Potato Leaf Area Index Estimation Using Multi-Sensor Unmanned Aerial Vehicle (UAV) Imagery and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiments
2.2. Data Collection
2.3. Image Process and Features Calculation
2.3.1. RGB-Based Features
2.3.2. LiDAR-Based Features
2.3.3. HSI-Based Features
2.4. Feature Search/Selection/Comparison Strategies
2.4.1. Grid Searching Bands and Fixed Bands
2.4.2. Combination of VIs from Different Data Sources
2.4.3. Statistical Features Selection and Combination with VIs
2.5. Machine Learning Model
2.6. Evaluation Metrics
3. Results
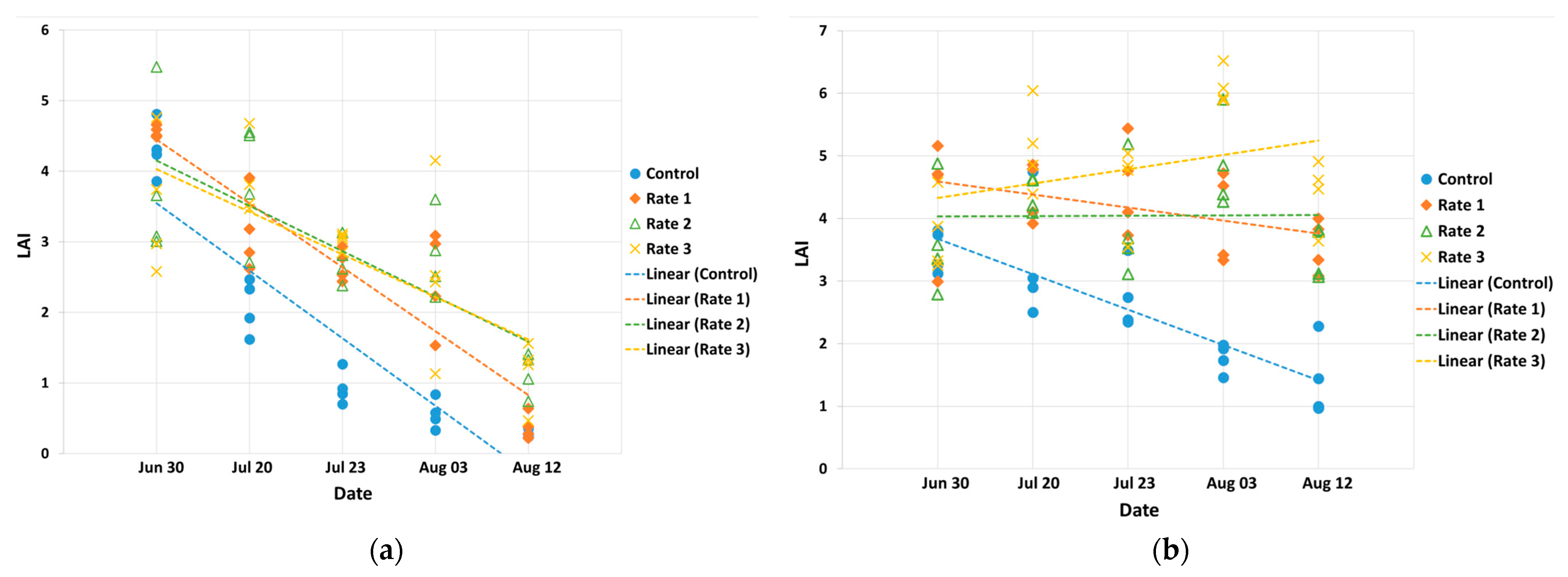
3.1. Ground Data Statistics
3.2. Comparison of VIs with Searched and Fixed Bands
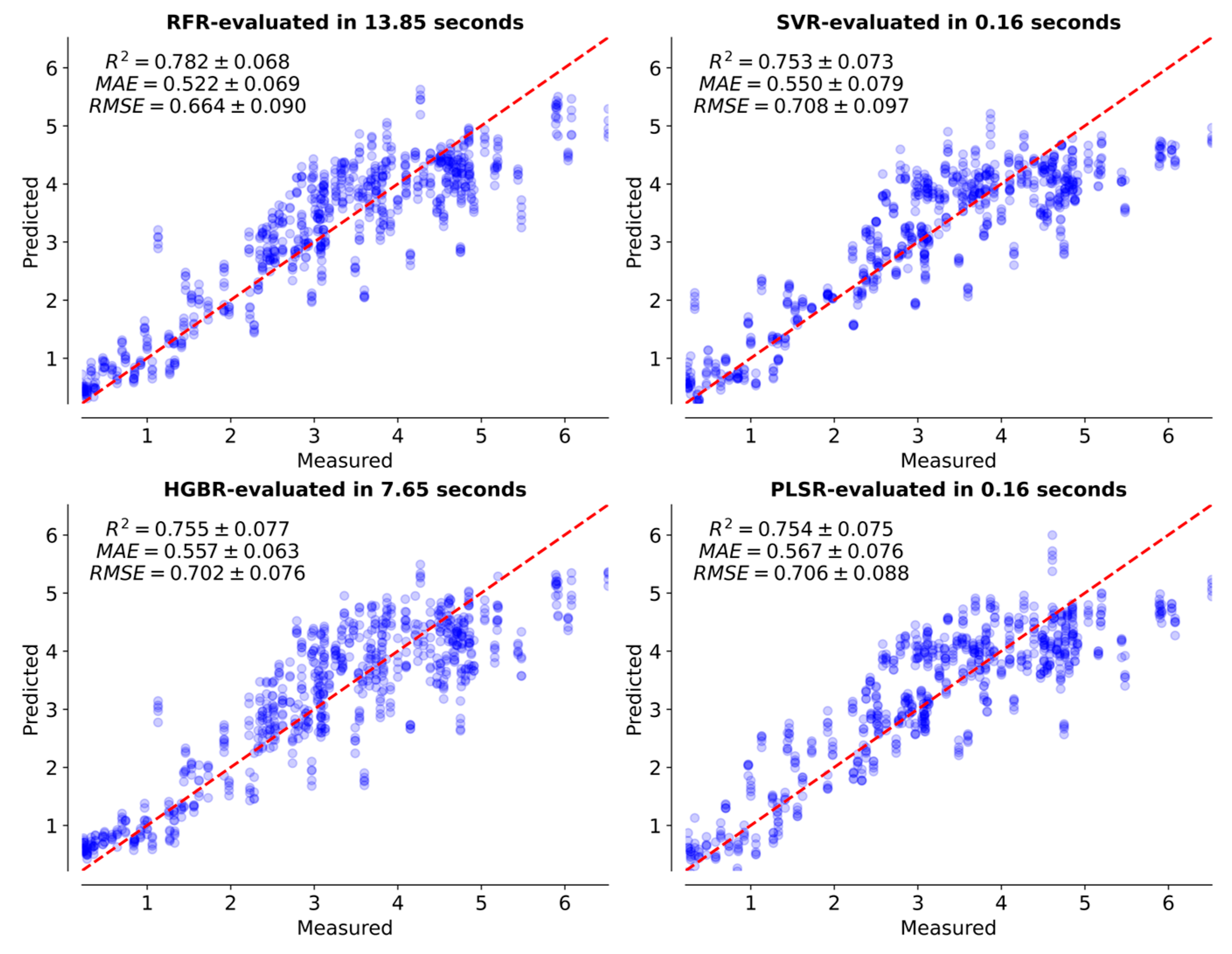
3.3. Combination of VIs from Different Data Sources
3.4. Combination of VIs and Selected Statistical Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ezekiel, R.; Singh, N.; Sharma, S.; Kaur, A. Beneficial Phytochemicals in Potato—A Review. Food Res. Int. 2013, 50, 487–496. [Google Scholar] [CrossRef]
- Campos, H.; Ortiz, O. (Eds.) The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Springer International Publishing: Cham, Switzerland, 2020; ISBN 978-3-030-28682-8. [Google Scholar]
- Devaux, A.; Goffart, J.-P.; Kromann, P.; Andrade-Piedra, J.; Polar, V.; Hareau, G. The Potato of the Future: Opportunities and Challenges in Sustainable Agri-Food Systems. Potato Res. 2021, 64, 681–720. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Liu, J.; Qian, B.; He, L.; Liu, J.; Wang, R.; Jing, Q.; Champagne, C.; McNairn, H.; Powers, J.; et al. Estimating Crop Biomass Using Leaf Area Index Derived from Landsat 8 and Sentinel-2 Data. ISPRS J. Photogramm. Remote Sens. 2020, 168, 236–250. [Google Scholar] [CrossRef]
- Simic Milas, A.; Romanko, M.; Reil, P.; Abeysinghe, T.; Marambe, A. The Importance of Leaf Area Index in Mapping Chlorophyll Content of Corn under Different Agricultural Treatments Using UAV Images. Int. J. Remote Sens. 2018, 39, 5415–5431. [Google Scholar] [CrossRef]
- Baez-Gonzalez, A.D.; Kiniry, J.R.; Maas, S.J.; Tiscareno, M.L.; Macias C., J.; Mendoza, J.L.; Richardson, C.W.; Salinas G., J.; Manjarrez, J.R. Large-Area Maize Yield Forecasting Using Leaf Area Index Based Yield Model. Agron. J. 2005, 97, 418–425. [Google Scholar] [CrossRef]
