Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province
Abstract
1. Introduction
2. Materials and Methods
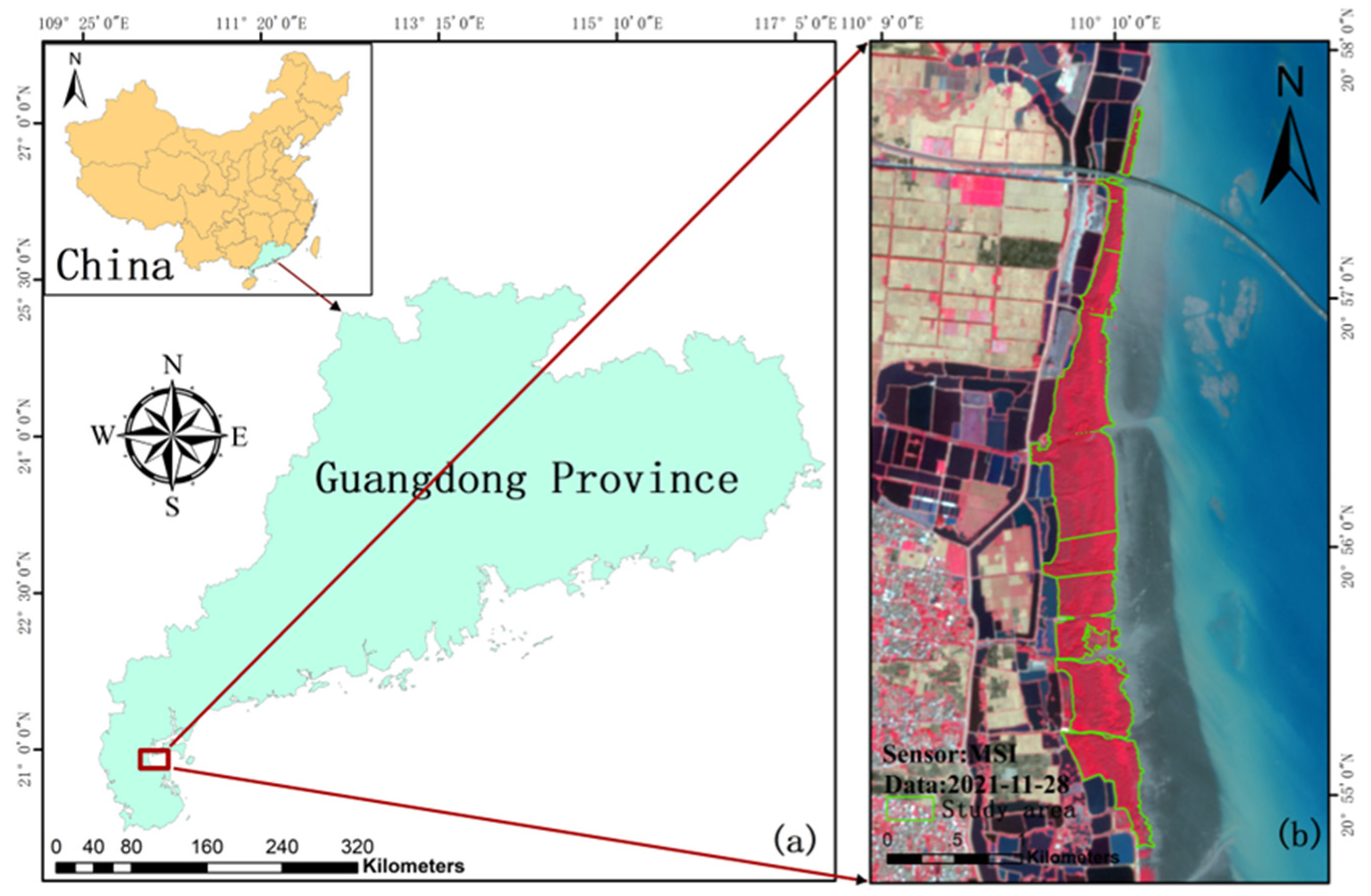
2.1. Study Area
2.2. Field Data
2.3. Remote Sensing Data and Pre-Processing
2.3.1. Optical Data and Pre-Processing
2.3.2. SAR Data Pre-Processing
2.4. Optical and SAR Data Conversion
| Variables | Definition | Source |
|---|---|---|
| DVI | [26] | |
| MSAVI | [23] | |
| GNDVI | [27] | |
| IRECI | [28] | |
| SAVI | [29] | |
| RVI | [30] | |
| NDI45 | [31] | |
| Mean | [32] | |
| Variance | ||
| Homogeneity | ||
| Contrast | ||
| Dissimilarity | ||
| Entropy | ||
| ASM | ||
| New band combination 1 | B8 + B4 | |
| New band combination 2 | 5(B9 − B4) |
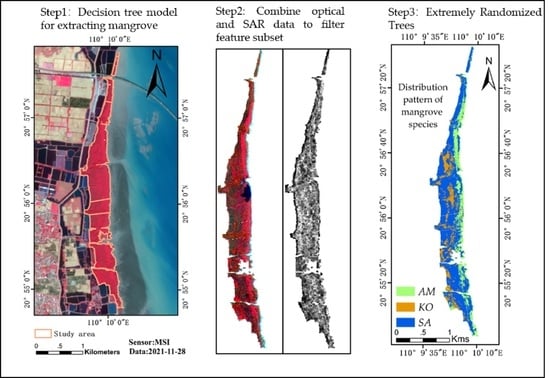
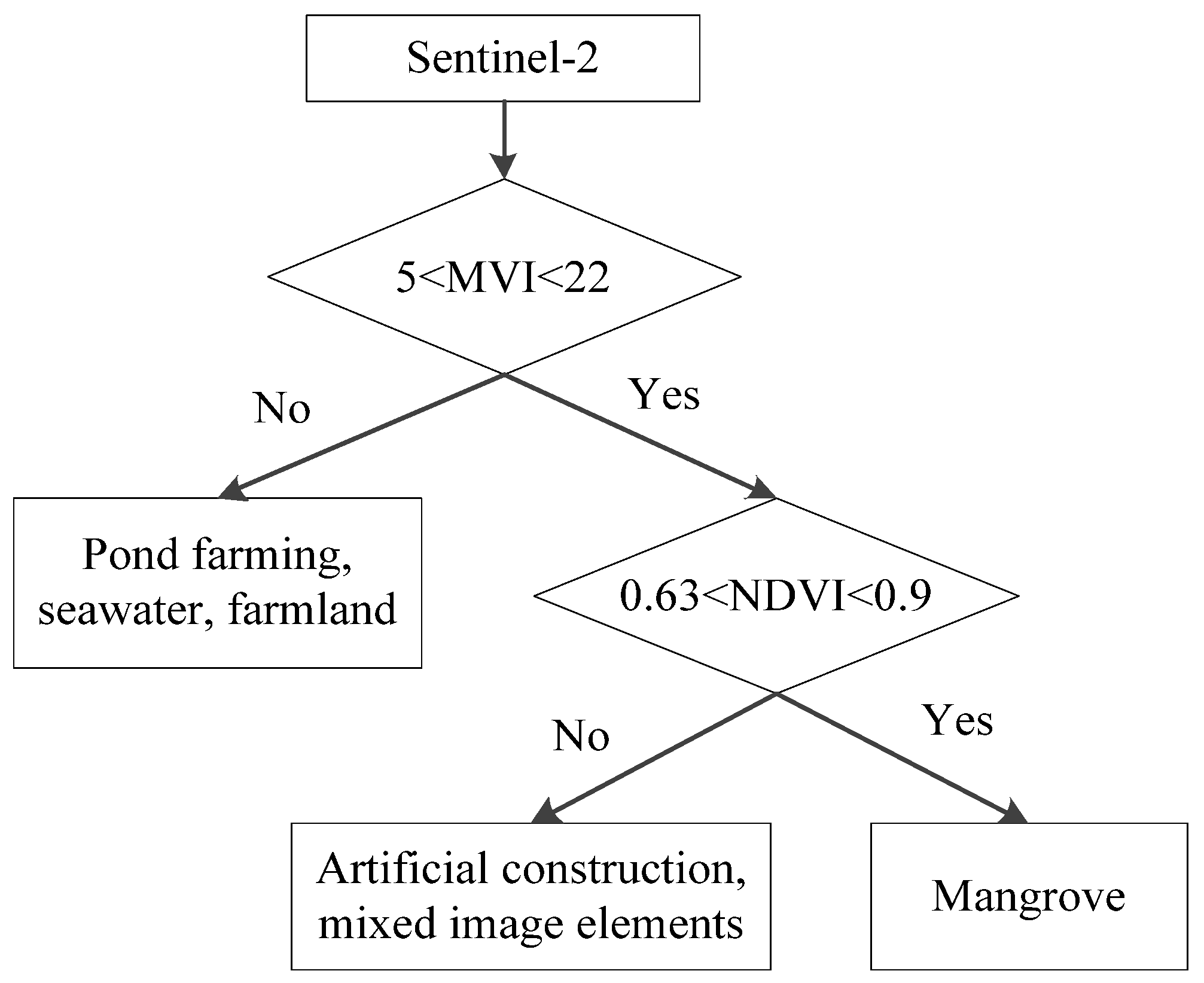
2.5. Mangroves and Non-Mangroves: Decision Trees
2.6. Mangrove Interspecies Classification: Extremely Randomized Trees
3. Results
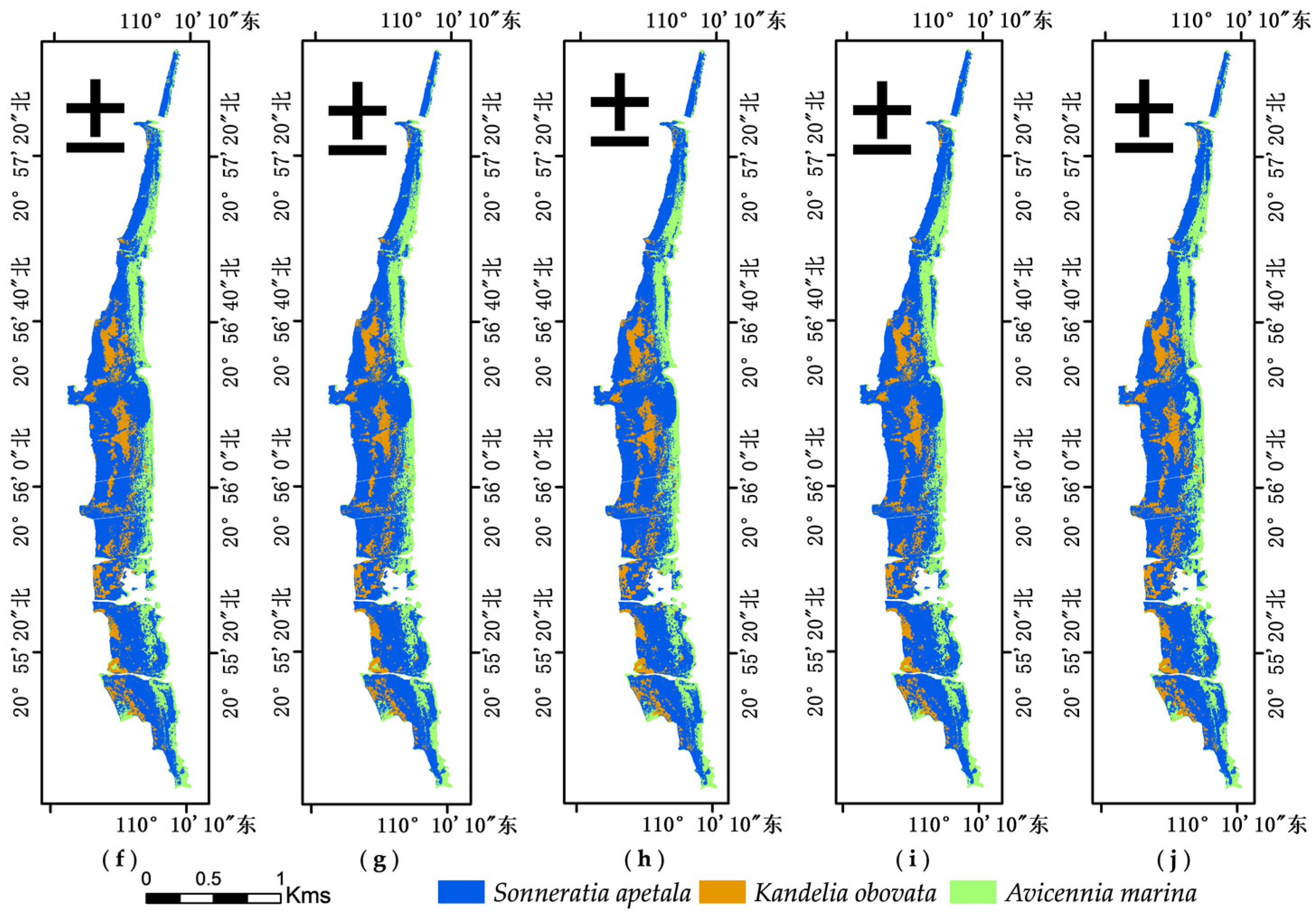
3.1. Species Classification Results from Different Data Sources
3.2. User Accuracy and Producer Accuracy
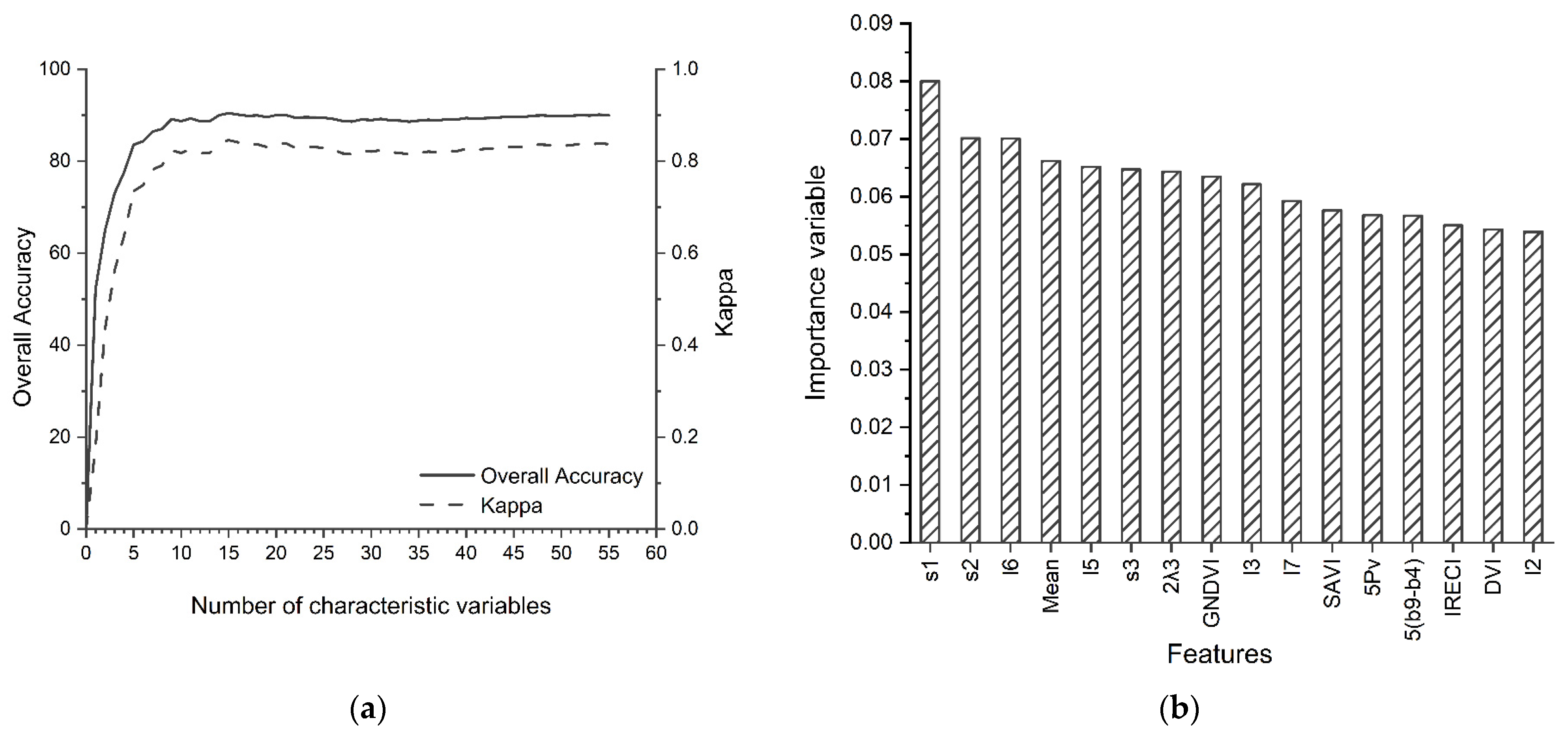
3.3. Data Feature Selection Results and Feature Importance
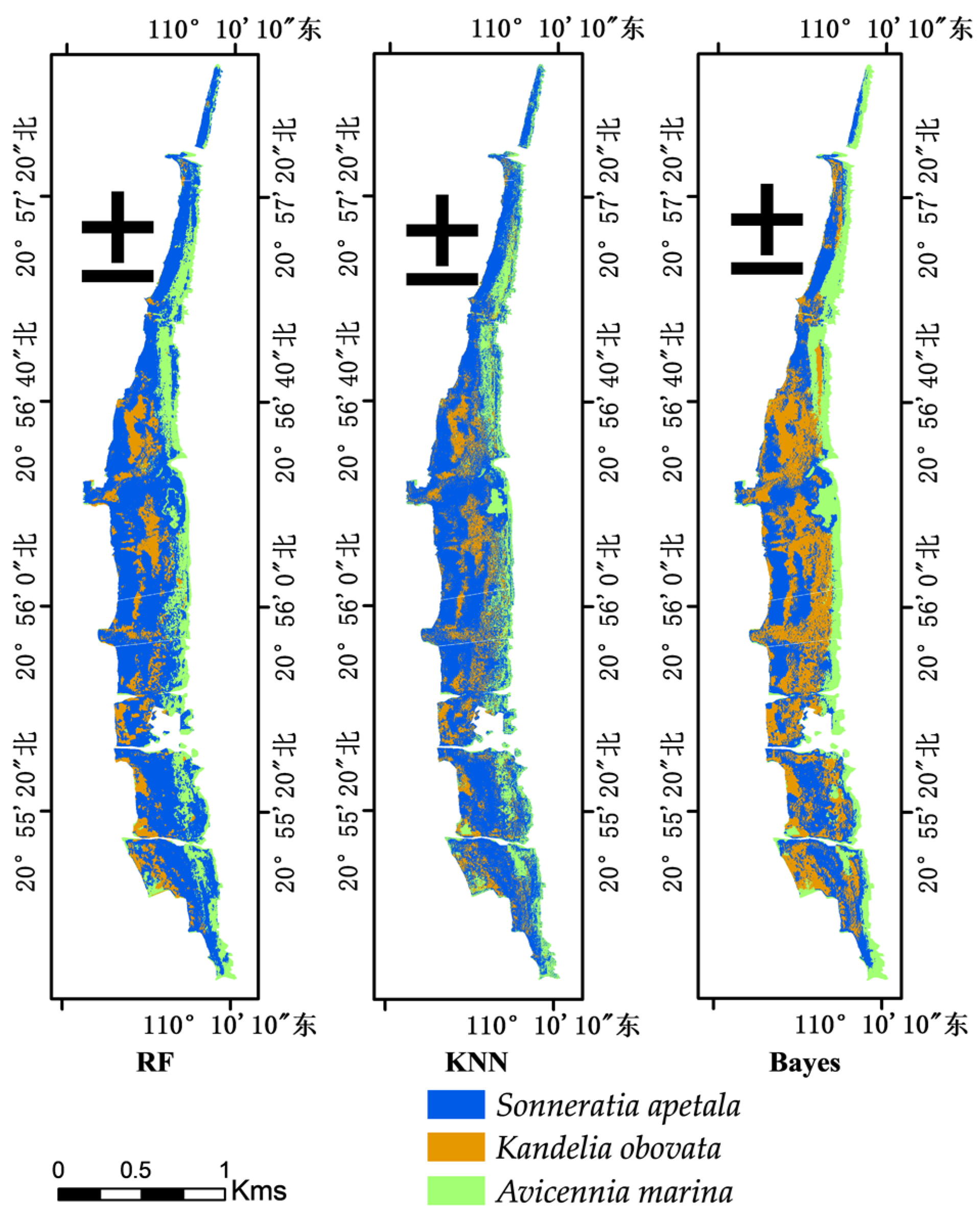
3.4. Classification Results of Different Methods Based on the Preferred Feature Set
4. Discussion
4.1. Effect of Spatial Resolution of Spectral Data on Species Classification
