Novel Hybrid Model to Estimate Leaf Carotenoids Using Multilayer Perceptron and PROSPECT Simulations
Abstract
:1. Introduction
2. Materials and Methods
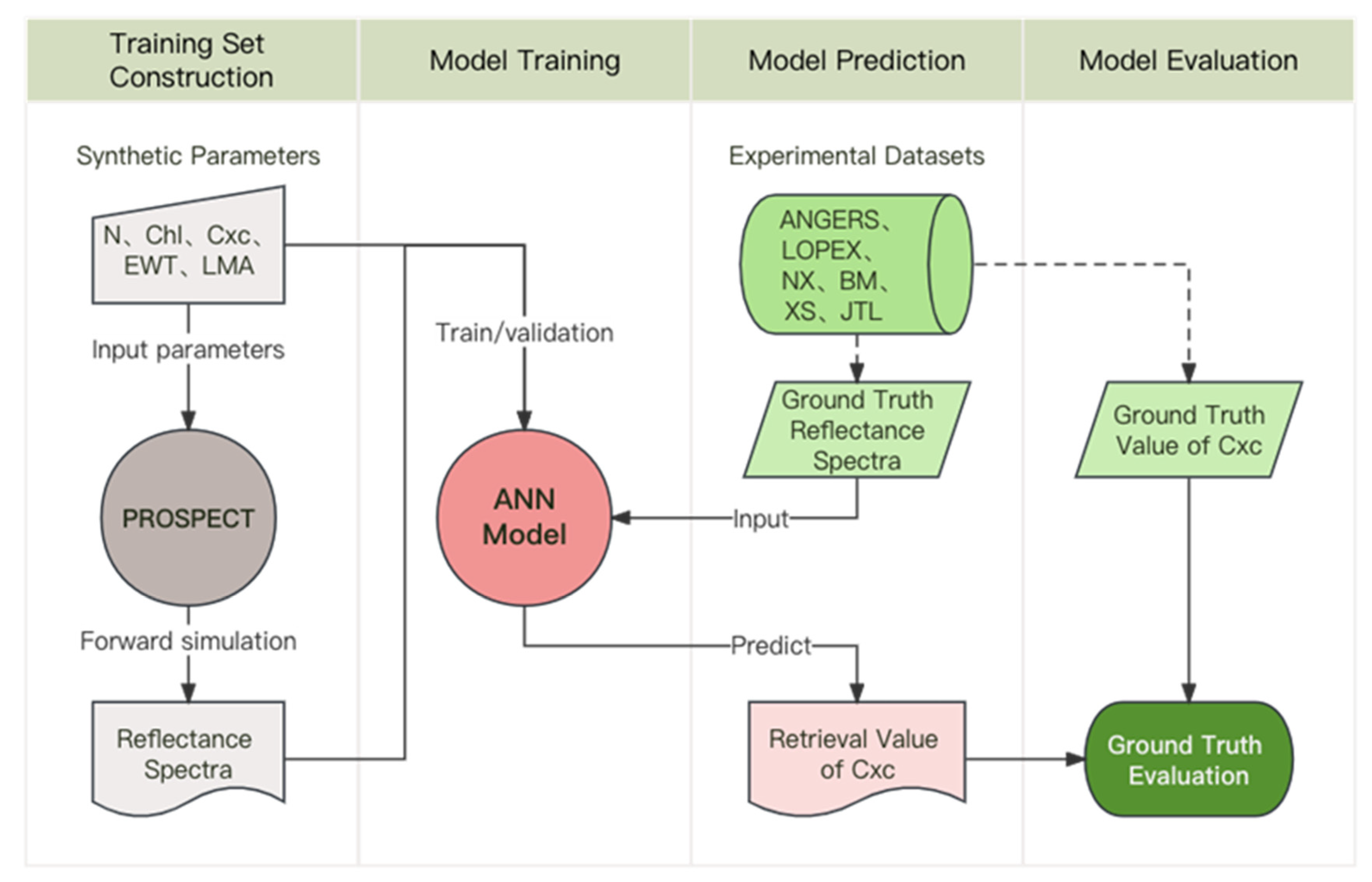
2.1. Training Set Construction: Simulated Data
2.2. Model Training: Multilayer Perceptron
2.2.1. Model Description
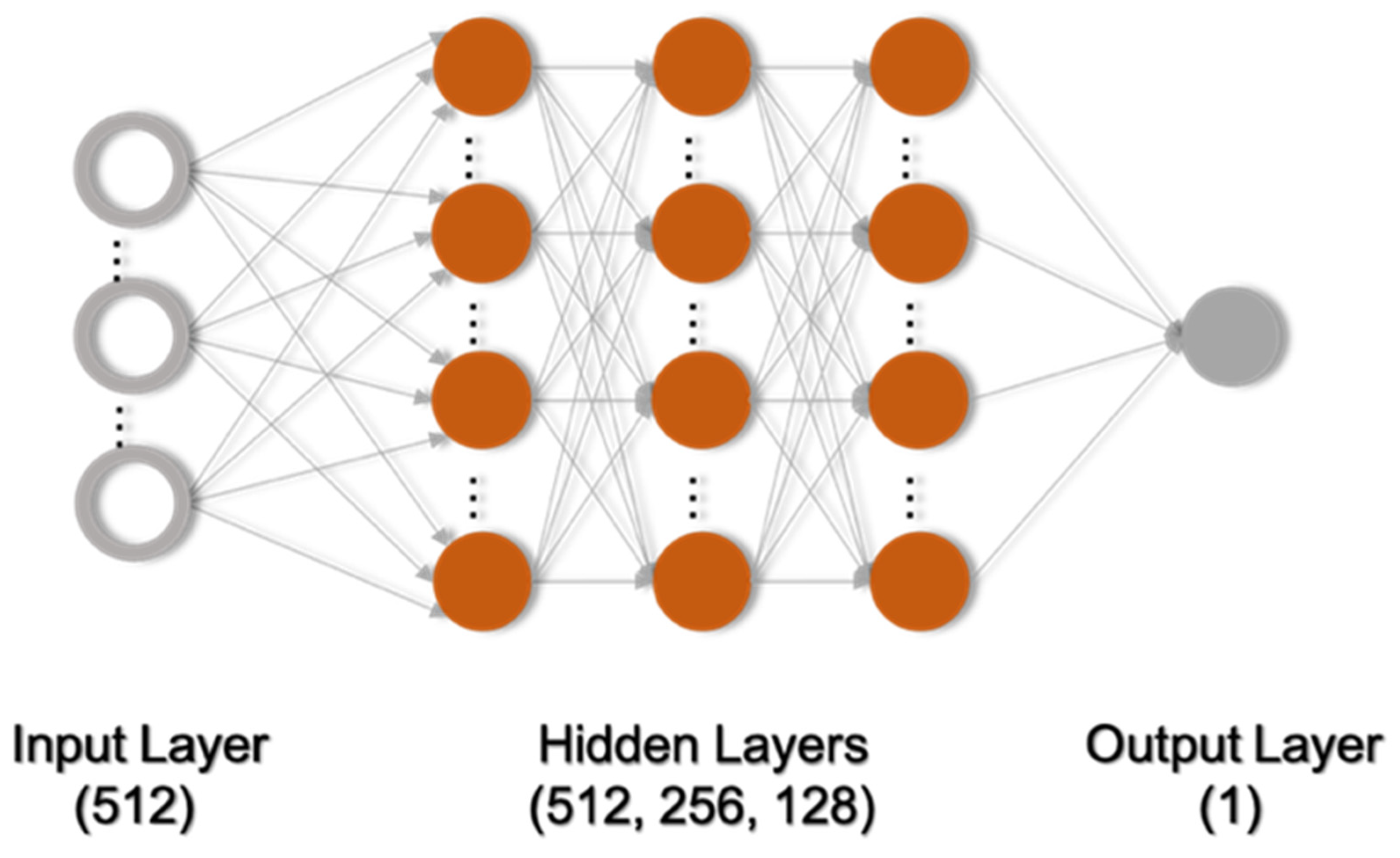
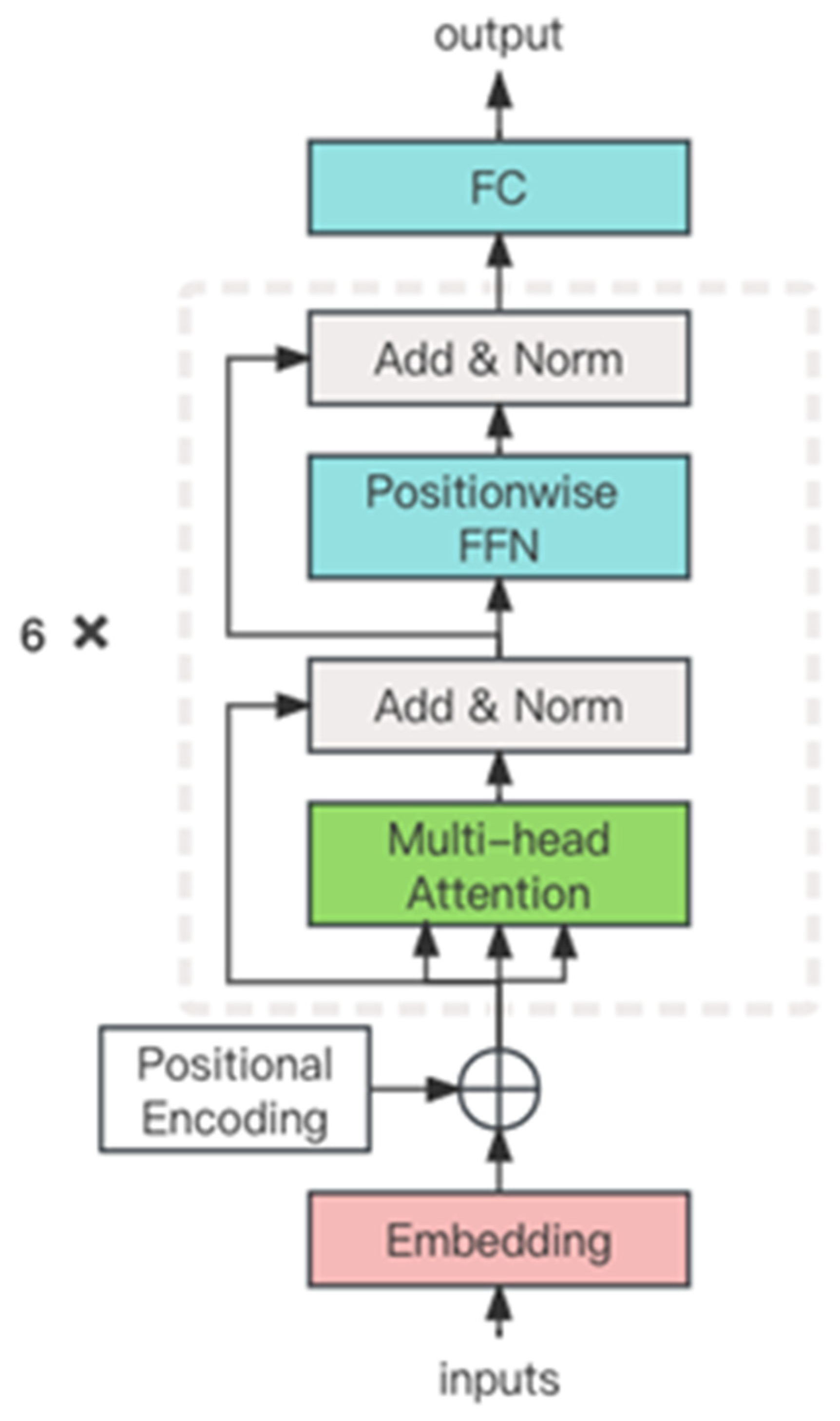
2.2.2. Model Architecture
2.3. Model Prediction: Experimental Datasets
2.4. Model Evaluation
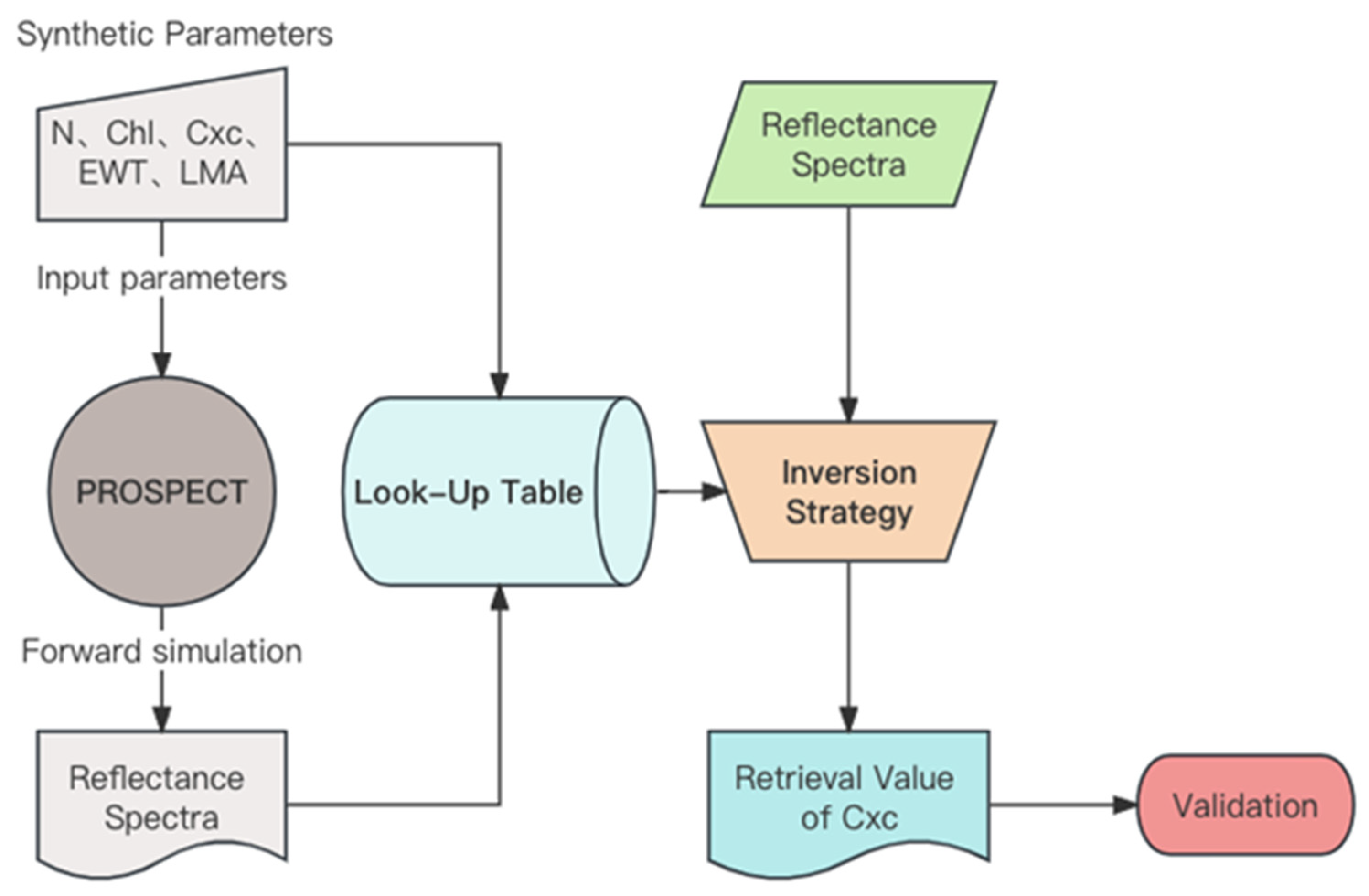
2.5. Alternative Inversion Methods for Comparison
3. Results
3.1. Performance of the MLP Model Using Simulated Data
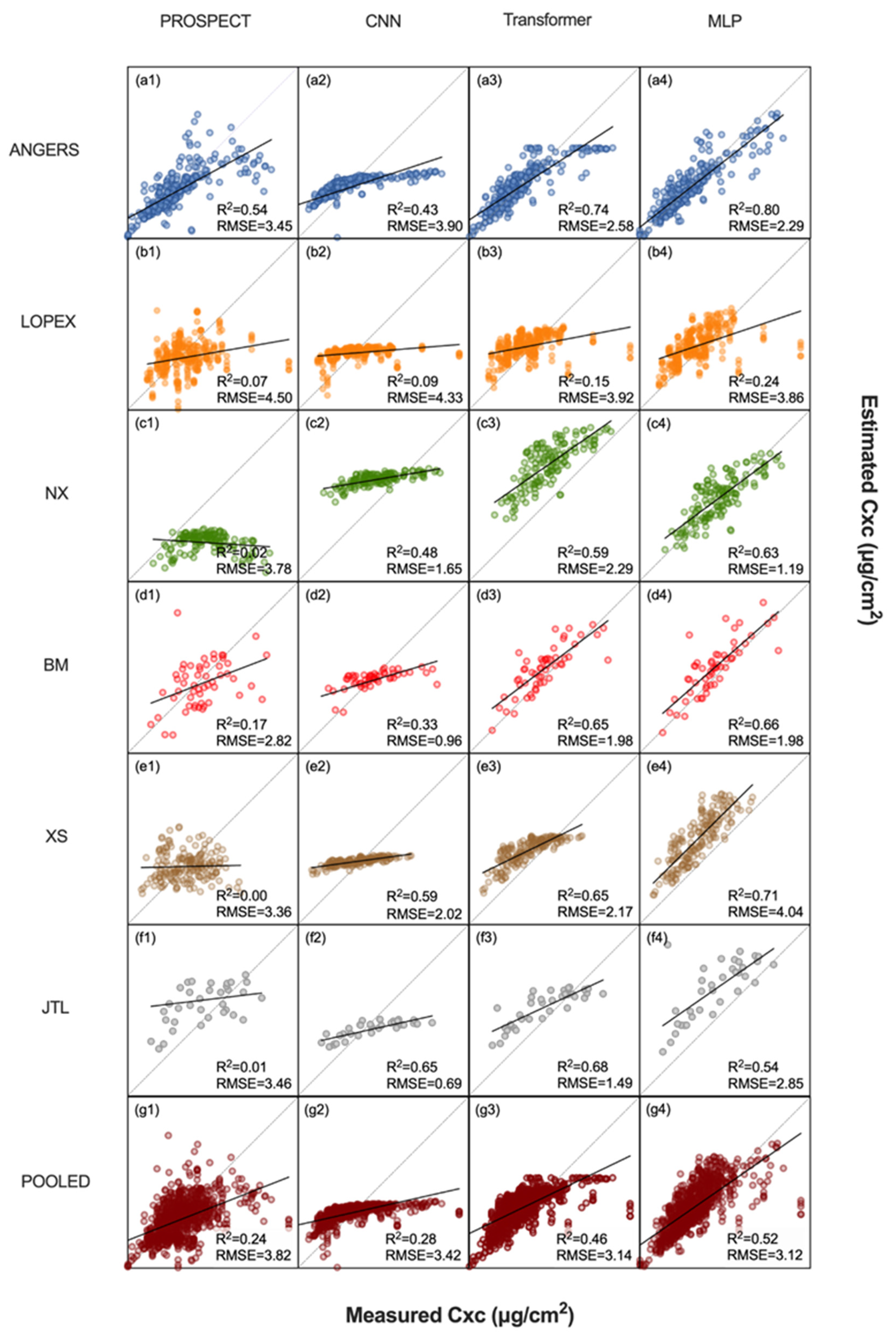
3.2. Performance of the MLP and Alternative Methods Using Experimental Datasets
4. Discussion
4.1. Comparison with Alternative Approaches
4.2. Cxc Retrieval Accuracy in Comparison with Previous Studies
4.3. Study Constraints and Future Prospects
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hagan, J.G.; Henn, J.J.; Osterman, W.H. Plant traits alone are good predictors of ecosystem properties when used carefully. Nat. Ecol. Evol. 2023, 7, 332–334. [Google Scholar] [CrossRef] [PubMed]
- Ross, J. The Radiation Regime and Architecture of Plant Stands; Springer Science & Business Media: Berlin, Germany, 1981. [Google Scholar]
- Demmig-Adams, B.; Adams, W.W. The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996, 1, 21–26. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Gilmore, A.M.; Iii, W.W.A. In vivo functions of carotenoids in higher plants. FASEB J. 1996, 10, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Zur, Y.; Chivkunova, O.B.; Mark, N.M. Assessing Carotenoid Content in Plant Leaves with Reflectance Spectroscopy. Photochem. Photobiol. 2002, 75, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Zarco-Tejada, P.J. Carotenoid content estimation in a heterogeneous conifer forest using narrow-band indices and PROSPECT+DART simulations. Remote Sens. Environ. 2012, 127, 298–315. [Google Scholar] [CrossRef]
- Zhang, J.; Han, W.; Huang, L.; Zhang, Z.; Ma, Y. Leaf Chlorophyll Content Estimation of Winter Wheat Based on Visible and Near-Infrared Sensors. Sensors 2016, 16, 437. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Buddenbaum, H.; Townsend, P.A. Imaging Spectroscopy of Forest Ecosystems: Perspectives for the Use of Space-borne Hyperspectral Earth Observation Systems. Surv. Geophys. 2019, 40, 553–588. [Google Scholar] [CrossRef]
