Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging
Abstract
1. Introduction
2. Materials and Methods
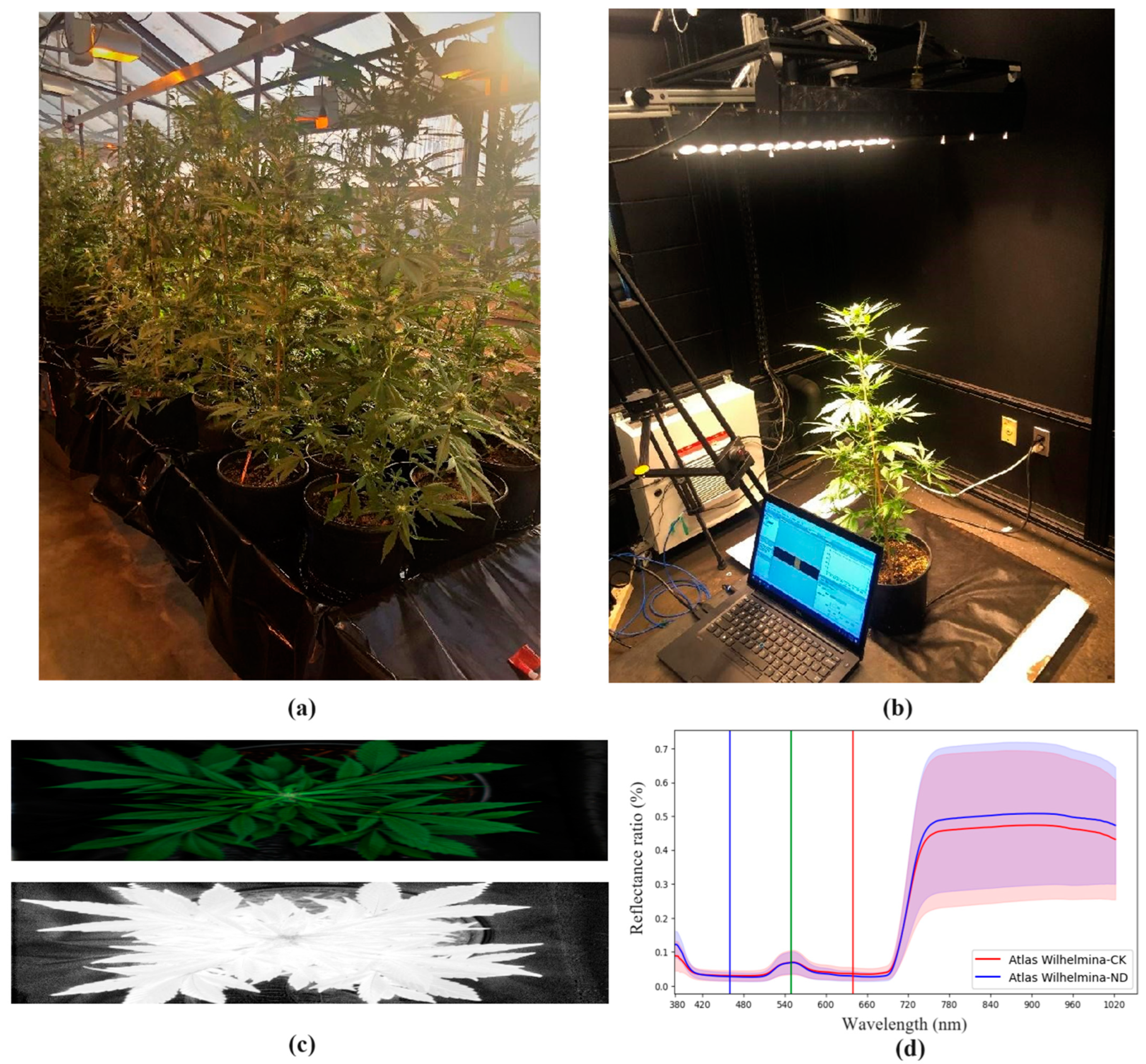
2.1. Experimental Greenhouse Setup and Hemp Cultivar Selection
2.2. Hyperspectral Data Acquisition and Preprocessing