- Duchemin, B.; Hadria, R.; Erraki, S.; Boulet, G.; Maisongrande, P.; Chehbouni, A.; Escadafal, R.; Ezzahar, J.; Hoedjes, J.C.B.; Kharrou, M.H.; et al. Monitoring Wheat Phenology and Irrigation in Central Morocco: On the Use of Relationships between Evapotranspiration, Crops Coefficients, Leaf Area Index and Remotely-Sensed Vegetation Indices. Agric. Water Manag. 2006, 79, 1–27. [Google Scholar] [CrossRef]
- Liu, J.; Pattey, E.; Jégo, G. Assessment of Vegetation Indices for Regional Crop Green LAI Estimation from Landsat Images over Multiple Growing Seasons. Remote Sens. Environ. 2012, 123, 347–358. [Google Scholar] [CrossRef]
- Kamenova, I.; Dimitrov, P. Evaluation of Sentinel-2 Vegetation Indices for Prediction of LAI, FAPAR and FCover of Winter Wheat in Bulgaria. Eur. J. Remote Sens. 2021, 54, 89–108. [Google Scholar] [CrossRef]
- Deng, F.; Chen, J.M.; Plummer, S.; Chen, M.; Pisek, J. Algorithm for Global Leaf Area Index Retrieval Using Satellite Imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2219–2229. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Wang, J.; Xiang, Y.; Zhao, X.; Song, J. Long-Time-Series Global Land Surface Satellite Leaf Area Index Product Derived from MODIS and AVHRR Surface Reflectance. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5301–5318. [Google Scholar] [CrossRef]
- Aslan, M.F.; Durdu, A.; Sabanci, K.; Ropelewska, E.; Gültekin, S.S. A Comprehensive Survey of the Recent Studies with UAV for Precision Agriculture in Open Fields and Greenhouses. Appl. Sci. 2022, 12, 1047. [Google Scholar] [CrossRef]
- Pandey, A.; Jain, K. An Intelligent System for Crop Identification and Classification from UAV Images Using Conjugated Dense Convolutional Neural Network. Comput. Electron. Agric. 2022, 192, 106543. [Google Scholar] [CrossRef]
- Bouguettaya, A.; Zarzour, H.; Kechida, A.; Taberkit, A.M. Deep Learning Techniques to Classify Agricultural Crops through UAV Imagery: A Review. Neural Comput. Appl. 2022, 34, 9511–9536. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A Review on the Use of Unmanned Aerial Vehicles and Imaging Sensors for Monitoring and Assessing Plant Stresses. Drones 2019, 3, 40. [Google Scholar] [CrossRef]