4.2. Influence of Spectral Bands on Species Classification in Multispectral Data
4.3. Contribution of SAR Data Features
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, J.; Liu, K.; Liu, L.; Zhu, Y.; Li, J.; He, Z. Identifying mangrove species using field close-range snapshot hyperspectral imaging and machine-learning techniques. Remote Sens. 2018, 10, 2047. [Google Scholar] [CrossRef]
- Friess, D.A.; Rogers, K.; Lovelock, C.E.; Krauss, K.W.; Hamilton, S.E.; Lee, S.Y.; Lucas, R.; Primavera, J.; Rajkaran, A.; Shi, S. The state of the world’s mangrove forests: Past, present, and future. Annu. Rev. Environ. Resour. 2019, 44, 89–115. [Google Scholar] [CrossRef]
- Ramírez-García, P.; Lopez-Blanco, J.; Ocaña, D. Mangrove vegetation assessment in the santiago river mouth, mexico, by means of supervised classification using landsat tm imagery. For. Ecol. Manag. 1998, 105, 217–229. [Google Scholar] [CrossRef]
- Ghosh, M.K.; Kumar, L.; Roy, C. Mapping long-term changes in mangrove species composition and distribution in the sundarbans. Forests 2016, 7, 305. [Google Scholar] [CrossRef]
- Myint, S.W.; Giri, C.P.; Wang, L.; Zhu, Z.; Gillette, S.C. Identifying mangrove species and their surrounding land use and land cover classes using an object-oriented approach with a lacunarity spatial measure. GIScience Remote Sens. 2008, 45, 188–208. [Google Scholar] [CrossRef]
