Quality Analysis of a High-Precision Kinematic Laser Scanning System for the Use of Spatio-Temporal Plant and Organ-Level Phenotyping in the Field
Abstract
1. Introduction
2. Materials and Methods
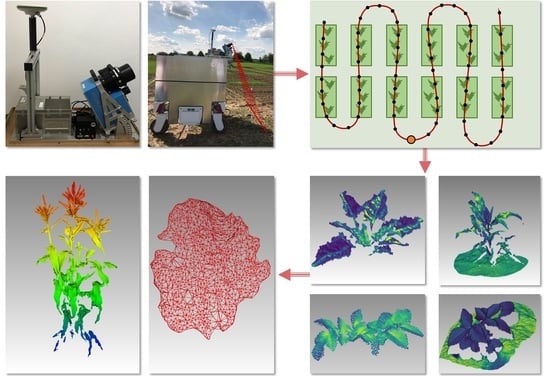
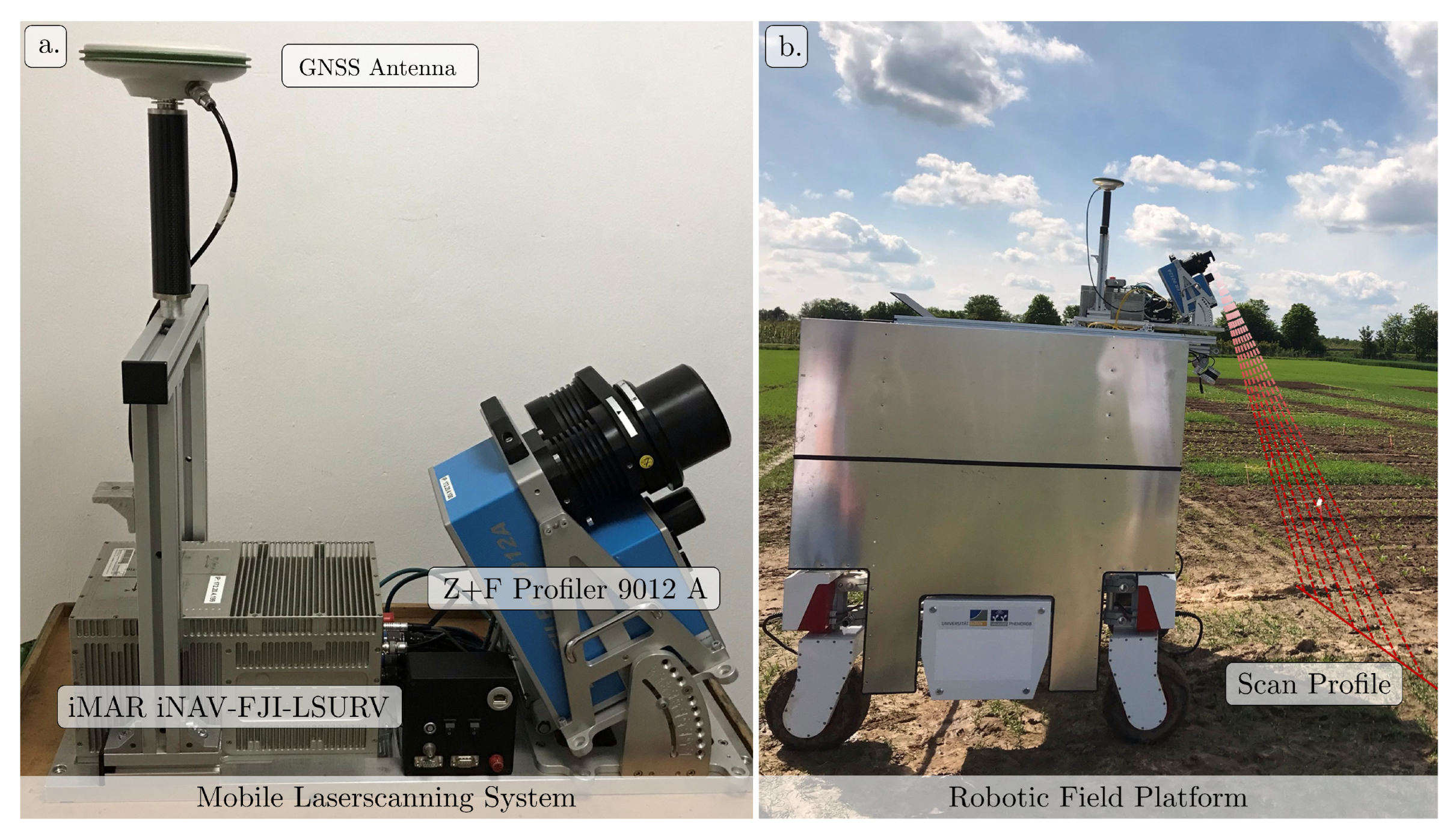
2.1. Mobile Laser Scanning System
2.2. Field Experiments
2.3. Evaluation Criteria in Kinematic Crop Laser Scanning
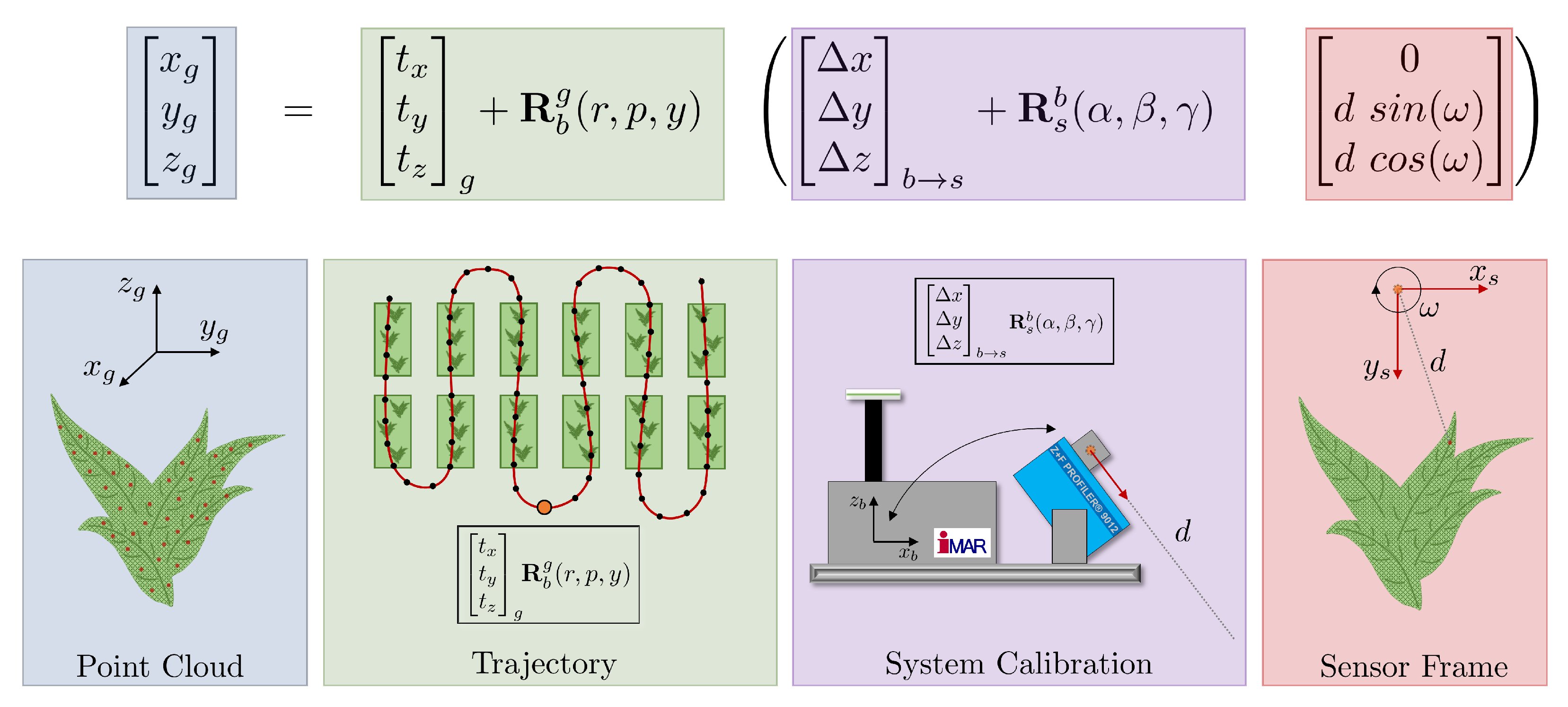
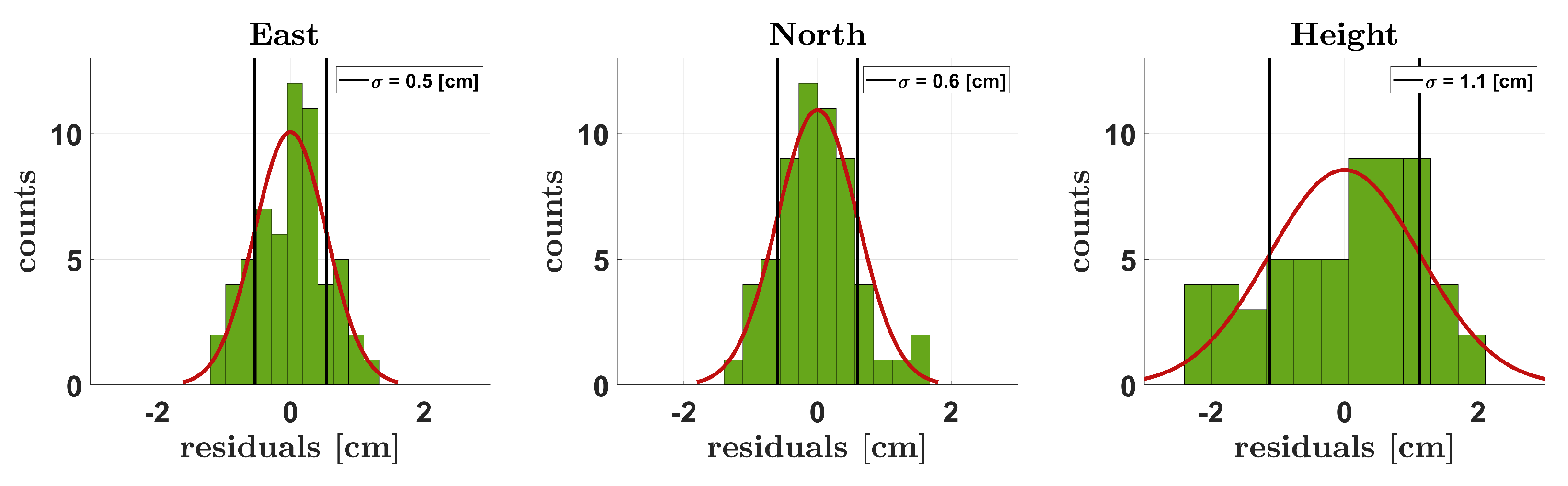
2.3.1. Georeferencing Accuracy
2.3.2. Point Cloud Quality
2.3.3. Time Consumption and Potential for Automation
2.3.4. Field Capability
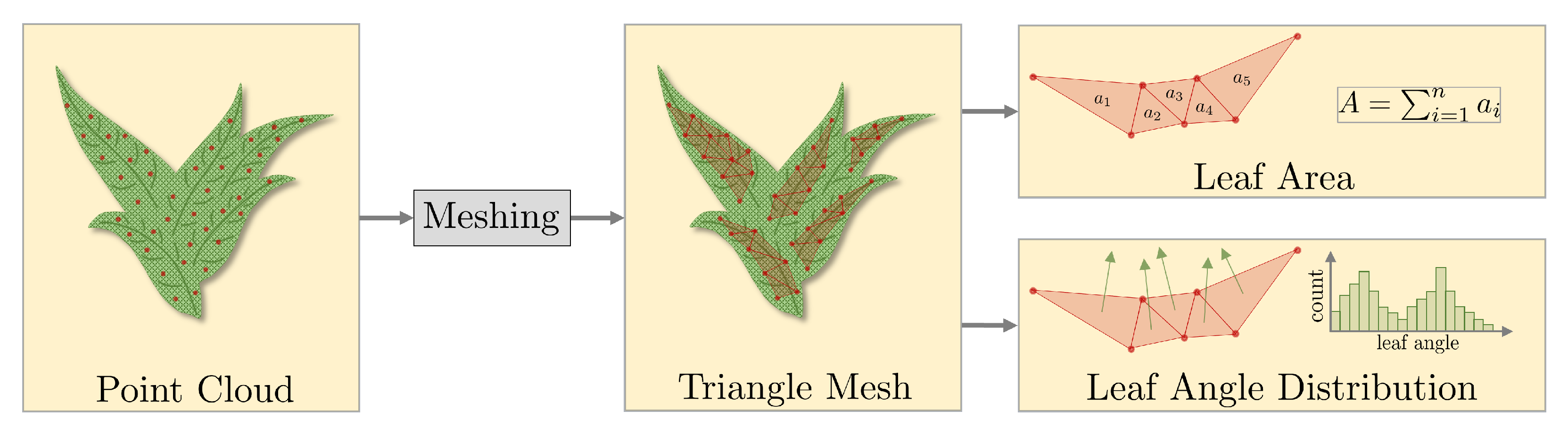
2.4. Extraction of Phenotyping Parameters
3. Results and Discussions
3.1. Georeferencing Accuracy
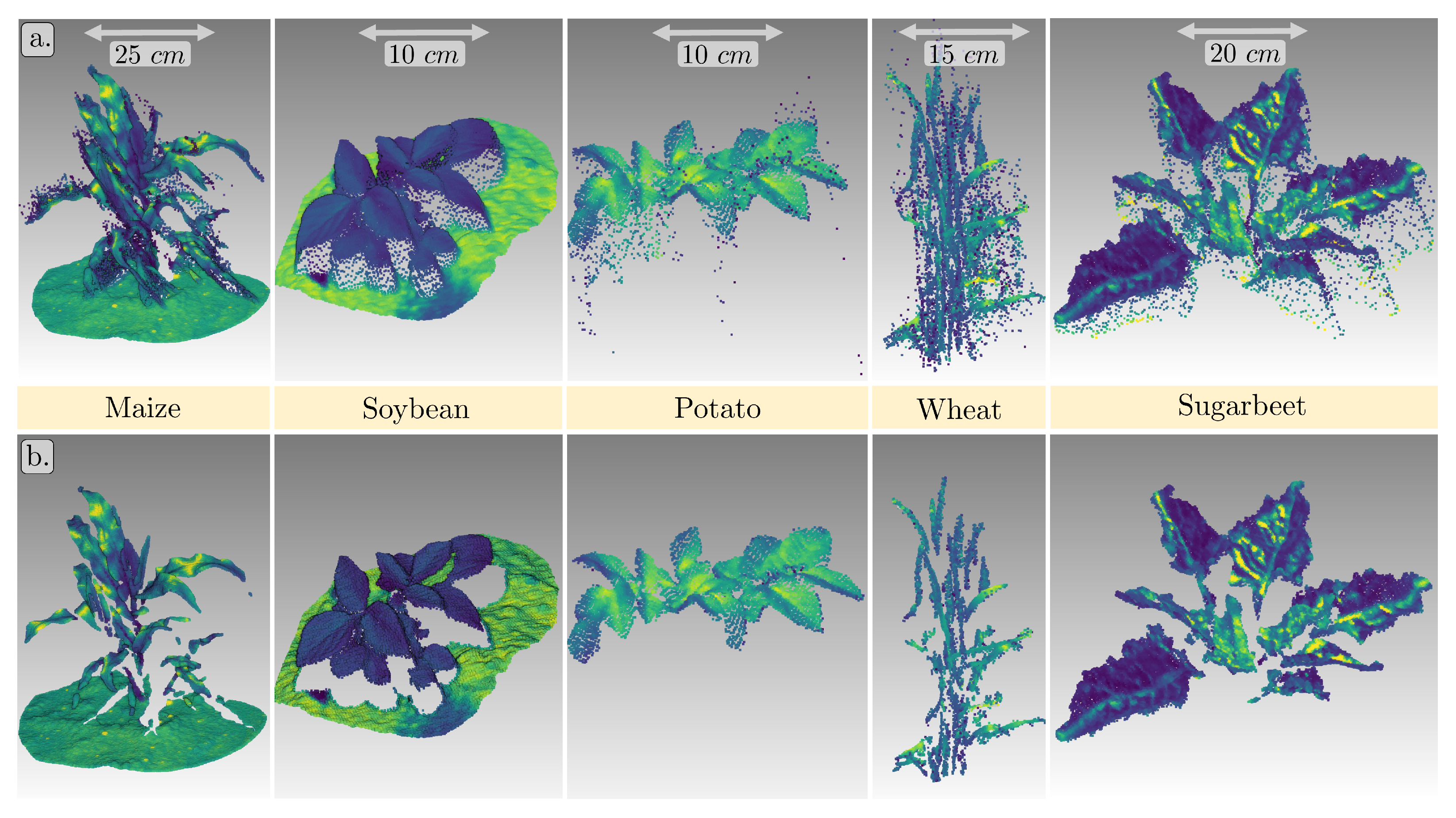
3.2. Point Cloud Quality
3.2.1. Point Precision
3.2.2. Spatial Resolution
3.2.3. Outliers
3.2.4. Completeness
3.3. Time Consumption and Potential for Automation
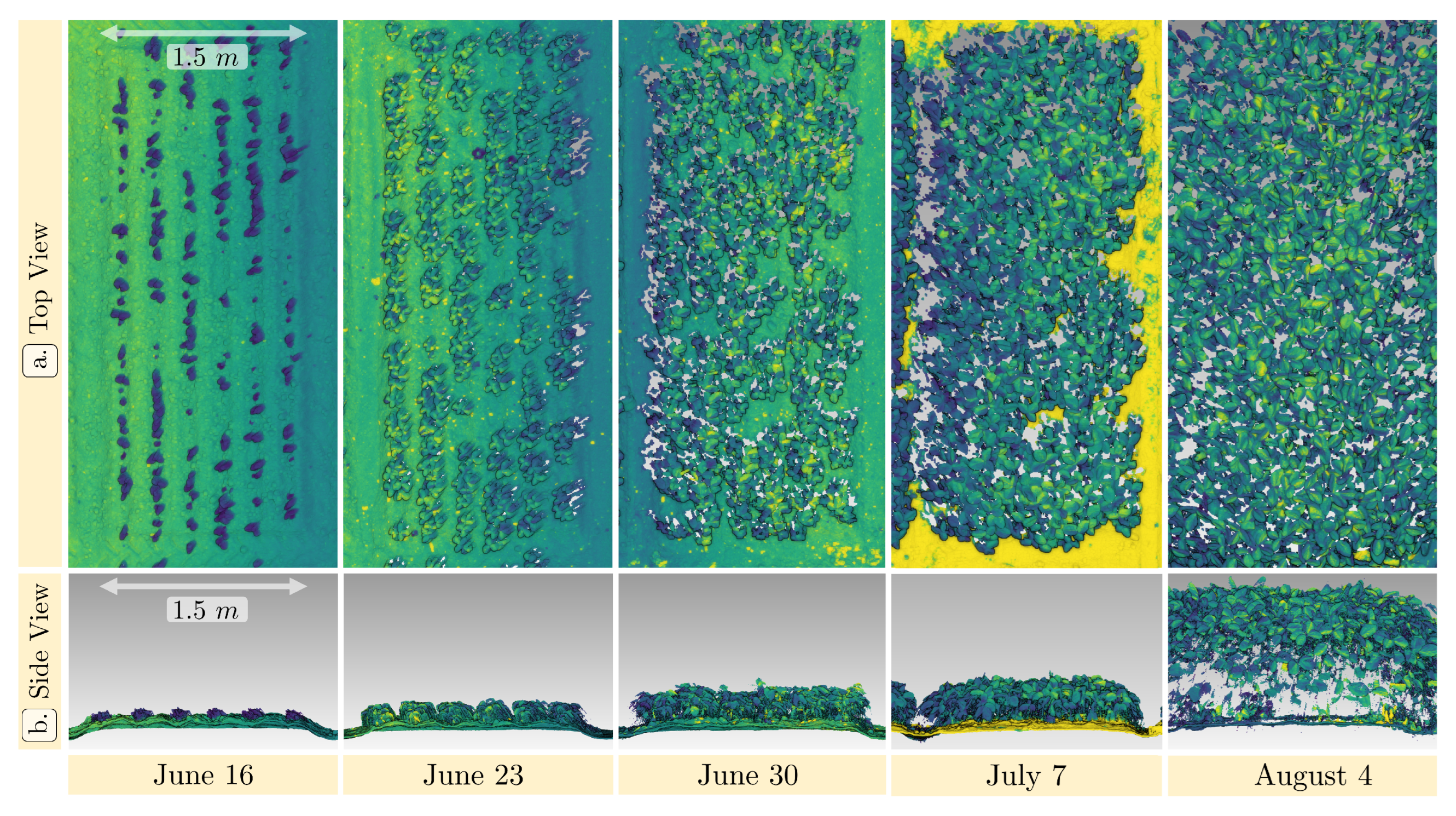
3.4. Field Capability
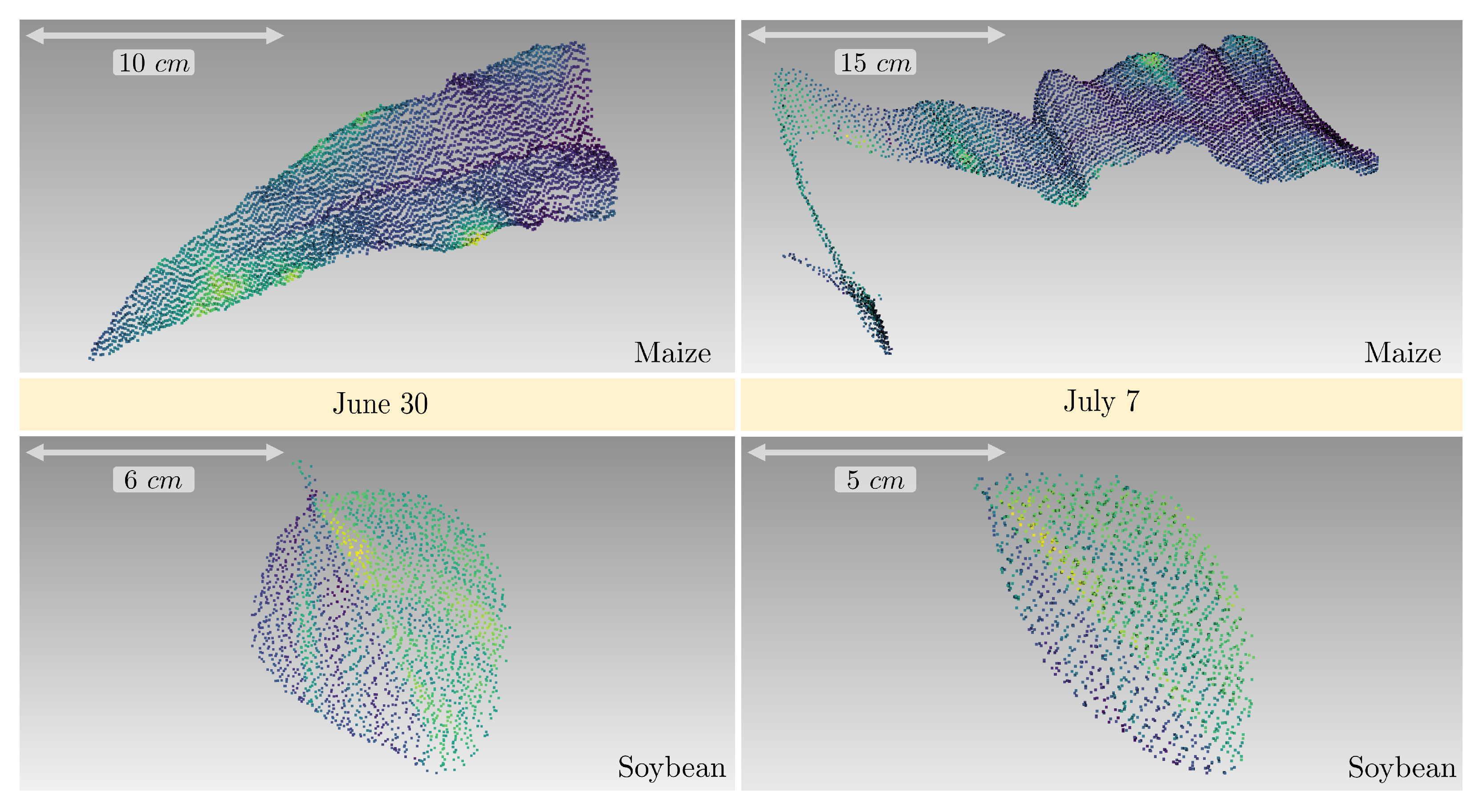
3.5. Phenotyping Parameter Extraction
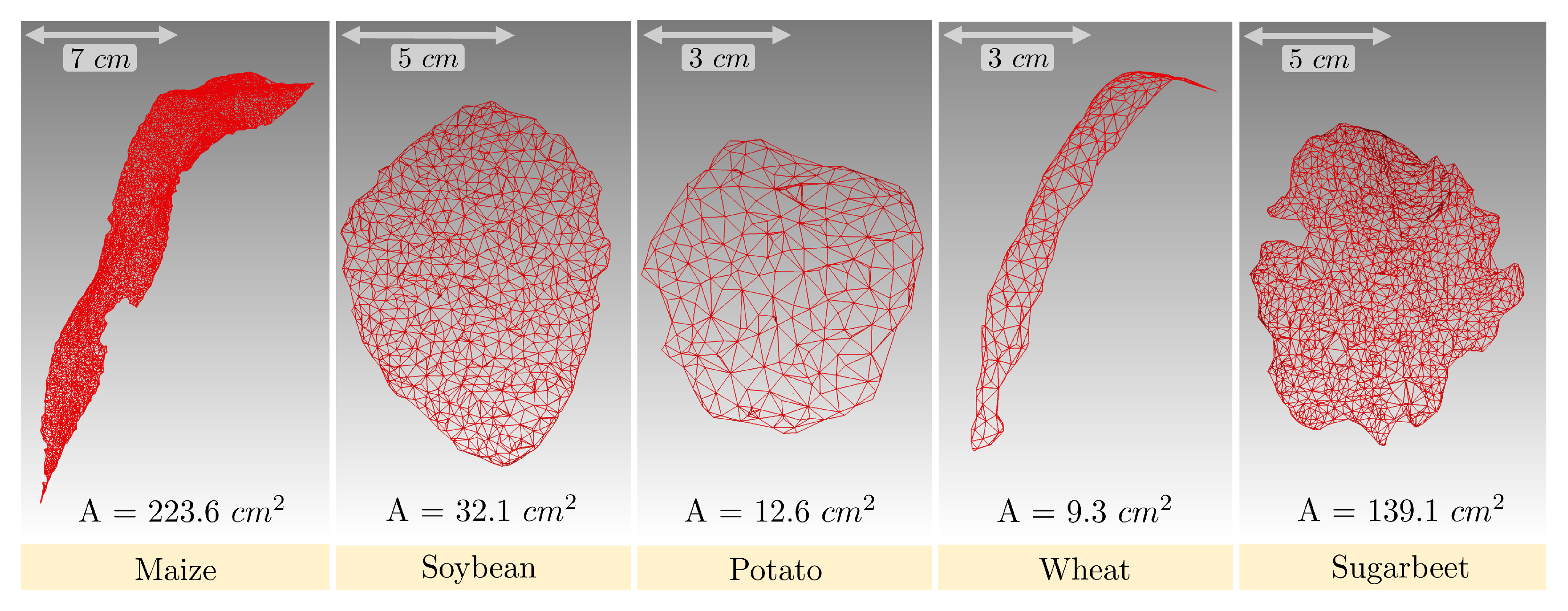
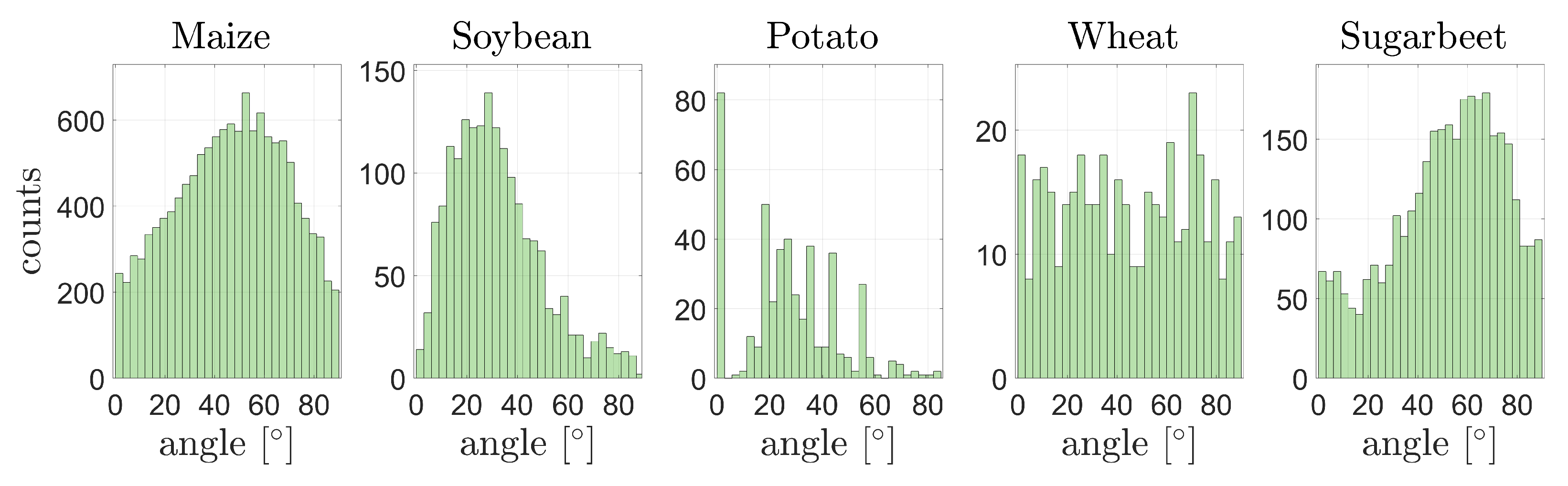
3.5.1. Leaf Area and Leaf Inclination Angle Distribution
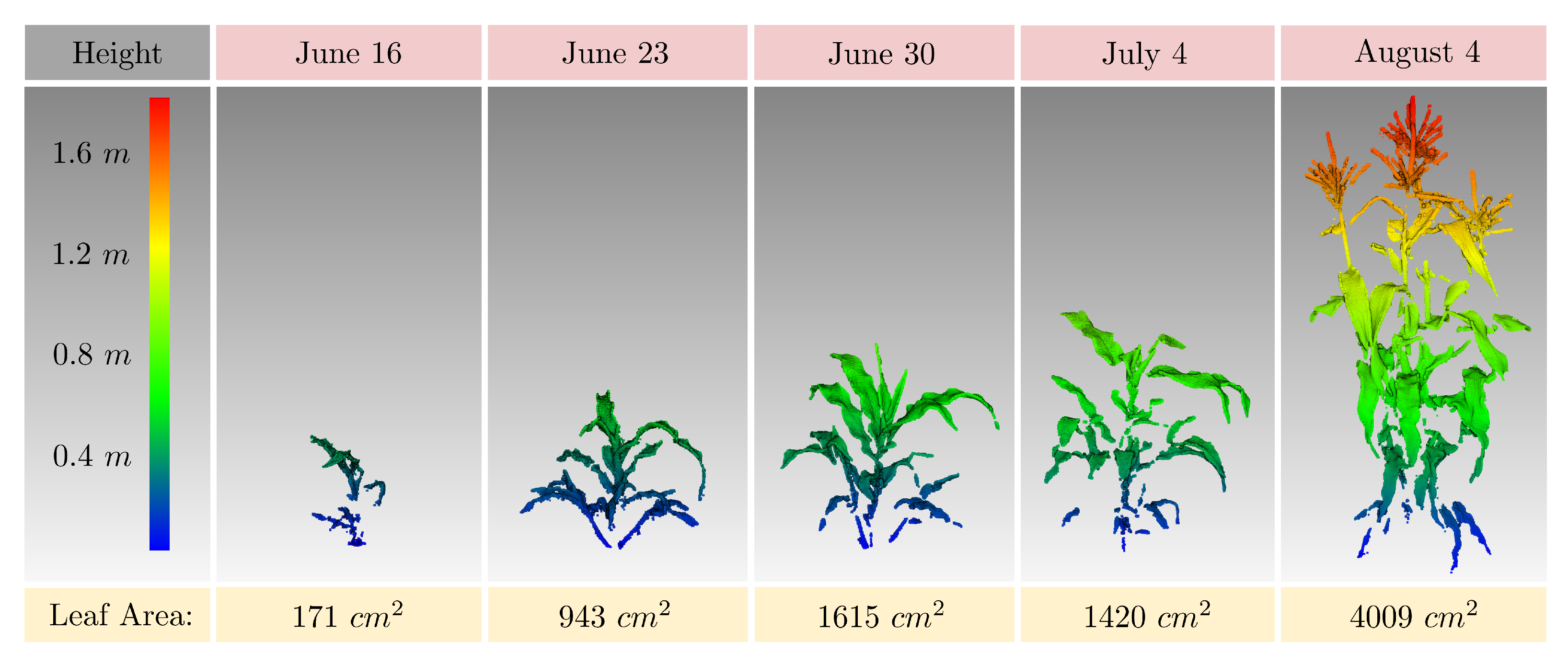
3.5.2. Spatio–Temporal Height and Leaf Area Estimation
4. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3D | Three-Dimensional |
| TLS | Terrestrial Laser Scanning |
| GNSS | Global Navigation Satellite System |
| IMU | Inertial Measurement Unit |
| LIDAR | Light Detection and Ranging |
| UTM | Universal Transverse Mercator |