- Feret, J.B.; Maire, G.L.; Jay, S.; Berveiller, D.; Lefèvre-Fonollosa, M. Estimating leaf mass per area and equivalent water thickness based on leaf optical properties: Potential and limitations of physical modeling and machine learning. Remote Sens. Environ. 2018, 231, 110959. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33, L11402. [Google Scholar] [CrossRef]
- Wan, L.; Tang, Z.; Zhang, J.; Chen, S.; Zhou, W.; Cen, H. Upscaling from leaf to canopy: Improved spectral indices for leaf biochemical traits estimation by minimizing the difference between leaf adaxial and abaxial surfaces. Field Crops Res. 2021, 274, 108330. [Google Scholar] [CrossRef]
- Malenovsky, Z.; Homolová, L.; Zurita-Milla, R.; Luke, P.; Kaplan, V.; Hanu, J.; Gastellu-Etchegorry, J.P.; Schaepman, M.E. Retrieval of spruce leaf chlorophyll content from airborne image data using continuum removal and radiative transfer. Remote Sens. Environ. 2013, 131, 85–102. [Google Scholar] [CrossRef]
- Jiao, Q.; Sun, Q.; Zhang, B.; Huang, W.; Ye, H.; Zhang, Z.; Zhang, X.; Qian, B. A Random Forest Algorithm for Retrieving Canopy Chlorophyll Content of Wheat and Soybean Trained with PROSAIL Simulations Using Adjusted Average Leaf Angle. Remote Sens. 2022, 14, 98. [Google Scholar] [CrossRef]
- Sonobe, R.; Hirono, Y.; Oi, A. Quantifying chlorophyll-a and b content in tea leaves using hyperspectral reflectance and deep learning. Remote Sens. Lett. 2020, 11, 933–942. [Google Scholar] [CrossRef]
- Barman, U. Deep Convolutional neural network (CNN) in tea leaf chlorophyll estimation: A new direction of modern tea farming in Assam, India. J. Appl. Nat. Sci. 2021, 13, 1059–1064. [Google Scholar] [CrossRef]
- Prilianti, K.R.; Onggara, I.C.; Adhiwibawa, M.A.; Brotosudarmo, T.H.; Anam, S.; Suryanto, A. Multispectral imaging and convolutional neural network for photosynthetic pigments prediction. In Proceedings of the 2018 5th International Conference on Electrical Engineering, Computer Science and Informatics (EECSI), Malang, Indonesia, 16–18 October 2018. [Google Scholar]
- Verrelst, J.; Alonso, L.; Camps-Valls, G.; Delegido, J.; Moreno, J. Retrieval of Vegetation Biophysical Parameters Using Gaussian Process Techniques. IEEE Trans. Geosci. Remote Sens. 2012, 50, 1832–1843. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Zhang, L.; Deng, M.; Hui, L. Estimation of crop LAI using hyperspectral vegetation indices and a hybrid inversion method. Remote Sens. Environ. 2015, 165, 123–134. [Google Scholar] [CrossRef]