- Xie, T.; Li, J.; Yang, C.; Jiang, Z.; Chen, Y.; Guo, L.; Zhang, J. Crop Height Estimation Based on UAV Images: Methods, Errors, and Strategies. Comput. Electron. Agric. 2021, 185, 106155. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, Z.; Ma, Y.; Du, Q.; Williams, P.; Drewry, J.; Luck, B. Alfalfa Yield Prediction Using UAV-Based Hyperspectral Imagery and Ensemble Learning. Remote Sens. 2020, 12, 2028. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley Using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Kou, J.; Duan, L.; Yin, C.; Ma, L.; Chen, X.; Gao, P.; Lv, X. Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images. Sustainability 2022, 14, 9259. [Google Scholar] [CrossRef]
- Xu, X.; Fan, L.; Li, Z.; Meng, Y.; Feng, H.; Yang, H.; Xu, B. Estimating Leaf Nitrogen Content in Corn Based on Information Fusion of Multiple-Sensor Imagery from UAV. Remote Sens. 2021, 13, 340. [Google Scholar] [CrossRef]
- Hammond, K.; Kerry, R.; Jensen, R.R.; Spackman, R.; Hulet, A.; Hopkins, B.G.; Yost, M.A.; Hopkins, A.P.; Hansen, N.C. Assessing Within-Field Variation in Alfalfa Leaf Area Index Using UAV Visible Vegetation Indices. Agronomy 2023, 13, 1289. [Google Scholar] [CrossRef]
- Cheng, Q.; Xu, H.; Fei, S.; Li, Z.; Chen, Z. Estimation of Maize LAI Using Ensemble Learning and UAV Multispectral Imagery under Different Water and Fertilizer Treatments. Agriculture 2022, 12, 1267. [Google Scholar] [CrossRef]
- Liao, K.; Li, Y.; Zou, B.; Li, D.; Lu, D. Examining the Role of UAV Lidar Data in Improving Tree Volume Calculation Accuracy. Remote Sens. 2022, 14, 4410. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating Forest Structural Attributes Using UAV-LiDAR Data in Ginkgo Plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Onishi, M.; Ise, T. Explainable Identification and Mapping of Trees Using UAV RGB Image and Deep Learning. Sci. Rep. 2021, 11, 903. [Google Scholar] [CrossRef]
- Li, B.; Xu, X.; Han, J.; Zhang, L.; Bian, C.; Jin, L.; Liu, J. The Estimation of Crop Emergence in Potatoes by UAV RGB Imagery. Plant Methods 2019, 15, 15. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Using 3D Point Clouds Derived from UAV RGB Imagery to Describe Vineyard 3D Macro-Structure. Remote Sens. 2017, 9, 111. [Google Scholar] [CrossRef]