| LAI | Leaf Area Index |
| LAD | Leaf Area Distribution |
| SOR | Statistical Outlier Filter |
References
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Van Eeuwijk, F.A.; Bustos-Korts, D.; Millet, E.J.; Boer, M.P.; Kruijer, W.; Thompson, A.; Malosetti, M.; Iwata, H.; Quiroz, R.; Kuppe, C.; et al. Modelling strategies for assessing and increasing the effectiveness of new phenotyping techniques in plant breeding. Plant Sci. 2019, 282, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Romero, A.; Vergara-Díaz, O.; Thierfelder, C.; Cairns, J.E.; Kefauver, S.C.; Araus, J.L. Phenotyping conservation agriculture management effects on ground and aerial remote sensing assessments of maize hybrids performance in Zimbabwe. Remote Sens. 2018, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.L.; Desai, S.V.; Guo, W.; Balasubramanian, V.N. Computer vision with deep learning for plant phenotyping in agriculture: A survey. arXiv 2020, arXiv:2006.11391. [Google Scholar]
- Khan, Z.; Rahimi-Eichi, V.; Haefele, S.; Garnett, T.; Miklavcic, S.J. Estimation of vegetation indices for high-throughput phenotyping of wheat using aerial imaging. Plant Methods 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Perich, G.; Hund, A.; Anderegg, J.; Roth, L.; Boer, M.P.; Walter, A.; Liebisch, F.; Aasen, H. Assessment of multi-image unmanned aerial vehicle based high-throughput field phenotyping of canopy temperature. Front. Plant Sci. 2020, 11, 150. [Google Scholar] [CrossRef]