- Giri, S.; Mukhopadhyay, A.; Hazra, S.; Mukherjee, S.; Roy, D.; Ghosh, S.; Ghosh, T.; Mitra, D. A study on abundance and distribution of mangrove species in indian sundarban using remote sensing technique. J. Coast. Conserv. 2014, 18, 359–367. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, L.; Wang, L. Evaluation of morphological texture features for mangrove forest mapping and species discrimination using multispectral ikonos imagery. IEEE Geosci. Remote Sens. Lett. 2009, 6, 393–397. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Lin, H.; Fang, C. Textural–spectral feature-based species classification of mangroves in mai po nature reserve from worldview-3 imagery. Remote Sens. 2016, 8, 24. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Wang, S.; Liu, H. Retrieval of mangrove aboveground biomass at the individual species level with worldview-2 images. Remote Sens. 2015, 7, 12192–12214. [Google Scholar] [CrossRef]
- Hirata, Y.; Tabuchi, R.; Patanaponpaiboon, P.; Poungparn, S.; Yoneda, R.; Fujioka, Y. Estimation of aboveground biomass in mangrove forests using high-resolution satellite data. J. For.-JPN 2014, 19, 34–41. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, H.; Wang, T.; Li, G.; Lin, H. Mangrove species discrimination from very high resolution imagery using gaussian markov random field model. Wetlands 2018, 38, 861–874. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Qiu, P.; Su, Y.; Guo, Q.; Wu, X. Artificial mangrove species mapping using pléiades-1: An evaluation of pixel-based and object-based classifications with selected machine learning algorithms. Remote Sens. 2018, 10, 294. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Qiu, P.; Su, Y.; Guo, Q.; Wang, R.; Sun, F.; Wu, X. Evaluating the performance of sentinel-2, landsat 8 and pléiades-1 in mapping mangrove extent and species. Remote Sens. 2018, 10, 1468. [Google Scholar] [CrossRef]
- Hati, J.P.; Samanta, S.; Chaube, N.R.; Misra, A.; Giri, S.; Pramanick, N.; Gupta, K.; Majumdar, S.D.; Chanda, A.; Mukhopadhyay, A.; et al. Mangrove classification using airborne hyperspectral aviris-ng and comparing with other spaceborne hyperspectral and multispectral data. Egypt. J. Remote Sens. Space Sci. 2021, 24, 273–281. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Myint, S.W.; Du, Z.; Li, Y.; Cao, J.; Liu, L.; Wu, Z. Integration of gf2 optical, gf3 sar, and uav data for estimating aboveground biomass of chia’s largest artificially planted mangroves. Remote Sens. 2020, 12, 2039. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, T.; Liu, M.; Jia, M.; Lin, H.; Chu, L.M.; Devlin, A.T. Potential of combining optical and dual polarimetric sar data for improving mangrove species discrimination using rotation forest. Remote Sens. 2018, 10, 467. [Google Scholar] [CrossRef]