- Yan, Y.; Yang, J.; Li, B.; Qin, C.; Ji, W.; Xu, Y.; Huang, Y. High-Resolution Mapping of Soil Organic Matter at the Field Scale Using UAV Hyperspectral Images with a Small Calibration Dataset. Remote Sens. 2023, 15, 1433. [Google Scholar] [CrossRef]
- Sun, Q.; Gu, X.; Chen, L.; Xu, X.; Wei, Z.; Pan, Y.; Gao, Y. Monitoring Maize Canopy Chlorophyll Density under Lodging Stress Based on UAV Hyperspectral Imagery. Comput. Electron. Agric. 2022, 193, 106671. [Google Scholar] [CrossRef]
- Tang, H.; Armston, J.; Hancock, S.; Marselis, S.; Goetz, S.; Dubayah, R. Characterizing Global Forest Canopy Cover Distribution Using Spaceborne Lidar. Remote Sens. Environ. 2019, 231, 111262. [Google Scholar] [CrossRef]
- ten Harkel, J.; Bartholomeus, H.; Kooistra, L. Biomass and Crop Height Estimation of Different Crops Using UAV-Based Lidar. Remote Sens. 2020, 12, 17. [Google Scholar] [CrossRef]
- Mulugeta Aneley, G.; Haas, M.; Köhl, K. LIDAR-Based Phenotyping for Drought Response and Drought Tolerance in Potato. Potato Res. 2022. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Zhang, Q.; Duan, R.; Liu, J.; Qin, Y.; Wang, X. Toward Multi-Stage Phenotyping of Soybean with Multimodal UAV Sensor Data: A Comparison of Machine Learning Approaches for Leaf Area Index Estimation. Remote Sens. 2023, 15, 7. [Google Scholar] [CrossRef]
- Yan, P.; Han, Q.; Feng, Y.; Kang, S. Estimating LAI for Cotton Using Multisource UAV Data and a Modified Universal Model. Remote Sens. 2022, 14, 4272. [Google Scholar] [CrossRef]
- Yang, Q.; Ye, H.; Huang, K.; Zha, Y.; Shi, L. Estimation of Leaf Area Index of Sugarcane Using Crop Surface Model Based on UAV Image. Trans. Chin. Soc. Agric. Eng. 2017, 33, 104–111. [Google Scholar]
- Li, S.; Yuan, F.; Ata-UI-Karim, S.T.; Zheng, H.; Cheng, T.; Liu, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cao, Q. Combining Color Indices and Textures of UAV-Based Digital Imagery for Rice LAI Estimation. Remote Sens. 2019, 11, 1763. [Google Scholar] [CrossRef]
- Feng, H.; Tao, H.; Li, Z.; Yang, G.; Zhao, C. Comparison of UAV RGB Imagery and Hyperspectral Remote-Sensing Data for Monitoring Winter Wheat Growth. Remote Sens. 2022, 14, 3811. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Long, H.; Yue, J.; Li, Z.; Yang, G.; Yang, X.; Fan, L. Estimation of Crop Growth Parameters Using UAV-Based Hyperspectral Remote Sensing Data. Sensors 2020, 20, 1296. [Google Scholar] [CrossRef]