2.3. Multivariate Analysis of Hyperspectral Data
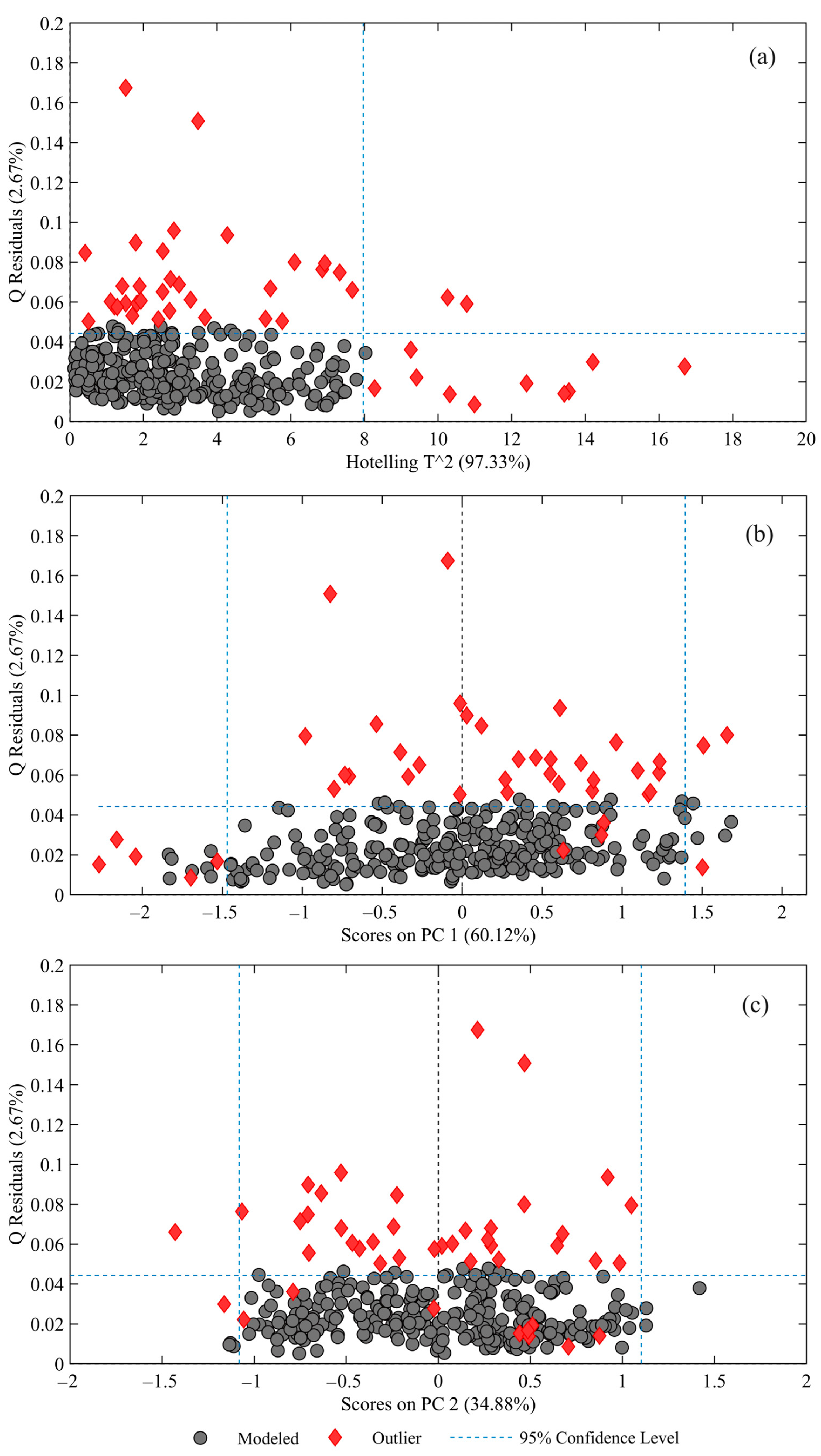
2.3.1. Optimizing Spectral Analysis through Robust Outlier Detection and Noise Reduction
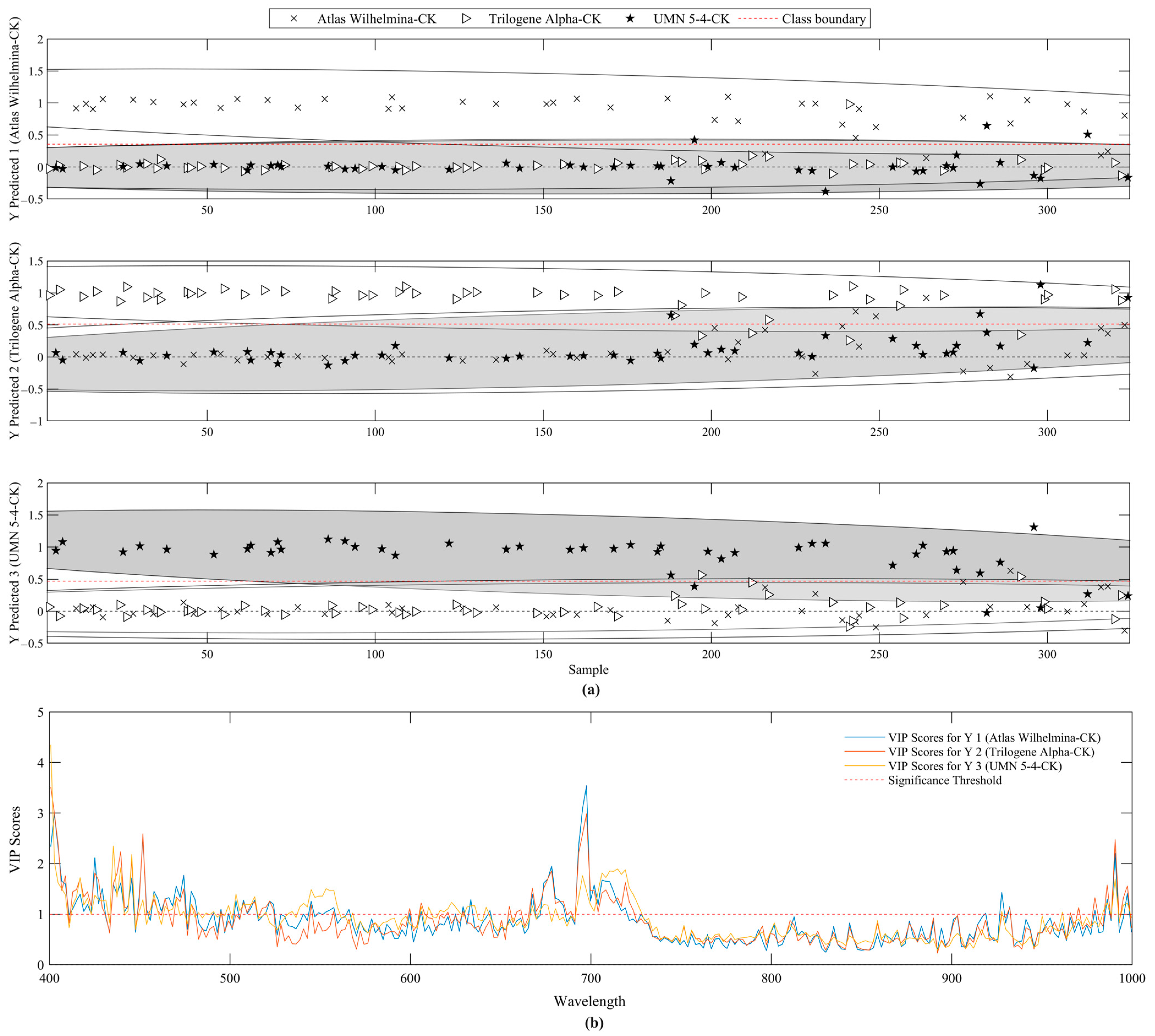
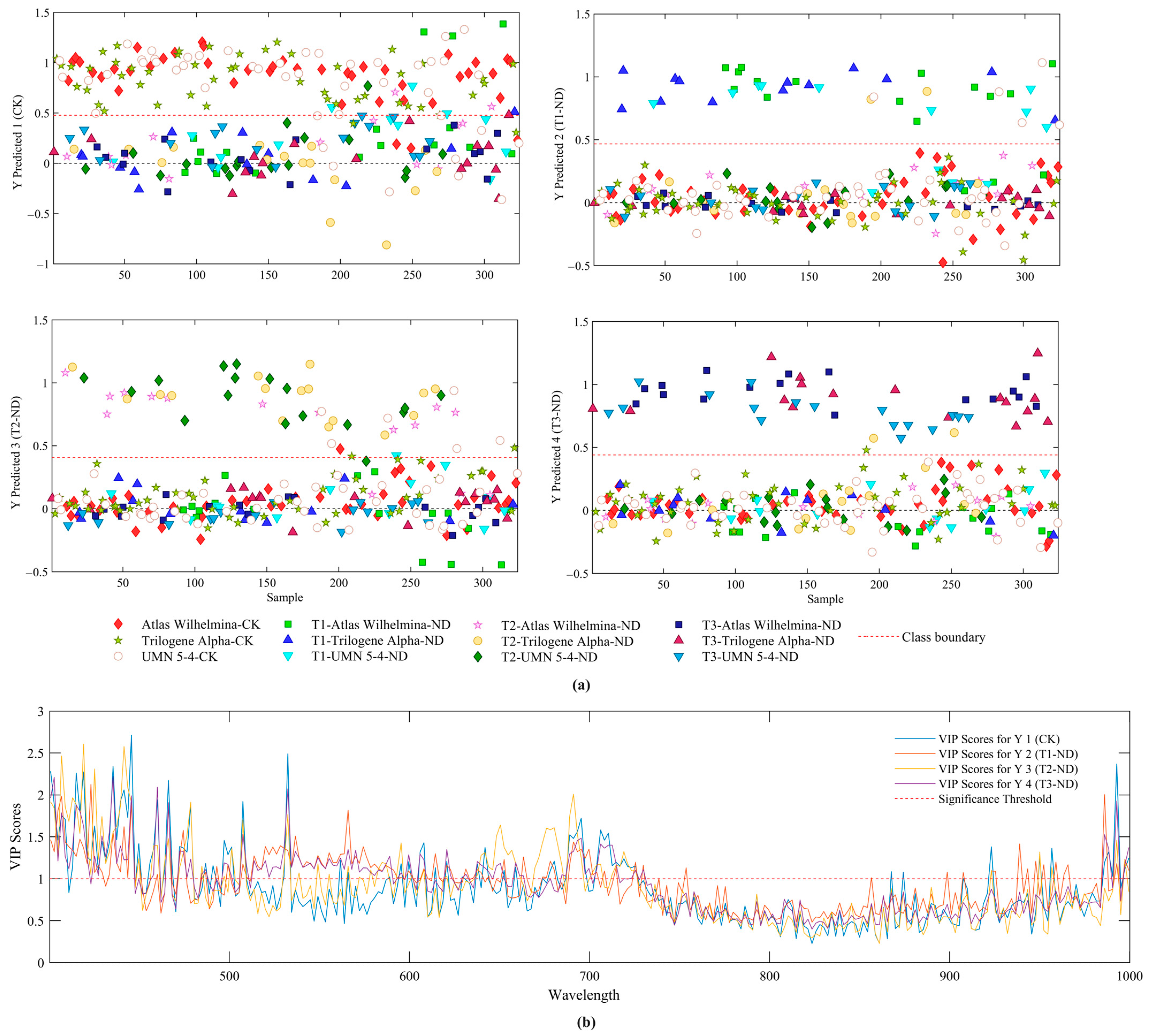