- Sun, J.; Shi, S.; Yang, J.; Chen, B.; Gong, W.; Du, L.; Mao, F.; Song, S. Estimating leaf chlorophyll status using hyperspectral lidar measurements by PROSPECT model inversion. Remote Sens. Environ. 2018, 212, 1–7. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT+ SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Wang, J.; Ping, C.; Song, J. Use of general regression neural networks for generating the GLASS Leaf Area Index Product from Time Series MODIS Surface Reflectance. IEEE Trans. Geosci. Remote Sens. 2014, 52, 209–223. [Google Scholar] [CrossRef]
- Yang, B.; He, Y.; Chen, W. A simple method for estimation of leaf dry matter content in fresh leaves using leaf scattering albedo. Glob. Ecol. Conserv. 2020, 23, e01201. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Soudani, K.; Berveiller, D.; Pontailler, J.-Y.; Bréda, N.; Genet, H.; Davi, H.; Dufrêne, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Wang, P. Quan Retrieval of Leaf Biochemical Parameters Using PROSPECT Inversion: A New Approach for Alleviating Ill-Posed Problems. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2499–2506. [Google Scholar]
- Croft, H.; Chen, J.; Wang, R.; Mo, G.; Luo, S.; Luo, X.; He, L.; Gonsamo, A.; Arabian, J.; Zhang, Y. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236, 111479. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Munoz-Mari, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Berger, K.; Caicedo, J.P.R.; Martino, L.; Wocher, M.; Verrelst, J. A Survey of Active Learning for Quantifying Vegetation Traits from Terrestrial Earth Observation Data. Remote Sens. 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- De Grave, C.; Verrelst, J.; Morcillo-Pallarés, P.; Pipia, L.; Rivera-Caicedo, J.P.; Amin, E.; Belda, S.; Moreno, J. Quantifying vegetation biophysical variables from the Sentinel-3/FLEX tandem mission: Evaluation of the synergy of OLCI and FLORIS data sources. Remote Sens. Environ. 2020, 251, 112101. [Google Scholar] [CrossRef] [PubMed]
- Brede, B.; Verrelst, J.; Gastellu-Etchegorry, J.-P.; Clevers, J.G.; Goudzwaard, L.; den Ouden, J.; Verbesselt, J.; Herold, M. Assessment of workflow feature selection on forest LAI prediction with sentinel-2A MSI, landsat 7 ETM+ and Landsat 8 OLI. Remote Sens. 2020, 12, 915. [Google Scholar] [CrossRef] [PubMed]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J. Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, J.; Liu, X.; Du, L.; Shi, S.; Sun, J.; Chen, B. Effect of different regression algorithms on the estimating leaf parameters based on selected characteristic wavelengths by using the PROSPECT model. Appl. Opt. 2019, 58, 9904–9913. [Google Scholar] [CrossRef] [PubMed]
- Camps-Valls, G.; Verrelst, J.; Munoz-Mari, J.; Laparra, V.; Mateo-Jimenez, F.; Gomez-Dans, J. A survey on Gaussian processes for earth-observation data analysis: A comprehensive investigation. IEEE Geosci. Remote Sens. Mag. 2016, 4, 58–78. [Google Scholar] [CrossRef]