- Kripa, M.K.; Lele, N.; Panda, M.; Kumar Das, S.; Nivas, A.H.; Divakaran, N.; Sawant, A.; Naik-Gaonkar, S.; Pattnaik, A.K.; Samal, R.N.; et al. Biodiversity assessment of indian mangroves using in situ observations and remotely sensed data. Biodiversity 2020, 21, 198–216. [Google Scholar] [CrossRef]
- Ma, C.; Ai, B.; Zhao, J.; Xu, X.; Huang, W. Change detection of mangrove forests in coastal guangdong during the past three decades based on remote sensing data. Remote Sens. 2019, 11, 921. [Google Scholar] [CrossRef]
- Wong, F.K.; Fung, T. Combining eo-1 hyperion and envisat asar data for mangrove species classification in mai po ramsar site, hong kong. Int. J. Remote Sens. 2014, 35, 7828–7856. [Google Scholar] [CrossRef]
- Gao, X.M.; Liu, H. The mangrove and its conservation in leizhou peninsula, china. J. For. Res. 2009, 20, 174–178. [Google Scholar] [CrossRef]
- Ren, H. Restoration of mangrove plantations and colonisation by native species in leizhou bay, south china. Ecol. Res. 2008, 23, 401–407. [Google Scholar] [CrossRef]
- Dobson, M.C.; Ulaby, F.T.; LeToan, T.; Beaudoin, A.; Kasischke, E.S.; Christensen, N. Dependence of radar backscatter on coniferous forest biomass. IEEE Trans. Geosci. Remote Sens. 1992, 30, 412–415. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Lucas, R.M.; Ticehurst, C.; Prosiy, C.; Melius, A.; Rosenqvist, A.; Lucas, R.M. The potential of l-band sar for quantifying mangrove characteristics and change: Case studies from the tropics. Aquat. Conserv. Mar. Freshw. Ecosyst. 2007, 17, 245–264. [Google Scholar] [CrossRef]
- Brown, I.; Mwansasu, S.; Westerberg, L.O. L-band polarimetric target decomposition of mangroves of the rufiji delta, tanzania. Remote Sens. 2016, 8, 140. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S.A. modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Daughtry, C.S.; Walthall, C.; Kim, M.; De Colstoun, E.B.; McMurtrey Iii, J. Estimating corn leaf chlorophyll concentration from leaf and canopy reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (savi). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Blackburn, G.A. Quantifying chlorophylls and caroteniods at leaf and canopy scales: An evaluation of some hyperspectral approaches. Remote Sens. Environ. 1998, 66, 273–285. [Google Scholar] [CrossRef]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of sentinel-2 red-edge bands for empirical estimation of green lai and chlorophyll content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Zhang, C.; Ren, G.; WU, P.; HU, Y.; Ma, Y.; Yan, Y.; Zhang, J. Mangrove species classification in Hainan bamen Bay based on GF optics and fully polarimetric SAR. J. Trop. Oceanogr. 2022, 41, 1–17. Available online: http://www.jto.ac.cn/CN/10.11978/2022096 (accessed on 14 July 2022).