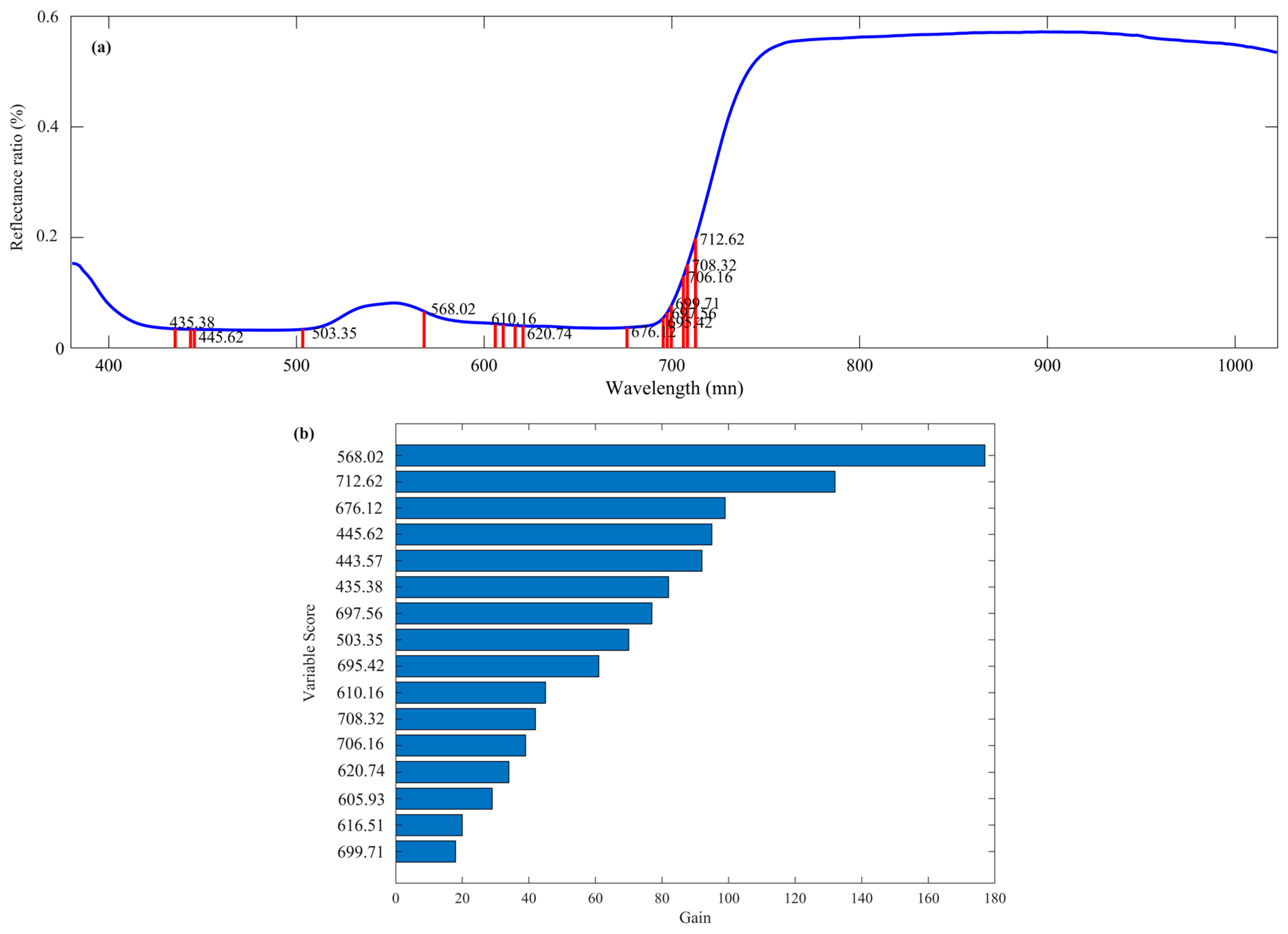
2.3.2. Wavelength Selection
2.3.3. Development of Classification Models
- Precision—The proportion of correctly classified positive samples out of all positive classifications. Higher values indicate greater effectiveness.