- Verrelst, J.; Berger, K.; Rivera-Caicedo, J.P. Intelligent Sampling for Vegetation Nitrogen Mapping Based on Hybrid Machine Learning Algorithms. IEEE Geosci. Remote Sens. Lett. 2020, 18, 2038–2042. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Qin, Z.; Zhao, S.; Di, L.; Zhang, C.; Deng, M.; Lin, H.; Zhang, L.; Wang, L.; Liu, Z. Estimating crop chlorophyll content with hyperspectral vegetation indices and the hybrid inversion method. Int. J. Remote Sens. 2016, 37, 2923–2949. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Smith, J.A. LAI inversion using a back-propagation neural network trained with a multiple scattering model. IEEE Trans. Geosci. Remote Sens. 1993, 31, 1102–1106. [Google Scholar] [CrossRef]
- Gong, P. Inverting a canopy reflectance model using a neural network. Int. J. Remote Sens. 1999, 20, 111–122. [Google Scholar] [CrossRef]
- Shi, R.H.; Sun, J. Estimating Leaf Biochemical Information from Leaf Reflectance Spectrum using Artificial Neural Network. In Proceedings of the International Conference on Machine Learning & Cybernetics, Hong Kong, China, 19–22 August 2007. [Google Scholar]
- Shi, S.; Xu, L.; Gong, W.; Chen, B.; Chen, B.; Qu, F.; Tang, X.; Sun, J.; Yang, J. A convolution neural network for forest leaf chlorophyll and carotenoid estimation using hyperspectral reflectance. Int. J. Appl. Earth Obs. Geoinf. 2022, 108, 102719. [Google Scholar] [CrossRef]
- Dorling, M. Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Leshno, M.; Lin, V.Y.; Pinkus, A.; Schocken, S. Original Contribution: Multilayer feedforward networks with a nonpolynomial activation function can approximate any function. Neural Netw. 1993, 6, 861–867. [Google Scholar] [CrossRef]
- Danner, M.; Berger, K.; Wocher, M.; Mauser, W.; Hank, T. Efficient RTM-based training of machine learning regression algorithms to quantify biophysical & biochemical traits of agricultural crops. ISPRS J. Photogramm. Remote Sens. 2021, 173, 278–296. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2012; Volume 25. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2017; Volume 30. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Féret, J.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Feret, J.B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.; Ustin, S.L.; Maire, G.L.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2011, 112, 3030–3043. [Google Scholar] [CrossRef]