- Ali, B.; Zhao, F.; Li, Z.; Zhao, Q.; Gong, J.; Wang, L.; Tong, P.; Jiang, Y.; Su, W.; Bao, Y.; et al. Sensitivity Analysis of Canopy Structural and Radiative Transfer Parameters to Reconstructed Maize Structures Based on Terrestrial LiDAR Data. Remote Sens. 2021, 13, 3751. [Google Scholar] [CrossRef]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize plant phenotyping: Comparing 3D laser scanning, multi-view stereo reconstruction, and 3D digitizing estimates. Remote Sens. 2019, 11, 63. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Takeda, F.; Kramer, E.A.; Ashrafi, H.; Hunter, J. 3D point cloud data to quantitatively characterize size and shape of shrub crops. Hortic. Res. 2019, 6, 1–17. [Google Scholar] [CrossRef]
- Young, S.N.; Kayacan, E.; Peschel, J.M. Design and field evaluation of a ground robot for high-throughput phenotyping of energy sorghum. Precis. Agric. 2019, 20, 697–722. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H. In-field high-throughput phenotyping of cotton plant height using LiDAR. Remote Sens. 2017, 9, 377. [Google Scholar] [CrossRef]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic Technologies for High-Throughput Plant Phenotyping: Contemporary Reviews and Future Perspectives. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Magney, T.S.; Vierling, L.A.; Eitel, J.U.; Huggins, D.R.; Garrity, S.R. Response of high frequency Photochemical Reflectance Index (PRI) measurements to environmental conditions in wheat. Remote Sens. Environ. 2016, 173, 84–97. [Google Scholar] [CrossRef]
- Dente, L.; Satalino, G.; Mattia, F.; Rinaldi, M. Assimilation of leaf area index derived from ASAR and MERIS data into CERES-Wheat model to map wheat yield. Remote Sens. Environ. 2008, 112, 1395–1407. [Google Scholar] [CrossRef]