- Sensitivity—Measures the model’s ability to correctly identify positive cases. A sensitivity of 1 means all positive cases are detected.
- Specificity—Evaluates how well the model identifies negative cases. A specificity of 1 means no false positives occur.
- F1 Score—The harmonic mean of precision and sensitivity. Provides overall measure of model accuracy accounting for both false positives and false negatives.
- Class Error—The proportion of misclassified samples out of all samples. Lower values are better.
3. Results and Discussion
3.1. Spectral Response of Hemp Samples under Nutritional Stress
3.2. Outlier Detection Using RPCA
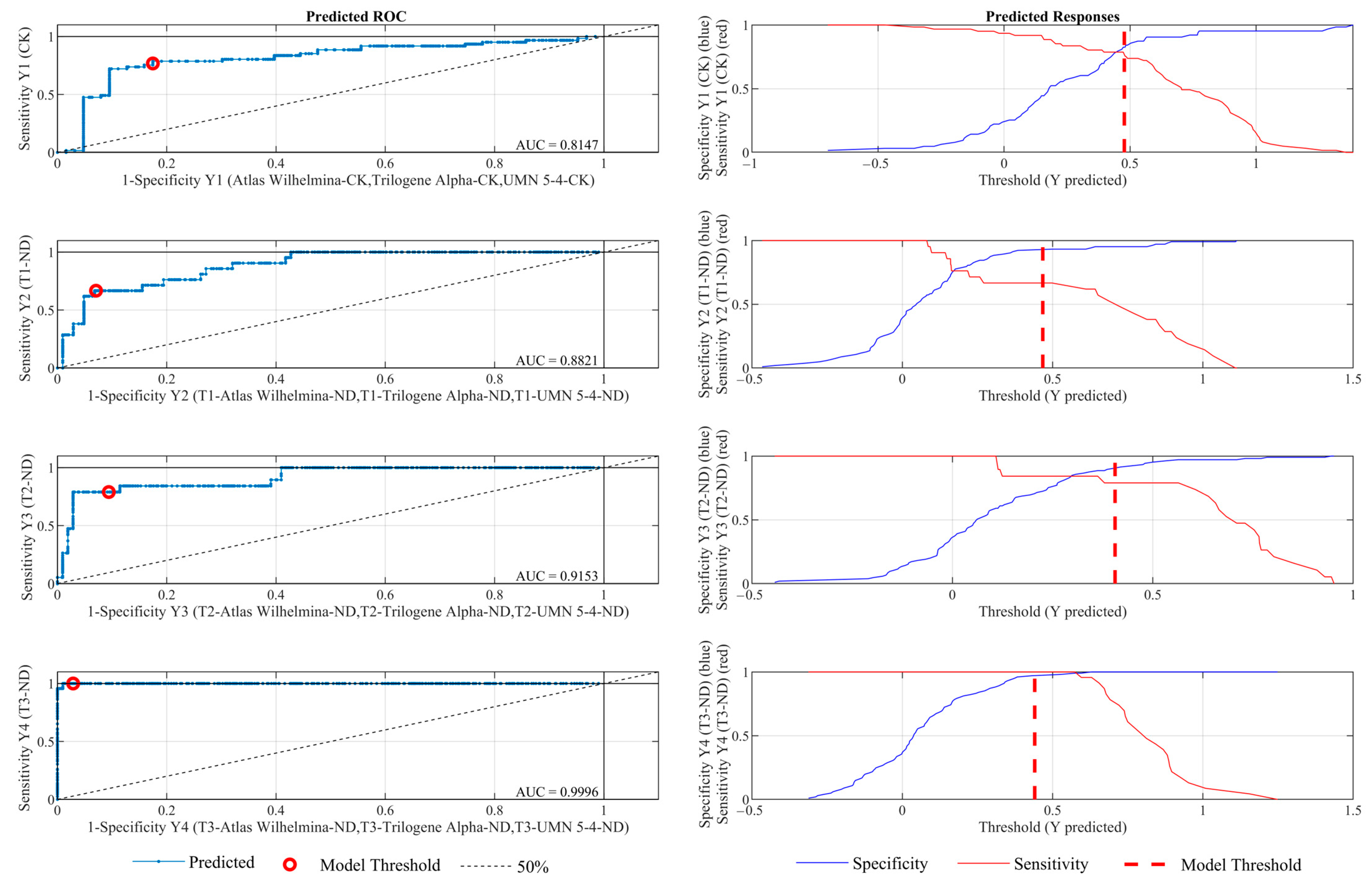
3.3. Temporal Classification of Nutrient Deficient Stress in Hemp Plants Using PLS-DA
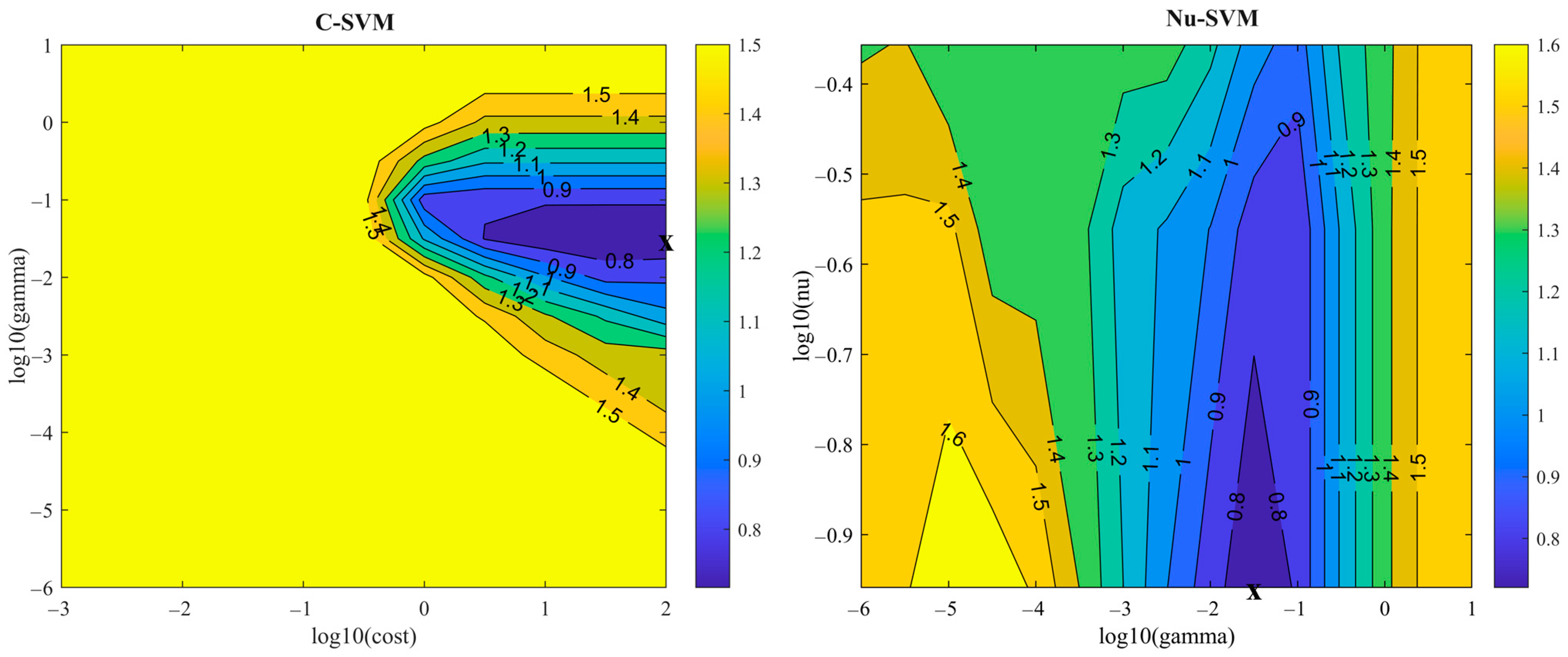
3.4. Enhancing Classification Performance Using SVM Models Combined with Wavelength Selection
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wylie, S.E.; Ristvey, A.G.; Fiorellino, N.M. Fertility Management for Industrial Hemp Production: Current Knowledge and Future Research Needs. GCB Bioenergy 2021, 13, 517–524. [Google Scholar] [CrossRef]
- Adesina, I.; Bhowmik, A.; Sharma, H.; Shahbazi, A. A Review on the Current State of Knowledge of Growing Conditions, Agronomic Soil Health Practices and Utilities of Hemp in the United States. Agriculture 2020, 10, 129. [Google Scholar] [CrossRef]
- Aubin, M.; Seguin, P.; Vanasse, A.; Tremblay, G.F.; Mustafa, A.F.; Charron, J. Industrial Hemp Response to Nitrogen, Phosphorus, and Potassium Fertilization. Crop Forage Turfgrass Manag. 2015, 1, 1–10. [Google Scholar] [CrossRef]
- Cockson, P.; Landis, H.; Smith, T.; Hicks, K.; Whipker, B.E. Characterization of Nutrient Disorders of Cannabis sativa. Appl. Sci. 2019, 9, 4432. [Google Scholar] [CrossRef]
- Payne, W.Z.; Kurouski, D. Raman-Based Diagnostics of Biotic and Abiotic Stresses in Plants. A Review. Front. Plant Sci. 2021, 11, 616672. [Google Scholar] [CrossRef] [PubMed]
- Sanaeifar, A.; Yang, C.; de la Guardia, M.; Zhang, W.; Li, X.; He, Y. Proximal Hyperspectral Sensing of Abiotic Stresses in Plants. Sci. Total Environ. 2023, 861, 160652. [Google Scholar] [CrossRef]
- Sanaeifar, A.; Zhang, W.; Chen, H.; Zhang, D.; Li, X.; He, Y. Study on Effects of Airborne Pb Pollution on Quality Indicators and Accumulation in Tea Plants Using Vis-NIR Spectroscopy Coupled with Radial Basis Function Neural Network. Ecotoxicol. Environ. Saf. 2022, 229, 113056. [Google Scholar] [CrossRef]
- Mohd Asaari, M.S.; Mishra, P.; Mertens, S.; Dhondt, S.; Inzé, D.; Wuyts, N.; Scheunders, P. Close-Range Hyperspectral Image Analysis for the Early Detection of Stress Responses in Individual Plants in a High-Throughput Phenotyping Platform. ISPRS J. Photogramm. Remote Sens. 2018, 138, 121–138. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern Imaging Techniques in Plant Nutrition Analysis: A Review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Lassalle, G. Monitoring Natural and Anthropogenic Plant Stressors by Hyperspectral Remote Sensing: Recommendations and Guidelines Based on a Meta-Review. Sci. Total Environ. 2021, 788, 147758. [Google Scholar] [CrossRef]