- Verger, A.; Baret, F.; Camacho, F. Optimal modalities for radiative transfer-neural network estimation of canopy biophysical characteristics: Evaluation over an agricultural area with CHRIS/PROBA observations. Remote Sens. Environ. 2011, 115, 415–426. [Google Scholar] [CrossRef]
- Sun, J.; Shi, S.; Yang, J.; Du, L.; Gong, W.; Chen, B.; Song, S. Analyzing the performance of PROSPECT model inversion based on different spectral information for leaf biochemical properties retrieval. ISPRS J. Photogramm. Remote Sens. 2018, 135, 74–83. [Google Scholar] [CrossRef]
- Féret, J.-B.; François, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Danner, M.; Wocher, M.; Mauser, W.; Hank, T. Model-based optimization of spectral sampling for the retrieval of crop variables with the PROSAIL model. Remote Sens. 2018, 10, 2063. [Google Scholar] [CrossRef]
- Barry, K.; Newnham, G.J. Quantification of chlorophyll and carotenoid pigments in eucalyptus foliage with the radiative transfer model PROSPECT 5 is affected by anthocyanin and epicuticular waxes. In Proceedings of the Geospatial Science Research Symposium, Melbourne, Australia, 10–12 December 2012. [Google Scholar]
- Jiang, J.; Comar, A.; Burger, P.; Bancal, P.; Weiss, M.; Baret, F. Estimation of leaf traits from reflectance measurements: Comparison between methods based on vegetation indices and several versions of the PROSPECT model. Plant Methods 2018, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Yu, D. Deep learning: Methods and applications. Found. Trends® Signal Process. 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Liu, T.; Yang, L.; Lunga, D. Change detection using deep learning approach with object-based image analysis—ScienceDirect. Remote Sens. Environ. 2021, 256, 112308. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Sun, J.; Shi, S.; Yang, J.; Gong, W.; Qiu, F.; Wang, L.; Du, L.; Chen, B. Wavelength selection of the multispectral lidar system for estimating leaf chlorophyll and water contents through the PROSPECT model. Agric. For. Meteorol. 2019, 266–267, 43–52. [Google Scholar] [CrossRef]
- Spafford, L.; le Maire, G.; MacDougall, A.; de Boissieu, F.; Féret, J.-B. Spectral subdomains and prior estimation of leaf structure improves PROSPECT inversion on reflectance or transmittance alone. Remote Sens. Environ. 2021, 252, 112176. [Google Scholar] [CrossRef]
- Klambauer, G.; Unterthiner, T.; Mayr, A.; Hochreiter, S. Self-normalizing neural networks. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Ma, R.; Letu, H.; Yang, K.; Wang, T.; Chen, L. Estimation of Surface Shortwave Radiation From Himawari-8 Satellite Data Based on a Combination of Radiative Transfer and Deep Neural Network. IEEE Trans. Geosci. Remote Sens. 2020, 58, 5304–5316. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Hosgood, B.; Jacquemoud, S.; Andreoli, G.; Verdebout, J.; Pedrini, G.; Schmuck, G. Leaf Optical Properties Experiment 93 (LOPEX93); The European Commission: Brussels, Belgium, 1995. [Google Scholar]