- Baloloy, A.B.; Blanco, A.C.; Ana, R.R.C.S.; Nadaoka, K. Development and application of a new mangrove vegetation index (mvi) for rapid and accurate mangrove mapping. ISPRS J. Photogramm. Remote Sens. 2020, 166, 95–117. [Google Scholar] [CrossRef]
- Shang, K.; Yao, Y.; Li, Y.; Yang, J.; Guo, X. Fusion of five satellite-derived products using extremely randomized trees to estimate terrestrial latent heat flux over europe. Remote Sens. 2020, 12, 687. [Google Scholar] [CrossRef]
- Eslami, E.; Salman, A.; Choi, Y.; Sayeed, A.; Lops, Y. A data ensemble approach for real-time air quality forecasting using extremely randomized trees and deep neural networks. Neural Comput. Appl. 2020, 32, 7563–7579. [Google Scholar] [CrossRef]
- Heddam, S.; Ptak, M.; Zhu, S. Modelling of daily lake surface water temperature from air temperature: Extremely randomized trees (ert) versus air2water, mars, m5tree, rf and mlpnn. J. Hydrol. 2020, 588, 125130. [Google Scholar] [CrossRef]
- Bunting, P.; Rosenqvist, A.; Lucas, R.M.; Rebelo, L.M.; Hilarides, L.; Thomas, N.; Hardy, A.; Itoh, T.; Shimada, M.; Finlayson, C.M. The global mangrove watch—A new 2010 global baseline of mangrove extent. Remote Sens. 2018, 10, 1669. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Skakun, S.; Kussul, N.; Shelestov, A.Y.; Lavreniuk, M.; Kussul, O. Efficiency assessment of multitemporal c-band radarsat-2 intensity and landsat-8 surface reflectance satellite imagery for crop classification in ukraine. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 9, 3712–3719. [Google Scholar] [CrossRef]
- Yang, Y.; Tian, Q.; Zhan, Y.; Tao, B.; Kaijian, X.U. Effects of spatial resolution and texture features on multi-spectral remote sensing classification. J. Geo-Inf. Sci. 2018, 20, 99–107. [Google Scholar] [CrossRef]
- Xu, K.; Tian, Q.; Yang, Y.; Xu, N. Response of spatial scale for land cover classification of remote sensing. Geo-Inf. Sci. 2018, 20, 246–253. [Google Scholar] [CrossRef]
- Xia, J.; Yokoya, N.; Pham, T.D. Probabilistic mangrove species mapping with multiple-source remote-sensing datasets using label distribution learning in xuan thuy national park, vietnam. Remote Sens. 2020, 12, 3834. [Google Scholar] [CrossRef]



| Species Name | Field Pictures | Remote Sensing Image | Interpretation Features |
|---|---|---|---|
| Avicennia marina |  |  | Avicennia marina is bright red in the remote sensing image, and the texture is smooth and fine |
| Kandelia obovata |  |  | The Kandelia obovata is dark red in the remote sensing image, with fine texture |
| Sonneratia apetala |  |  | The Sonneratia apetala is dark red in the remote sensing image, with rough texture |
| Satellite | Date | Bands | Spatial Resolution (m) |
|---|---|---|---|
| GaoFen-1 | 15 May 2021 | 4 | 2 |
| GaoFen-3 | 2 February 2021 | 4 | 8 |
| GaoFen-3 | 13 May 2022 | 4 | 8 |
| Sentinel-2 | 28 November 2021 | 12 | 10 |
| Landsat-9 | 4 May 2022 | 7 | 15 |
| Serial No. | Data Used | Bands/Indices Used | Resolution (m) |
|---|---|---|---|
| 1 | Sentinel-2 | original 12 bands | 10 |
| 2 | Sentinel-2, Gaofen-1 | 12 bands of Sentinel-2 after fusion | 2 |
| 3 | Landsat-9 | original 7 bands | 15 |
| 4 | Landsat-9, Gaofen-1 | 7 bands of Landsat-9 after fusion | 2 |
| 5 | Sentinel-2, Landsat-9 Gaofen-1 | 12 bands of Sentinel-2 after fusion, 7 bands of Landsat-9 after fusion | 2 |
| 6 | Sentinel-2, Landsat-9 Gaofen-1 | 12 bands of Sentinel-2 after fusion, 7 bands of Landsat-9 after fusion, 9 indices, 7 texture features | 2 |
| 7 | Sentinel-2, Landsat-9 Gaofen-1, Gaofen-3 | 12 bands of Sentinel-2 after fusion, 7 bands of Landsat-9 after fusion, 9 indices, 7 texture features, backscattering coefficients HH, HV, VH, and VV, and polarization decomposition characteristics in February | 2 |
| 8 | Sentinel-2, Landsat-9 Gaofen-1, Gaofen-3 | 12 bands of Sentinel-2 after fusion, 7 bands of Landsat-9 after fusion, 9 indices, 7 texture features, backscattering coefficients HH, HV, VH, and VV, and polarization decomposition characteristics in May | 2 |