- Weksler, S.; Rozenstein, O.; Haish, N.; Moshelion, M.; Walach, R.; Ben-Dor, E. A Hyperspectral-Physiological Phenomics System: Measuring Diurnal Transpiration Rates and Diurnal Reflectance. Remote Sens. 2020, 12, 1493. [Google Scholar] [CrossRef]
- Siedliska, A.; Baranowski, P.; Pastuszka-Woźniak, J.; Zubik, M.; Krzyszczak, J. Identification of Plant Leaf Phosphorus Content at Different Growth Stages Based on Hyperspectral Reflectance. BMC Plant Biol. 2021, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Osco, L.P.; Ramos, A.P.M.; Pinheiro, M.M.F.; Moriya, É.A.S.; Imai, N.N.; Estrabis, N.; Ianczyk, F.; de Araújo, F.F.; Liesenberg, V.; de Castro Jorge, L.A.; et al. A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sens. 2020, 12, 906. [Google Scholar] [CrossRef]
- Holmes, W.S.; Po-Leen Ooi, M.; Kuang, Y.C.; Simpkin, R.; Lopez-Ubiria, I.; Vidiella, A.; Blanchon, D.; Gupta, G.S.; Demidenko, S. Classifying Cannabis sativa Flowers, Stems and Leaves Using Statistical Machine Learning with Near-Infrared Hyperspectral Reflectance Imaging. In Proceedings of the 2020 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Dubrovnik, Croatia, 25–28 May 2020; IEEE: New York, NY, USA, 2020; pp. 1–6. [Google Scholar]
- Pereira, J.F.Q.; Pimentel, M.F.; Amigo, J.M.; Honorato, R.S. Detection and Identification of Cannabis sativa L. Using near Infrared Hyperspectral Imaging and Machine Learning Methods. A Feasibility Study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 237, 118385. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Young, S.; Linder, E.; Whipker, B.; Suchoff, D. Hyperspectral Imaging With Machine Learning to Differentiate Cultivars, Growth Stages, Flowers, and Leaves of Industrial Hemp (Cannabis sativa L.). Front. Plant Sci. 2022, 12, 810113. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Young, S.; Li, X.; Linder, E.; Suchoff, D. Hyperspectral Imaging with Chemometrics for Non-Destructive Determination of Cannabinoids in Floral and Leaf Materials of Industrial Hemp (Cannabis sativa L.). Comput. Electron. Agric. 2022, 202, 107387. [Google Scholar] [CrossRef]
- Qin, J.; Monje, O.; Nugent, M.R.; Finn, J.R.; O’Rourke, A.E.; Wilson, K.D.; Fritsche, R.F.; Baek, I.; Chan, D.E.; Kim, M.S. A Hyperspectral Plant Health Monitoring System for Space Crop Production. Front. Plant Sci. 2023, 14, 1133505. [Google Scholar] [CrossRef] [PubMed]
- Htitiou, A.; Boudhar, A.; Lebrini, Y.; Hadria, R.; Lionboui, H.; Benabdelouahab, T. A Comparative Analysis of Different Phenological Information Retrieved from Sentinel-2 Time Series Images to Improve Crop Classification: A Machine Learning Approach. Geocarto Int. 2022, 37, 1426–1449. [Google Scholar] [CrossRef]
- Sanaeifar, A.; Li, X.; He, Y.; Huang, Z.; Zhan, Z. A Data Fusion Approach on Confocal Raman Microspectroscopy and Electronic Nose for Quantitative Evaluation of Pesticide Residue in Tea. Biosyst. Eng. 2021, 210, 206–222. [Google Scholar] [CrossRef]