- Li, D.; Cheng, T.; Jia, M.; Zhou, K.; Lu, N.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W. PROCWT: Coupling PROSPECT with continuous wavelet transform to improve the retrieval of foliar chemistry from leaf bidirectional reflectance spectra. Remote Sens. Environ. 2018, 206, 1–14. [Google Scholar] [CrossRef]
- Qiu, F.; Chen, J.M.; Ju, W.; Wang, J.; Zhang, Q.; Fang, M. Improving the PROSPECT Model to Consider Anisotropic Scattering of Leaf Internal Materials and Its Use for Retrieving Leaf Biomass in Fresh Leaves. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3119–3136. [Google Scholar] [CrossRef]
- Xu, M.; Liu, R.; Chen, J.M.; Liu, Y.; Wolanin, A.; Croft, H.; He, L.; Shang, R.; Ju, W.; Zhang, Y.; et al. A 21-Year Time Series of Global Leaf Chlorophyll Content Maps From MODIS Imagery. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–13. [Google Scholar] [CrossRef]
- Rivera, J.P.; Verrelst, J.; Leonenko, G.; Moreno, J. Multiple Cost Functions and Regularization Options for Improved Retrieval of Leaf Chlorophyll Content and LAI through Inversion of the PROSAIL Model. Remote Sens. 2013, 5, 3280–3304. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Y.; Li, L. Study of the Application of Deep Convolutional Neural Networks (CNNs) in Processing Sensor Data and Biomedical Images. Sensors 2019, 19, 3584. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, H.; Shi, C.; Liu, J. Chapter 2—The basics of deep learning. In Deep Learning on Edge Computing Devices; Zhou, X., Liu, H., Shi, C., Liu, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 19–36. [Google Scholar]
- Fang, W.; Luo, H.; Xu, S.; Love, P.E.; Lu, Z.; Ye, C. Automated text classification of near-misses from safety reports: An improved deep learning approach. Adv. Eng. Inform. 2020, 44, 101060. [Google Scholar] [CrossRef]
- Zhu, J.; Xia, Y.; Wu, L.; He, D.; Qin, T.; Zhou, W.; Li, H.; Liu, T.-Y. Incorporating bert into neural machine translation. arXiv 2020, arXiv:2002.06823. [Google Scholar]
- Amatriain, X. Transformer models: An introduction and catalog. arXiv 2023, arXiv:2302.07730. [Google Scholar]
- Han, X.; Wang, Y.-T.; Feng, J.-L.; Deng, C.; Chen, Z.-H.; Huang, Y.-A.; Su, H.; Hu, L.; Hu, P.-W. A survey of transformer-based multimodal pre-trained modals. Neurocomputing 2023, 515, 89–106. [Google Scholar]