- Gao, L.; Yang, G.; Yu, H.; Xu, B.; Zhao, X.; Dong, J.; Ma, Y. Retrieving Winter Wheat Leaf Area Index Based on Unmanned Aerial…: Ingenta Connect. Trans. Chin. Soc. Agric. Eng. 2016, 32, 113–120. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.; Chen, P. Comparing Different Methods for Wheat LAI Inversion Based on Hyperspectral Data. Agriculture 2022, 12, 1353. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Q.; Yi, X.; Ma, L.; Zhang, L.; Huang, C.; Zhang, Z.; Lv, X. Estimation of Cotton Leaf Area Index (LAI) Based on Spectral Transformation and Vegetation Index. Remote Sens. 2022, 14, 136. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Xi, X.; Nie, S.; Fan, X.; Chen, H.; Yang, X.; Peng, D.; Lin, Y.; Zhou, G. Combining Hyperspectral Imagery and LiDAR Pseudo-Waveform for Predicting Crop LAI, Canopy Height and above-Ground Biomass. Ecol. Indic. 2019, 102, 801–812. [Google Scholar] [CrossRef]
- Yue, J.; Feng, H.; Jin, X.; Yuan, H.; Li, Z.; Zhou, C.; Yang, G.; Tian, Q. A Comparison of Crop Parameters Estimation Using Images from UAV-Mounted Snapshot Hyperspectral Sensor and High-Definition Digital Camera. Remote Sens. 2018, 10, 1138. [Google Scholar] [CrossRef]
- van der Meij, B.; Kooistra, L.; Suomalainen, J.; Barel, J.M.; De Deyn, G.B. Remote Sensing of Plant Trait Responses to Field-Based Plant–Soil Feedback Using UAV-Based Optical Sensors. Biogeosciences 2017, 14, 733–749. [Google Scholar] [CrossRef]
- Bradford, B.Z.; Colquhoun, J.B.; Chapman, S.A.; Gevens, A.J.; Groves, R.L.; Heider, D.J.; Nice, G.R.W.; Ruark, M.D.; Wang, Y. Commercial Vegetable Production in Wisconsin—2023; University of Wisconsin–Madison: Madison, WI, USA, 2023. [Google Scholar]
- Zhang, W.; Qi, J.; Wan, P.; Wang, H.; Xie, D.; Wang, X.; Yan, G. An Easy-to-Use Airborne LiDAR Data Filtering Method Based on Cloth Simulation. Remote Sens. 2016, 8, 501. [Google Scholar] [CrossRef]
- Arnqvist, J.; Freier, J.; Dellwik, E. Robust Processing of Airborne Laser Scans to Plant Area Density Profiles. Biogeosciences 2020, 17, 5939–5952. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Deering, D.W.; Schell, J.A.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA: Washington, DC, USA, 1974.