- Luo, X.; Gouda, M.; Perumal, A.B.; Huang, Z.; Lin, L.; Tang, Y.; Sanaeifar, A.; He, Y.; Li, X.; Dong, C. Using Surface-Enhanced Raman Spectroscopy Combined with Chemometrics for Black Tea Quality Assessment during Its Fermentation Process. Sens. Actuators B Chem. 2022, 373, 132680. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, Q.X.; Sun, G.; Ding, C. Double Robust Principal Component Analysis. Neurocomputing 2020, 391, 119–128. [Google Scholar] [CrossRef]
- Yu, H.D.; Yun, Y.H.; Zhang, W.; Chen, H.; Liu, D.; Zhong, Q.; Chen, W.; Chen, W. Three-Step Hybrid Strategy towards Efficiently Selecting Variables in Multivariate Calibration of near-Infrared Spectra. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 224, 117376. [Google Scholar] [CrossRef] [PubMed]
- López-Maestresalas, A.; Lopez-Molina, C.; Oliva-Lobo, G.A.; Jarén, C.; Ruiz de Galarreta, J.I.; Peraza-Alemán, C.M.; Arazuri, S. Evaluation of Near-Infrared Hyperspectral Imaging for the Assessment of Potato Processing Aptitude. Front. Nutr. 2022, 9, 999877. [Google Scholar] [CrossRef] [PubMed]
- Sanaeifar, A.; Zhu, F.; Sha, J.; Li, X.; He, Y.; Zhan, Z. Rapid Quantitative Characterization of Tea Seedlings under Lead-Containing Aerosol Particles Stress Using Vis-NIR Spectra. Sci. Total Environ. 2022, 802, 149824. [Google Scholar] [CrossRef] [PubMed]
- Vieira, L.S.; Assis, C.; de Queiroz, M.E.L.R.; Neves, A.A.; de Oliveira, A.F. Building Robust Models for Identification of Adulteration in Olive Oil Using FT-NIR, PLS-DA and Variable Selection. Food Chem. 2021, 345, 128866. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, C.; Cao, F.; Liu, F.; Luo, S.; Tang, Y.; He, Y. Detection of Sclerotinia Stem Rot on Oilseed Rape (Brassica napus L.) Leaves Using Hyperspectral Imaging. Sensors 2018, 18, 1764. [Google Scholar] [CrossRef] [PubMed]
- Sanaeifar, A.; Bakhshipour, A.; De La Guardia, M. Prediction of Banana Quality Indices from Color Features Using Support Vector Regression. Talanta 2016, 148, 54–61. [Google Scholar] [CrossRef]
- Aissou, G.; Benouadah, S.; El Alami, H.; Kaabouch, N. Instance-Based Supervised Machine Learning Models for Detecting GPS Spoofing Attacks on UAS. In Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 26–29 January 2022; pp. 208–214. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral Image Analysis Techniques for the Detection and Classification of the Early Onset of Plant Disease and Stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef]
- Zubler, A.V.; Yoon, J.Y. Proximal Methods for Plant Stress Detection Using Optical Sensors and Machine Learning. Biosensors 2020, 10, 193. [Google Scholar] [CrossRef]
- Mishra, P.; Asaari, M.S.M.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close Range Hyperspectral Imaging of Plants: A Review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- Zhu, K.; Sun, Z.; Zhao, F.; Yang, T.; Tian, Z.; Lai, J.; Zhu, W.; Long, B. Relating Hyperspectral Vegetation Indices with Soil Salinity at Different Depths for the Diagnosis Ofwinter Wheat Salt Stress. Remote Sens. 2021, 13, 250. [Google Scholar] [CrossRef]