- Aboutalebi, M.; Torres-Rua, A.F.; McKee, M.; Kustas, W.P.; Nieto, H.; Alsina, M.M.; White, A.; Prueger, J.H.; McKee, L.; Alfieri, J.; et al. Incorporation of unmanned aerial vehicle (UAV) point cloud products into remote sensing evapotranspiration models. Remote Sens. 2019, 12, 50. [Google Scholar] [CrossRef]
- Simic Milas, A.; Romanko, M.; Reil, P.; Abeysinghe, T.; Marambe, A. The importance of leaf area index in mapping chlorophyll content of corn under different agricultural treatments using UAV images. Int. J. Remote Sens. 2018, 39, 5415–5431. [Google Scholar] [CrossRef]
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: The best is yet to come. Trends Plant Sci. 2013, 18, 428–439. [Google Scholar] [CrossRef]
- Agegnehu, G.; Ghizaw, A.; Sinebo, W. Yield performance and land-use efficiency of barley and faba bean mixed cropping in Ethiopian highlands. Eur. J. Agron. 2006, 25, 202–207. [Google Scholar] [CrossRef]
- Friedli, M.; Kirchgessner, N.; Grieder, C.; Liebisch, F.; Mannale, M.; Walter, A. Terrestrial 3D laser scanning to track the increase in canopy height of both monocot and dicot crop species under field conditions. Plant Methods 2016, 12, 1–15. [Google Scholar] [CrossRef]
- Su, Y.; Wu, F.; Ao, Z.; Jin, S.; Qin, F.; Liu, B.; Pang, S.; Liu, L.; Guo, Q. Evaluating maize phenotype dynamics under drought stress using terrestrial lidar. Plant Methods 2019, 15, 1–16. [Google Scholar] [CrossRef]