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a Two-Band Enhanced Vegetation Index without a Blue Band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Dong, T.; Liu, J.; Shang, J.; Qian, B.; Ma, B.; Kovacs, J.M.; Walters, D.; Jiao, X.; Geng, X.; Shi, Y. Assessment of Red-Edge Vegetation Indices for Crop Leaf Area Index Estimation. Remote Sens. Environ. 2019, 222, 133–143. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating Chlorophyll Content from Hyperspectral Vegetation Indices: Modeling and Validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS Terrestrial Chlorophyll Index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Gitelson, A.A.; Viña, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote Estimation of Canopy Chlorophyll Content in Crops. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- Tian, Y.-C.; Gu, K.-J.; Chu, X.; Yao, X.; Cao, W.-X.; Zhu, Y. Comparison of Different Hyperspectral Vegetation Indices for Canopy Leaf Nitrogen Concentration Estimation in Rice. Plant Soil 2014, 376, 193–209. [Google Scholar] [CrossRef]
- Liang, L.; Huang, T.; Di, L.; Geng, D.; Yan, J.; Wang, S.; Wang, L.; Li, L.; Chen, B.; Kang, J. Influence of Different Bandwidths on LAI Estimation Using Vegetation Indices. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 1494–1502. [Google Scholar] [CrossRef]
- Zhang, F.; Hassanzadeh, A.; Kikkert, J.; Pethybridge, S.J.; van Aardt, J. Evaluation of Leaf Area Index (LAI) of Broadacre Crops Using UAS-Based LiDAR Point Clouds and Multispectral Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 4027–4044. [Google Scholar] [CrossRef]
- Jayaraj, P. Estimation of Leaf Area Index (Lai) in Maize Planting Experiments Using Lidar and Hyperspectral Data Acquired from a Uav Platform. Master’s Thesis, Purdue University, West Lafayette, IN, USA, 2023. [Google Scholar]
- Dilmurat, K.; Sagan, V.; Moose, S. Ai-driven maize yield forecasting using unmanned aerial vehicle-based hyperspectral and lidar data fusion. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, V-3–2022, 193–199. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Huang, Y.; Yang, T.; Li, J.; Zhu, K.; Zhang, J.; Yang, B.; Shao, C.; Peng, J.; et al. Optimization of Multi-Source UAV RS Agro-Monitoring Schemes Designed for Field-Scale Crop Phenotyping. Precis. Agric. 2021, 22, 1768–1802. [Google Scholar] [CrossRef]
- Barbosa, B.D.S.; Ferraz, G.A.e.S.; Costa, L.; Ampatzidis, Y.; Vijayakumar, V.; dos Santos, L.M. UAV-Based Coffee Yield Prediction Utilizing Feature Selection and Deep Learning. Smart Agric. Technol. 2021, 1, 100010. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, D.; Wu, Z.; Song, H.; Zhang, X. Prediction of Buckwheat Maturity in UAV-RGB Images Based on Recursive Feature Elimination Cross-Validation: A Case Study in Jinzhong, Northern China. Plants 2022, 11, 3257. [Google Scholar] [CrossRef] [PubMed]
- Aslan, M.F. Comparative Analysis of CNN Models and Bayesian Optimization-Based Machine Learning Algorithms in Leaf Type Classification. Balk. J. Electr. Comput. Eng. 2023, 11, 13–24. [Google Scholar] [CrossRef]