- Sawinski, K.; Mersmann, S.; Robatzek, S.; Böhmer, M. Guarding the Green: Pathways to Stomatal Immunity. Mol. Plant-Microbe Interact. 2013, 26, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Wang, L.; Park, B.; Kang, R.; Chen, Q. Simultaneous Quantification of Chemical Constituents in Matcha with Visible-near Infrared Hyperspectral Imaging Technology. Food Chem. 2021, 350, 129141. [Google Scholar] [CrossRef] [PubMed]
- Ao, R.; Mu, X.; He, J. Enhance Tensor RPCA-Based Mahalanobis Distance Method for Hyperspectral Anomaly Detection. IEEE Geosci. Remote Sens. Lett. 2022, 19, 6008305. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, B.; Wang, T.; Bonni, A.; Zhao, G. Robust Principal Component Analysis for Accurate Outlier Sample Detection in RNA-Seq Data. BMC Bioinform. 2020, 21, 269. [Google Scholar] [CrossRef] [PubMed]
- da Mata, M.M.; Rocha, P.D.; de Farias, I.K.T.; da Silva, J.L.B.; Medeiros:, E.P.; Silva, C.S.; da Silva Simões, S. Distinguishing Cotton Seed Genotypes by Means of Vibrational Spectroscopic Methods (NIR and Raman) and Chemometrics. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 266, 120399. [Google Scholar] [CrossRef]
- Zeng, J.; Ping, W.; Sanaeifar, A.; Xu, X.; Luo, W.; Sha, J.; Huang, Z.; Huang, Y.; Liu, X.; Zhan, B.; et al. Quantitative Visualization of Photosynthetic Pigments in Tea Leaves Based on Raman Spectroscopy and Calibration Model Transfer. Plant Methods 2021, 17, 4. [Google Scholar] [CrossRef]
- Zhu, X.; Cheng, Y.; Wu, J.; Hu, R.; Cui, X. Steady-State Process Fault Detection for Liquid Rocket Engines Based on Convolutional Auto-Encoder and One-Class Support Vector Machine. IEEE Access 2020, 8, 3144–3158. [Google Scholar] [CrossRef]
- Fonseca Cacho, J.R.; Taghva, K. OCR Post Processing Using Support Vector Machines. In Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1229, pp. 694–713. [Google Scholar]
- Asefa, B.G.; Tsige, F.; Mehdi, M.; Kore, T.; Lakew, A. Rapid Classification of Tef [Eragrostis Tef (Zucc.) Trotter] Grain Varieties Using Digital Images in Combination with Multivariate Technique. Smart Agric. Technol. 2023, 3, 100097. [Google Scholar] [CrossRef]
- Rubio-Delgado, J.; Pérez, C.J.; Vega-Rodríguez, M.A. Predicting Leaf Nitrogen Content in Olive Trees Using Hyperspectral Data for Precision Agriculture. Precis. Agric. 2021, 22, 1–21. [Google Scholar] [CrossRef]
- Yamashita, H.; Sonobe, R.; Hirono, Y.; Morita, A.; Ikka, T. Dissection of Hyperspectral Reflectance to Estimate Nitrogen and Chlorophyll Contents in Tea Leaves Based on Machine Learning Algorithms. Sci. Rep. 2020, 10, 17360. [Google Scholar] [CrossRef] [PubMed]
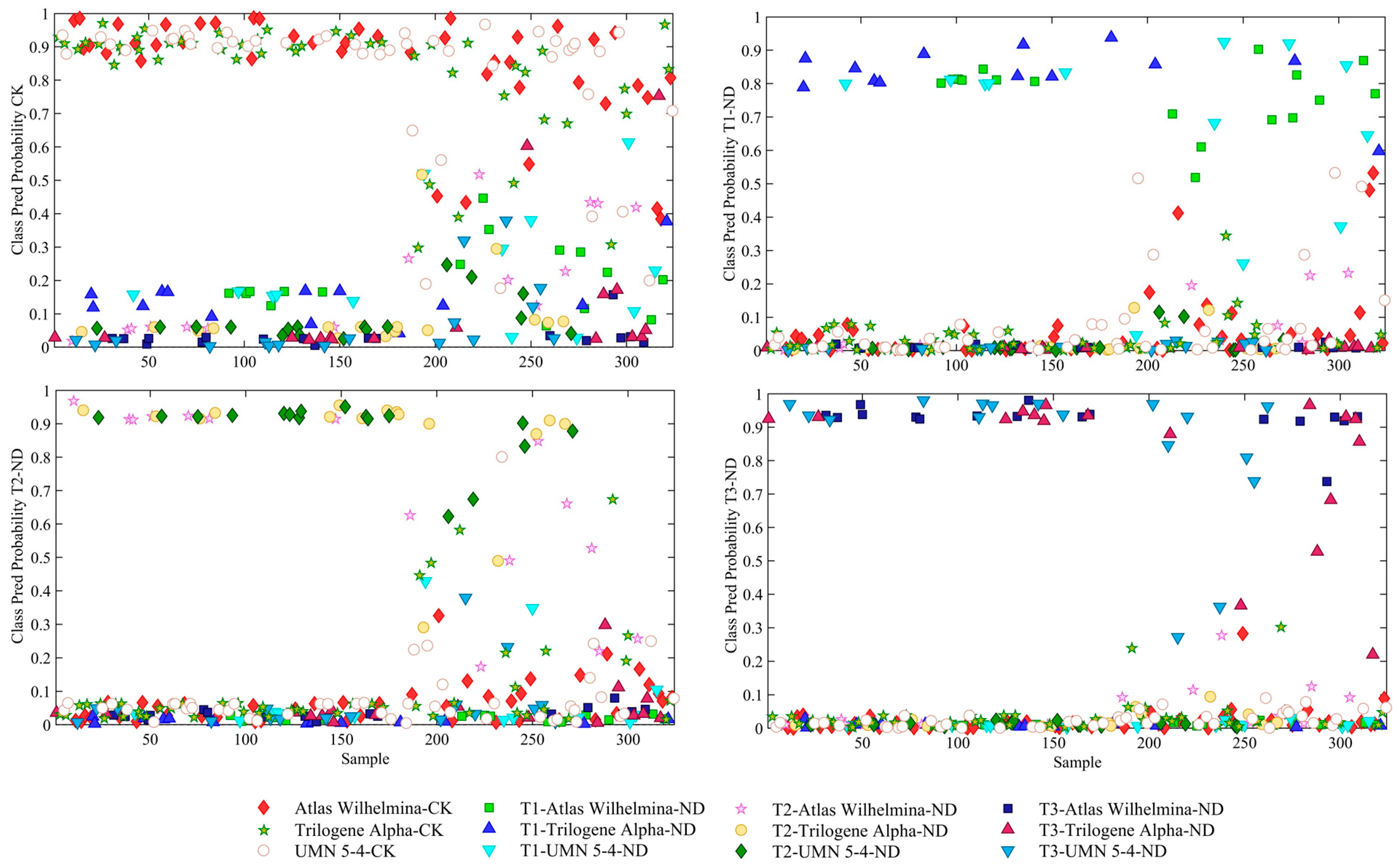
| Variety/Nutrient Deficiency Stage | T1 | T2 | T3 |
|---|---|---|---|
| Atlas Wilhelmina | CK (18), ND (18) | CK (18), ND (18) | CK (18), ND (18) |
| Trilogene Alpha | CK (18), ND (18) | CK (18), ND (18) | CK (18), ND (18) |
| UMN 5-4 | CK (18), ND (18) | CK (18), ND (18) | CK (18), ND (18) |
| Class | Sensitivity | Specificity | Class Error | Precision | F1 | |
|---|---|---|---|---|---|---|
| CK | Train | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 |
| Validation | 0.883 | 0.848 | 0.135 | 0.850 | 0.866 | |