- Jay, S.; Rabatel, G.; Hadoux, X.; Moura, D.; Gorretta, N. In-field crop row phenotyping from 3D modeling performed using Structure from Motion. Comput. Electron. Agric. 2015, 110, 70–77. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Wang, Y.; Fan, J.; Wang, C.; Gou, W.; Guo, X. MVS-Pheno: A portable and low-cost phenotyping platform for maize shoots using multiview stereo 3D reconstruction. Plant Phenomics 2020, 2020, 1848437. [Google Scholar] [CrossRef]
- Lou, L.; Liu, Y.; Sheng, M.; Han, J.; Doonan, J.H. A cost-effective automatic 3D reconstruction pipeline for plants using multi-view images. In Proceedings of the Conference Towards Autonomous Robotic Systems, Birmingham, UK, 1–3 September 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 221–230. [Google Scholar]
- Abd Rabbou, M.; El-Rabbany, A. Tightly coupled integration of GPS precise point positioning and MEMS-based inertial systems. GPS Solut. 2015, 19, 601–609. [Google Scholar] [CrossRef]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-based high-throughput phenotyping for maize plant using 3D LiDAR point cloud generated with a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef]
- Gage, J.L.; Richards, E.; Lepak, N.; Kaczmar, N.; Soman, C.; Chowdhary, G.; Gore, M.A.; Buckler, E.S. In-field whole-plant maize architecture characterized by subcanopy rovers and latent space phenotyping. Plant Phenome J. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Iqbal, J.; Xu, R.; Halloran, H.; Li, C. Development of a multi-purpose autonomous differential drive mobile robot for plant phenotyping and soil sensing. Electronics 2020, 9, 1550. [Google Scholar] [CrossRef]