- Sanh, V.; Debut, L.; Chaumond, J.; Wolf, T. DistilBERT, a distilled version of BERT: Smaller, faster, cheaper and lighter. arXiv 2019, arXiv:1910.01108. [Google Scholar]
- Moutik, O.; Sekkat, H.; Tigani, S.; Chehri, A.; Saadane, R.; Tchakoucht, T.A.; Paul, A. Convolutional neural networks or vision transformers: Who will win the race for action recognitions in visual data? Sensors 2023, 23, 734. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Zhang, W.; Lei, W.; Zhang, W.; Chen, X.; Wang, Y. Remote sensing image instance segmentation network with transformer and multi-scale feature representation. Expert Syst. Appl. 2023, 234, 121007. [Google Scholar]
- Guo, K.; Zeng, S.; Yu, J.; Wang, Y.; Yang, H. A Survey of FPGA Based Neural Network Accelerator. arXiv 2017, arXiv:1712.08934. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Verbraeken, J.; Wolting, M.; Katzy, J.; Kloppenburg, J.; Verbelen, T.; Rellermeyer, J.S. A survey on distributed machine learning. ACM Comput. Surv. 2020, 53, 1–33. [Google Scholar]
- Chen, T.; Moreau, T.; Jiang, Z.; Zheng, L.; Yan, E.; Shen, H.; Cowan, M.; Wang, L.; Hu, Y.; Ceze, L. {TVM}: An automated {End-to-End} optimizing compiler for deep learning. In Proceedings of the 13th USENIX Symposium on Operating Systems Design and Implementation (OSDI 18), Carlsbad, CA, USA, 8–10 October 2018. [Google Scholar]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflectance modeling: The SAIL model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar]
- Atzberger, C. Development of an invertible forest reflectance model: The INFOR-model. In A Decade of Trans, Proceedings of the 20th EARSeL Symposium, Dresden, Germany, 14–16 June 2000; The European Commission: Brussels, Belgium, 2000. [Google Scholar]
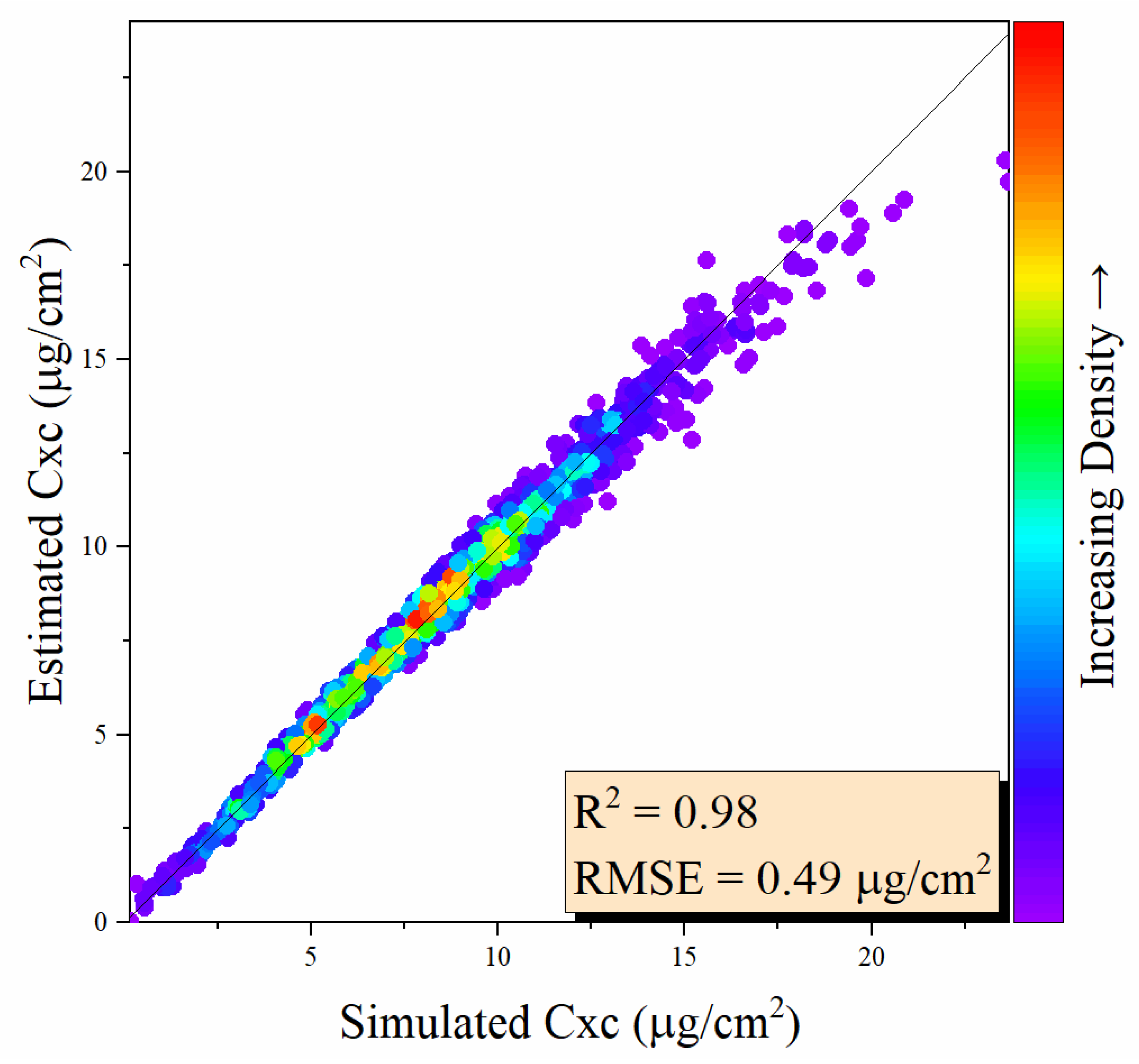
| Parameter | Unit | Min | Max | Mean | Std. |
|---|---|---|---|---|---|
| Leaf structure parameter (N) | - | 1.1 | 2.3 | 1.60 | 0.30 |
| Total chlorophyll content (Chl) | μg/cm2 | 0.3 | 106.72 | 32.81 | 18.87 |
| Carotenoid content (Cxc) | μg/cm2 | 0.04 | 25.3 | 8.51 | 3.92 |
| Equivalent water thickness (EWT) | cm | 0.0043 | 0.0713 | 0.0129 | 0.0073 |
| Leaf mass per area (LMA) | g/cm2 | 0.0008 | 0.0331 | 0.0077 | 0.0035 |
| LOPEX | ANGERS | XS | NX | BM | JTL | |
|---|---|---|---|---|---|---|
| Year | 1996 | 2003 | 2014 | 2014 | 2015 | 2015 |
| Number of samples | 330 | 276 | 175 | 140 | 54 | 35 |
| Number of species | 46 | 43 | 2 | 1 | 8 | 1 |
| Spectral range (nm) | 400–2500 | 400–2450 | 400–2300 | 400–2300 | 400–2300 | 400–2300 |
| Chlorophyll (μg/cm2) | ||||||
| Min | 1.29 | 0.78 | 16.75 | 20.12 | 1.41 | 30.10 |
| Max | 119.87 | 106.72 | 93.77 | 71.72 | 80.82 | 83.88 |