| 9 | Sentinel-2, Landsat-9 Gaofen-1, Gaofen-3 | 12 bands of Sentinel-2 after fusion, 7 bands of Landsat-9 after fusion, 9 indices, 7 texture features, backscattering coefficients HH, HV, VH, and VV, and polarization decomposition characteristics in February and May | 2 |
| 10 | Preferred subset of features | Sentinel-2 deep blue band, blue band, green band, DVI, 5 (b9-b4), Landsad-9’s green, NIR, shortwave IR1, shortwave IR2 band, mean, GNDVI, IRECI, SAVI, 2λ3, 5 Pv | 2 |
| Serial No. | ERT | OA | Kappa | |||
|---|---|---|---|---|---|---|
| SA | KO | AM | ||||
| 1 | PA | 0.69 | 0.69 | 0.59 | 66.59% | 0.45 |
| UA | 0.76 | 0.55 | 0.6 | |||
| 2 | PA | 0.84 | 0.86 | 0.85 | 84.91% | 0.75 |
| UA | 0.89 | 0.78 | 0.84 | |||
| 3 | PA | 0.6 | 0.55 | 0.63 | 59.68% | 0.34 |
| UA | 0.66 | 0.47 | 0.61 | |||
| 4 | PA | 0.81 | 0.79 | 0.8 | 80.45% | 0.68 |
| UA | 0.84 | 0.75 | 0.8 | |||
| 5 | PA | 0.85 | 0.86 | 0.86 | 85.49% | 0.76 |
| UA | 0.88 | 0.8 | 0.86 | |||
| 6 | PA | 0.84 | 0.86 | 0.87 | 85.10% | 0.75 |
| UA | 0.9 | 0.77 | 0.83 | |||
| 7 | PA | 0.86 | 0.9 | 0.9 | 87.95% | 0.8 |
| UA | 0.93 | 0.81 | 0.86 | |||
| 8 | PA | 0.86 | 0.9 | 0.92 | 88.51% | 0.81 |
| UA | 0.93 | 0.81 | 0.86 | |||
| 9 | PA | 0.88 | 0.92 | 0.93 | 90% | 0.83 |
| UA | 0.94 | 0.83 | 0.88 | |||
| 10 | PA | 0.89 | 0.91 | 0.91 | 90.13% | 0.84 |
| UA | 0.93 | 0.86 | 0.90 | |||
| Classifier | RF | KNN | Bayes | ||||
|---|---|---|---|---|---|---|---|
| PA | UA | PA | UA | PA | UA | ||
| SA | 0.88 | 0.91 | 0.72 | 0.77 | 0.8 | 0.51 | |
| KO | 0.89 | 0.84 | 0.67 | 0.63 | 0.47 | 0.81 | |
| AM | 0.89 | 0.88 | 0.72 | 0.65 | 0.63 | 0.61 | |
| OA | 88.47% | 70.74% | 61.26% | ||||
| Kappa | 0.81 | 0.52 | 0.42 | ||||
| Sample True Value | Classification Result | |||
|---|---|---|---|---|
| SA | AM | KO | Total | |
| SA | 87.01961569 | 6.293972504 | 6.029115196 | 99.34270339 |
| AM | 5.690461859 | 15.28142336 | 0.742151633 | 21.71403685 |
| KO | 9.016527601 | 1.381555296 | 23.62586764 | 34.02395053 |
| total | 101.7266051 | 22.95695116 | 30.39713447 | 155.0806908 |
| Sample True Value | Classification Result | |||
|---|---|---|---|---|
| SA | AM | KO | Total | |
| SA | 86.77964846 | 6.562051645 | 6.000983684 | 99.34268379 |
| AM | 5.515079054 | 15.48237008 | 0.716586494 | 21.71403562 |
| KO | 8.979577959 | 1.43338697 | 23.61098254 | 34.02394747 |
| total | 101.2743055 | 23.47780869 | 30.32855272 | 155.0806669 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERT | OA | 66.59% | 84.91% | 59.68% | 80.45% | 85.49% | 85.10% | 87.95% | 88.51% | 90.00% | 90.13% |
| Kappa | 0.45 | 0.75 | 0.34 | 0.68 | 0.76 | 0.75 | 0.80 | 0.81 | 0.83 | 0.84 | |
| RF | OA | 64.35% | 83.72% | 59.94% | 79.35% | 84.36% | 84.43% | 87.28% | 87.37% | 88.91% | 88.47% |
| Kappa | 0.42 | 0.73 | 0.34 | 0.67 | 0.75 | 0.75 | 0.79 | 0.79 | 0.82 | 0.81 | |
| KNN | OA | 57.32% | 77.36% | 59.68% | 77.19% | 81.01% | 75.38% | 75.39% | 75.38% | 75.38% | 70.74% |
| Kappa | 0.28 | 0.63 | 0.32 | 0.63 | 0.69 | 0.60 | 0.60 | 0.60 | 0.60 | 0.52 | |
| Bayes | OA | 57.32% | 55.42% | 46.42% | 57.72% | 58.12% | 58.57% | 59.51% | 58.09% | 59.05% | 61.26% |
| Kappa | 0.28 | 0.32 | 0.22 | 0.35 | 0.38 | 0.38 | 0.40 | 0.38 | 0.39 | 0.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Tan, L.; Fan, J. Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province. Remote Sens. 2023, 15, 1386. https://doi.org/10.3390/rs15051386
Wang X, Tan L, Fan J. Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province. Remote Sensing. 2023; 15(5):1386. https://doi.org/10.3390/rs15051386
Chicago/Turabian StyleWang, Xinzhe, Linlin Tan, and Jianchao Fan. 2023. "Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province" Remote Sensing 15, no. 5: 1386. https://doi.org/10.3390/rs15051386
APA StyleWang, X., Tan, L., & Fan, J. (2023). Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province. Remote Sensing, 15(5), 1386. https://doi.org/10.3390/rs15051386