- Groves, P.D. Principles of GNSS, inertial, and multisensor integrated navigation systems, [Book review]. IEEE Aerosp. Electron. Syst. Mag. 2015, 30, 26–27. [Google Scholar] [CrossRef]
- Heinz, E.; Holst, C.; Kuhlmann, H.; Klingbeil, L. Design and evaluation of a permanently installed plane-based calibration field for mobile laser scanning systems. Remote Sens. 2020, 12, 555. [Google Scholar] [CrossRef]
- Schweitzer, J.; Schwieger, V. Modeling of quality for engineering geodesy processes in civil engineering. J. Appl. Geod. 2011, 5, 13–22. [Google Scholar] [CrossRef]
- Balangé, L.; Zhang, L.; Schwieger, V. First Step Towards the Technical Quality Concept for Integrative Computational Design and Construction. In Proceedings of the Contributions to International Conferences on Engineering Surveying; Springer: Cham, Switzerland, 2021; pp. 118–127. [Google Scholar]
- Dreier, A.; Janßen, J.; Kuhlmann, H.; Klingbeil, L. Quality Analysis of Direct Georeferencing in Aspects of Absolute Accuracy and Precision for a UAV-Based Laser Scanning System. Remote Sens. 2021, 13, 3564. [Google Scholar] [CrossRef]
- Chaudhry, S.; Salido-Monzú, D.; Wieser, A. Simulation of 3D laser scanning with phase-based EDM for the prediction of systematic deviations. In Proceedings of the Modeling Aspects in Optical Metrology VII, Munich, Germany, 24–26 June 2019; SPIE: Bellingham, WA, USA, 2019; Volume 11057, pp. 92–104. [Google Scholar]
- Tang, P.; Akinci, B.; Huber, D. Quantification of edge loss of laser scanned data at spatial discontinuities. Autom. Constr. 2009, 18, 1070–1083. [Google Scholar] [CrossRef]