| Test | 0.787 | 0.841 | 0.185 | 0.828 | 0.807 | |
| T1-ND | Train | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 |
| Validation | 0.773 | 0.985 | 0.045 | 0.895 | 0.829 | |
| Test | 0.667 | 0.932 | 0.113 | 0.667 | 0.667 | |
| T2-ND | Train | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 |
| Validation | 0.759 | 0.969 | 0.071 | 0.846 | 0.800 | |
| Test | 0.684 | 0.924 | 0.113 | 0.619 | 0.650 | |
| T3-ND | Train | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 |
| Validation | 0.964 | 0.969 | 0.032 | 0.871 | 0.915 | |
| Test | 1.000 | 0.990 | 0.008 | 0.958 | 0.979 | |
| Wavelength Selection Method | Number of Wavelengths | SVM Type | SVM Optimal Parameters | Class | Sensitivity | Specificity | Class Error | Precision | F1 |
|---|---|---|---|---|---|---|---|---|---|
| VIP | 70 | C-SVM | Cost = 100 Gamma = 0.00031623 | CK | 0.836 | 0.778 | 0.194 | 0.785 | 0.810 |
| T1-ND | 0.571 | 0.942 | 0.121 | 0.667 | 0.615 | ||||
| T2-ND | 0.684 | 0.952 | 0.089 | 0.722 | 0.703 | ||||
| T3-ND | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 | ||||
| Nu-SVM | Nu = 0.275 Gamma = 0.0001 | CK | 0.852 | 0.778 | 0.185 | 0.788 | 0.819 | ||
| T1-ND | 0.571 | 0.961 | 0.105 | 0.750 | 0.649 | ||||
| T2-ND | 0.737 | 0.952 | 0.081 | 0.737 | 0.737 | ||||
| T3-ND | 1.000 | 1.000 | 0.000 | 1.000 | 1.000 | ||||
| iPLS | 16 | C-SVM | Cost = 100 Gamma = 0.031623 | CK | 0.852 | 0.841 | 0.153 | 0.839 | 0.846 |
| T1-ND | 0.857 | 0.951 | 0.065 | 0.783 | 0.818 | ||||
| T2-ND | 0.789 | 0.952 | 0.073 | 0.750 | 0.769 | ||||
| T3-ND | 0.826 | 1.000 | 0.032 | 1.000 | 0.905 | ||||
| Nu-SVM | Nu = 0.11 Gamma = 0.031623 | CK | 0.820 | 0.841 | 0.169 | 0.833 | 0.826 | ||
| T1-ND | 0.857 | 0.942 | 0.073 | 0.750 | 0.800 | ||||
| T2-ND | 0.789 | 0.943 | 0.081 | 0.714 | 0.750 | ||||
| T3-ND | 0.826 | 1.000 | 0.032 | 1.000 | 0.905 |
| Actual Class | |||||
|---|---|---|---|---|---|
| CK | T1-ND | T2-ND | T3-ND | ||
| Predicted as | CK | 52 | 3 | 4 | 3 |
| T1-ND | 5 | 18 | 0 | 0 | |
| T2-ND | 4 | 0 | 15 | 1 | |
| T3-ND | 0 | 0 | 0 | 19 | |
| Unassigned | 0 | 0 | 0 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanaeifar, A.; Yang, C.; Min, A.; Jones, C.R.; Michaels, T.E.; Krueger, Q.J.; Barnes, R.; Velte, T.J. Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging. Remote Sens. 2024, 16, 187. https://doi.org/10.3390/rs16010187
Sanaeifar A, Yang C, Min A, Jones CR, Michaels TE, Krueger QJ, Barnes R, Velte TJ. Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging. Remote Sensing. 2024; 16(1):187. https://doi.org/10.3390/rs16010187
Chicago/Turabian StyleSanaeifar, Alireza, Ce Yang, An Min, Colin R. Jones, Thomas E. Michaels, Quinton J. Krueger, Robert Barnes, and Toby J. Velte. 2024. "Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging" Remote Sensing 16, no. 1: 187. https://doi.org/10.3390/rs16010187
APA StyleSanaeifar, A., Yang, C., Min, A., Jones, C. R., Michaels, T. E., Krueger, Q. J., Barnes, R., & Velte, T. J. (2024). Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging. Remote Sensing, 16(1), 187. https://doi.org/10.3390/rs16010187