- Tan, L.; Lu, J.; Jiang, H. Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering 2021, 3, 542–558. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Zhou, H. Deep Learning Based Multi-Temporal Crop Classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Feng, L.; Ma, Y.; Du, Q. A New Attention-Based CNN Approach for Crop Mapping Using Time Series Sentinel-2 Images. Comput. Electron. Agric. 2021, 184, 106090. [Google Scholar] [CrossRef]




| Name | Seasonal Total N Rate | Planting | Emergence (Hilling) | Tuber Initiation | Fertigation Date | |||
|---|---|---|---|---|---|---|---|---|
| 23 April | 12 May | 2 June | 30 June | 10 July | 20 July | 30 July | ||
| C | 37 | 37 | - | - | - | - | - | - |
| R1 | 287 | 37 | 85 | 165 | - | - | - | - |
| R2 | 287 | 37 | 85 | 30 | 34 | 34 | 34 | 34 |
| R3 | 392 | 37 | 85 | 134 | 34 | 34 | 34 | 34 |
| Sensors | Description |
|---|---|
| RGB camera | Sony Cyber-shot DSC-RX1R II 42 MP full-frame sensor 35 mm F2 lens |
| LiDAR unit | Velodyne VLP-16 100 m range 905 nm infra-red (IR) lasers Dual Returns |
| Hyperspectral scanner | Headwall Nano-Hyperspec 274 bands with 2.2 nm spectral resolution (B1-B274) visible-near-infrared range (400–1000 nm) |
| Sensors | Name | Definition |
|---|---|---|
| LiDAR | MaxPlantHeight | Hmax − Hmin |
| H50th, H75th, H90th, H95th | The 50th, 75th, 90th, and 95th percentile height values. | |
| Canopy Volume | Sum (Hallpoints) | |
| Canopy Cover | Numbernon-ground/Numberallpoints | |
| Plant Area Index | plant area/ground surface area | |
| RGB | R | Mean (DNR) |
| G | Mean (DNG) | |
| B | Mean (DNB) | |
| Normalized_R | Mean (DNR/(DNR + DNG + DNB)) | |
| Normalized_G | Mean (DNG/(DNR + DNG + DNB)) | |
| Normalized_B | Mean (DNB/(DNR+ DNG + DNB)) | |
| Hyperspectral | NDVI | (RNIR − RRED)/(RNIR + RRED) |
| EVI2 | 2.5 × (RNIR − RRED)/(1 + 2.4 × RNIR + RRED) | |
| CIrededge | RNIR/RRED-EDGE − 1 | |
| CIgreen | RNIR/RGREEN − 1 | |
| MSRrededge | (RNIR/RRED-EDGE − 1)/(RNIR/RRED-EDGE + 1)1/2 | |
| MTCI | (RNIR − RRED-EDGE)/(RRED-EDGE − RRED) | |
| Mean | Mean value of each band | |
| Std | Standard deviation of each band |
| Model | Hyperparameter | Explanation |
|---|---|---|
| SVR | c | squared L2 penalty |
| gamma | gamma in RBF kernel | |
| RFR | n_estimators | number of trees |
| max_features | the number of features to consider when looking for the best split | |
| min_samples_leaf | The minimum number of samples required to be at a leaf node | |
| random_state | Pseudo-random number generator to control the sub-sampling in the binning process | |
| HGBR | max_iter | The maximum number of iterations of the boosting process |
| learning_rate | The learning rate, shrinkage | |
| max_leaf_nodes | The maximum number of leaves for each tree | |
| random_state | Pseudo-random number generator to control the subsampling in the binning process | |
| PLSR | n_components | Number of components to keep |
