.014 | ns | ns | 0.022 |
| Hypercholesterolemia, n (%) | 8 (67) | 0 (0) | 0.005 | 9 (75) | 1 (8) | 0.005 | 10 (83) | 1 (8) | 0.003 | ns | ns | ns |
| Arterial hypertension, n (%) | 12 (100) | 0 (0) | <0.001 | 11 (92) | 1 (8) | 0.004 | 10 (83) | 4 (33) | ns | ns | ns | 0.010 |
| Hypertriglyceridemia, n (%) | 1 (8) | 2 (17) | ns | 2 (17) | 0 (0) | ns | 0 (0) | 0 (0) | ns | ns | ns | ns |
| Low-Fat Control Diet (n = 12) | p2 | MedDiet + EVOO (n = 12) | p2 | MedDiet + nuts (n = 12) | p2 | p 03 | p 53 | p Δ3 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Food/component (g/day) | Baseline | 5 Years | Baseline | 5 Years | Baseline | 5 Years | ||||||
| Carbohydrates | 230.4 (46.7) | 267.9 (60.1) | ns | 239.4 (82.1) | 248.7 (76.2) | ns | 224.9 (52.3) | 240.8 (44.9) | ns | ns | ns | ns |
| Proteins | 89.9 (23.9) | 96.8 (16.1) | ns | 88.9 (16.0) | 88.7 (14.8) | ns | 91.5 (16.0) | 95.0 (18.9) | ns | ns | ns | ns |
| Fat | 100.5 (24.2) | 97.2 (19.5) | ns | 108.2 (30.3) | 117.3 (25.6) | ns | 108.9 (19.4) | 124.1 (25.1) | ns | ns | 0.017 | ns |
| MUFA | 51.8 (11.8) | 51.3 (11.0) | ns | 56.6 (16.4) | 63.9 (10.6) | ns | 55.3 (12.4) | 64.6 (13.0) | ns | ns | 0.014 | ns |
| SFA | 23.3 (7.2) | 22.5 (7.3) | ns | 26.7 (8.9) | 25.0 (6.8) | ns | 24.8 (5.3) | 28.8 (8.1) | ns | ns | ns | ns |
| PU FA | 15.8 (5.5) | 15.5 (5.0) | ns | 16.8 (6.3) | 16.9 (4.2) | ns | 18.6 (4.7) | 19.7 (4.1) | ns | ns | ns | ns |
| Vegetables | 302.1 (90.2) | 352.2 (104.2) | ns | 302.3 (117.8) | 370.2 (58.2) | ns | 298.9 (71.1) | 372.7 (129.1) | ns | ns | ns | ns |
| Fish | 73.5 (32.6) | 93.6 (37.5) | ns | 90.2 (33.8) | 82.6 (25.7) | ns | 91.2 (33.5) | 100.4 (39.9) | ns | ns | ns | ns |
| Fruits | 291.6 (145.1) | 431.4 (237.1) | 0.009 | 319.8 (181.9) | 459.3 (109.9) | ns | 367.2 (163.2) | 500.8 (186.7) | 0.034 | ns | ns | ns |
| Legumes | 17.9 (9.9) | 14.7 (6.4) | ns | 16.0 (4.7) | 19.6 (7.5) | ns | 15.5 (3.5) | 20.4 (5.8) | 0.019 | ns | ns | 0.034 |
| Nuts | 12.2 (18.2) | 5.5 (8.8) | ns | 9.9 (16.4) | 11.3 (8.3) | ns | 20.5 (15.1) | 28.2 (14.2) | ns | ns | <0.001 | ns |
| EVOO | 29.6 (26.3) | 32.9 (26.2) | ns | 39.6 (24.7) | 66.4 (8.1) | 0.003 | 25.0 (30.0) | 33.8 (32.4) | ns | ns | 0.002 | 0.049 |
| Rest of olive oils | 16.7 (24.6) | 16.7 (24.6) | ns | 10.0 (15.8) | 0 (0) | 0.049 | 26.3 (27.6) | 25.8 (32.6) | ns | ns | ns | ns |
| Red meat | 63.2 (42.5) | 59.8 (38.1) | ns | 71.7 (42.3) | 39.5 (28.0) | 0.045 | 57.5 (29.8) | 54.8 (32.6) | ns | ns | ns | ns |
| White meat | 54.9 (28.6) | 63.0 (25.4) | ns | 39.5 (22.4) | 36.7 (26.8) | ns | 43.1 (23.2) | 47.7 (26.4) | ns | ns | ns | ns |
| Pastries | 35.3 (18.2) | 36.9 (27.3) | ns | 43.4 (24.5) | 29.8 (21.4) | ns | 23.9 (21.1) | 37.6 (27.6) | ns | ns | ns | ns |
| Total energy (kcal/day) | 2310 (420) | 2412 (475) | ns | 2378 (651) | 2470 (513) | ns | 2349 (432) | 2535 (452) | ns | ns | ns | ns |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arpón, A.; Milagro, F.I.; Razquin, C.; Corella, D.; Estruch, R.; Fitó, M.; Marti, A.; Martínez-González, M.A.; Ros, E.; Salas-Salvadó, J.; et al. Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids. Nutrients 2018, 10, 15. https://doi.org/10.3390/nu10010015
Arpón A, Milagro FI, Razquin C, Corella D, Estruch R, Fitó M, Marti A, Martínez-González MA, Ros E, Salas-Salvadó J, et al. Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids. Nutrients. 2018; 10(1):15. https://doi.org/10.3390/nu10010015
Chicago/Turabian StyleArpón, Ana, Fermín I. Milagro, Cristina Razquin, Dolores Corella, Ramón Estruch, Montserrat Fitó, Amelia Marti, Miguel A. Martínez-González, Emilio Ros, Jordi Salas-Salvadó, and et al. 2018. "Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids" Nutrients 10, no. 1: 15. https://doi.org/10.3390/nu10010015
APA StyleArpón, A., Milagro, F. I., Razquin, C., Corella, D., Estruch, R., Fitó, M., Marti, A., Martínez-González, M. A., Ros, E., Salas-Salvadó, J., Riezu-Boj, J.-I., & Martínez, J. A. (2018). Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids. Nutrients, 10(1), 15. https://doi.org/10.3390/nu10010015