| Mean | 56.92 | 33.88 | 50.86 | 44.03 | 40.08 | 56.09 |
| Std. | 21.08 | 21.71 | 15.54 | 11.21 | 15.51 | 15.93 |
| Carotenoids (μg/cm2) | ||||||
| Min | 4.11 | 0.00 | 3.84 | 3.94 | 4.44 | 6.77 |
| Max | 27.41 | 25.28 | 17.23 | 12.82 | 16.67 | 15.22 |
| Mean | 12.35 | 8.66 | 9.90 | 8.04 | 9.88 | 10.74 |
| Std. | 4.86 | 5.07 | 2.86 | 1.87 | 2.65 | 2.33 |
| Water (g/m2) | ||||||
| Min | 2.93 | 43.93 | 58.99 | 84.43 | 64.99 | 143.79 |
| Max | 655.26 | 339.96 | 144.76 | 168.78 | 206.11 | 312.01 |
| Mean | 115.31 | 116.20 | 102.90 | 117.21 | 115.27 | 213.60 |
| Std. | 79.16 | 48.63 | 15.86 | 16.09 | 29.14 | 46.98 |
| Dry matter (g/m2) | ||||||
| Min | 17.07 | 16.55 | 55.37 | 8.74 | 49.04 | 66.95 |
| Max | 157.32 | 331.04 | 166.24 | 56.10 | 145.58 | 185.91 |
| Mean | 52.76 | 52.43 | 100.88 | 33.24 | 81.75 | 122.60 |
| Std. | 24.68 | 36.69 | 24.69 | 5.99 | 21.84 | 26.86 |
| Reference | Method | RMSE | R2 | Ref | Trans |
|---|---|---|---|---|---|
| [50] | PROSPECT-5 | 4.22 | \ | √ | √ |
| [49] | PROSPECT-D | 3.81 | \ | √ | √ |
| PROSPECT-5 | 6.90 | \ | √ | √ | |
| [52] | PROSPECT-5 | 3.22 | 0.74 | √ | √ |
| 3.88 | 0.49 | √ | \ | ||
| [70] | PROSPECT-5B | 3.38 | \ | √ | √ |
| 3.65 | \ | √ | \ | ||
| PROCWT-S3 | 1.84 | \ | √ | √ | |
| 2.50 | \ | √ | \ | ||
| [40] | PROSPECT-5 + CNN | 2.60 | 0.74 | √ | \ |
| Present study | MLP | 2.29 | 0.80 | √ | \ |
| Reference | Method | RMSE | R2 | Ref | Trans |
|---|---|---|---|---|---|
| [50] | PROSPECT-5 | 5.35 | \ | √ | √ |
| [52] | PROSPECT-5 | 4.27 | 0.17 | √ | √ |
| 5.08 | 0.06 | √ | \ | ||
| Present study | MLP | 3.86 | 0.24 | √ | \ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, W.; Sun, J.; Zhang, Z.; Zhang, K.; Qiu, F.; Xu, J. Novel Hybrid Model to Estimate Leaf Carotenoids Using Multilayer Perceptron and PROSPECT Simulations. Remote Sens. 2023, 15, 4997. https://doi.org/10.3390/rs15204997
Hao W, Sun J, Zhang Z, Zhang K, Qiu F, Xu J. Novel Hybrid Model to Estimate Leaf Carotenoids Using Multilayer Perceptron and PROSPECT Simulations. Remote Sensing. 2023; 15(20):4997. https://doi.org/10.3390/rs15204997
Chicago/Turabian StyleHao, Weilin, Jia Sun, Zichao Zhang, Kan Zhang, Feng Qiu, and Jin Xu. 2023. "Novel Hybrid Model to Estimate Leaf Carotenoids Using Multilayer Perceptron and PROSPECT Simulations" Remote Sensing 15, no. 20: 4997. https://doi.org/10.3390/rs15204997
APA StyleHao, W., Sun, J., Zhang, Z., Zhang, K., Qiu, F., & Xu, J. (2023). Novel Hybrid Model to Estimate Leaf Carotenoids Using Multilayer Perceptron and PROSPECT Simulations. Remote Sensing, 15(20), 4997. https://doi.org/10.3390/rs15204997