| tol | The tolerance used as convergence criteria in the power method |
| VI | Green (nm) | Red (nm) | Red Edge (nm) | NIR (nm) | r | Ref. | |
|---|---|---|---|---|---|---|---|
| Searched bands | NDVI | - | 677.968 | - | 826.143 | 0.830 | - |
| EVI2 | - | 708.93 | - | 773.065 | 0.823 | - | |
| CIrededge | - | - | 726.622 | 901.337 | 0.736 | - | |
| CIgreen | 551.908 | - | - | 914.606 | 0.585 | - | |
| MSRrededge | - | - | 717.776 | 901.337 | 0.759 | - | |
| MTCI | - | 631.525 | 713.353 | 797.393 | 0.549 | ||
| Fixed bands | NDVI | - | 670 (669.121) | - | 800 (799.604) | 0.820 | [43] |
| EVI2 | - | 670 (669.121) | - | 800 (799.604) | 0.736 | [43] | |
| CIrededge | - | - | 710 (708.930) | 800 (799.604) | 0.565 | [55] | |
| CIgreen | 550 (549.696) | - | - | 800 (799.604) | 0.519 | [55] | |
| MSRrededge | - | - | 705 (704.507) | 750 (750.95) | 0.548 | [52] | |
| MTCI | - | 681 (680.179) | 708 (708.93) | 753 (753.161) | 0.505 | [53] |
| Sources | Evaluation Model | R2 | RMSE | MAE | Evaluation Time (s) |
|---|---|---|---|---|---|
| RGB | RFR | 0.668 | 0.826 | 0.649 | 4.93 |
| SVR | 0.726 | 0.751 | 0.592 | 0.19 | |
| HGBR | 0.627 | 0.876 | 0.685 | 1.75 | |
| PLSR | 0.638 | 0.862 | 0.698 | 0.07 | |
| LiDAR | RFR | 0.666 | 0.824 | 0.643 | 4.93 |
| SVR | 0.552 | 0.958 | 0.750 | 0.10 | |
| HGBR | 0.633 | 0.865 | 0.683 | 1.75 | |
| PLSR | 0.640 | 0.856 | 0.693 | 0.07 | |
| HSI | RFR | 0.766 | 0.688 | 0.539 | 2.20 |
| SVR | 0.762 | 0.694 | 0.544 | 0.15 | |
| HGBR | 0.764 | 0.691 | 0.554 | 2.68 | |
| PLSR | 0.738 | 0.729 | 0.598 | 0.09 |
| Selection Methods | Base Model | Evaluation Model | Number of Features | R2 | RMSE | Evaluation Time (s) |
|---|---|---|---|---|---|---|
| RFECV | RFR | RFR | 44 | 0.766 | 0.686 | 2.34 |
| SVR | SVR | 40 | 0.763 | 0.694 | 0.10 | |
| SVR | HGBR | 40 | 0.746 | 0.713 | 1.66 | |
| RFR | PLSR | 44 | 0.747 | 0.717 | 0.07 | |
| - | - | RFR | 548 | 0.759 | 0.697 | 2.76 |
| - | SVR | 548 | 0.763 | 0.695 | 0.23 | |
| - | HGBR | 548 | 0.750 | 0.712 | 14.70 | |
| - | PLSR | 548 | 0.741 | 0.721 | 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Zhou, J.; Fan, J.; Wang, Y.; Zhang, Z. Potato Leaf Area Index Estimation Using Multi-Sensor Unmanned Aerial Vehicle (UAV) Imagery and Machine Learning. Remote Sens. 2023, 15, 4108. https://doi.org/10.3390/rs15164108
Yu T, Zhou J, Fan J, Wang Y, Zhang Z. Potato Leaf Area Index Estimation Using Multi-Sensor Unmanned Aerial Vehicle (UAV) Imagery and Machine Learning. Remote Sensing. 2023; 15(16):4108. https://doi.org/10.3390/rs15164108
Chicago/Turabian StyleYu, Tong, Jing Zhou, Jiahao Fan, Yi Wang, and Zhou Zhang. 2023. "Potato Leaf Area Index Estimation Using Multi-Sensor Unmanned Aerial Vehicle (UAV) Imagery and Machine Learning" Remote Sensing 15, no. 16: 4108. https://doi.org/10.3390/rs15164108
APA StyleYu, T., Zhou, J., Fan, J., Wang, Y., & Zhang, Z. (2023). Potato Leaf Area Index Estimation Using Multi-Sensor Unmanned Aerial Vehicle (UAV) Imagery and Machine Learning. Remote Sensing, 15(16), 4108. https://doi.org/10.3390/rs15164108