- Kowalczyk, K.; Rapinski, J. Investigating the error sources in reflectorless EDM. J. Surv. Eng. 2014, 140, 06014002. [Google Scholar] [CrossRef]
- Brunner, F. Modelling of atmospheric effects on terrestrial geodetic measurements. In Geodetic Refraction; Springer: Berlin/Heidelberg, Germany, 1984; pp. 143–162. [Google Scholar]
- Bernardini, F.; Mittleman, J.; Rushmeier, H.; Silva, C.; Taubin, G. The ball-pivoting algorithm for surface reconstruction. IEEE Trans. Vis. Comput. Graph. 1999, 5, 349–359. [Google Scholar] [CrossRef]
- Chebrolu, N.; Magistri, F.; Läbe, T.; Stachniss, C. Registration of spatio-temporal point clouds of plants for phenotyping. PLoS ONE 2021, 16, e0247243. [Google Scholar] [CrossRef]
- Magistri, F.; Chebrolu, N.; Stachniss, C. Segmentation-based 4D registration of plants point clouds for phenotyping. In Proceedings of the 2020 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), IEEE, Las Vegas, NV, USA, 25–29 October 2020; pp. 2433–2439. [Google Scholar]




| Month | Day | Sugar Beet | Maize | Soybean | Potato | Wheat and Bean (Mixed) | Targets |
|---|---|---|---|---|---|---|---|
| June | 2 | x | x | ||||
| June | 11 | x | |||||
| June | 16 | x | x | x | x | x | |
| June | 23 | x | x | x | x | ||
| June | 30 | x | x | ||||
| July | 7 | x | x | x | x | x | x |
| August | 4 | x | x | x | x | ||
| August | 12 | x | x | x | x | ||
| September | 3 | x | x |
| Value | Maize | Soybean | Potato | Wheat | Sugar Beet | |
|---|---|---|---|---|---|---|
| Mean Resolution | 0.99 | 1.02 | 1.44 | 1.48 | 1.54 | mm |
| Min/Max | 0.33/3.67 | 0.32/2.51 | 1.0/3.0 | 0.65/3.85 | 0.45/2.92 | mm |
| Maize | Soybean | Potato | Wheat | Sugar Beet | ||
|---|---|---|---|---|---|---|
| Number of points before filtering | 168,575 | 38,402 | 7785 | 11,959 | 32,321 | |
| Number of points after filtering | 149,690 | 35,409 | 6735 | 7939 | 27,451 | |
| Number of removed points | 18,885 | 2993 | 1050 | 4019 | 4870 | |
| Percentage of removed points | 11.2 | 7.8 | 13.5 | 33.61 | 15.1 | % |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esser, F.; Klingbeil, L.; Zabawa, L.; Kuhlmann, H. Quality Analysis of a High-Precision Kinematic Laser Scanning System for the Use of Spatio-Temporal Plant and Organ-Level Phenotyping in the Field. Remote Sens. 2023, 15, 1117. https://doi.org/10.3390/rs15041117
Esser F, Klingbeil L, Zabawa L, Kuhlmann H. Quality Analysis of a High-Precision Kinematic Laser Scanning System for the Use of Spatio-Temporal Plant and Organ-Level Phenotyping in the Field. Remote Sensing. 2023; 15(4):1117. https://doi.org/10.3390/rs15041117
Chicago/Turabian StyleEsser, Felix, Lasse Klingbeil, Lina Zabawa, and Heiner Kuhlmann. 2023. "Quality Analysis of a High-Precision Kinematic Laser Scanning System for the Use of Spatio-Temporal Plant and Organ-Level Phenotyping in the Field" Remote Sensing 15, no. 4: 1117. https://doi.org/10.3390/rs15041117
APA StyleEsser, F., Klingbeil, L., Zabawa, L., & Kuhlmann, H. (2023). Quality Analysis of a High-Precision Kinematic Laser Scanning System for the Use of Spatio-Temporal Plant and Organ-Level Phenotyping in the Field. Remote Sensing, 15(4), 1117. https://doi.org/10.3390/rs15041117