New Methodology for Intertidal Seaweed Biomass Estimation Using Multispectral Data Obtained with Unoccupied Aerial Vehicles
Abstract
1. Introduction
2. Materials and Methods
2.1. Seaweed Species-Based NDVI/Biomass Relationship
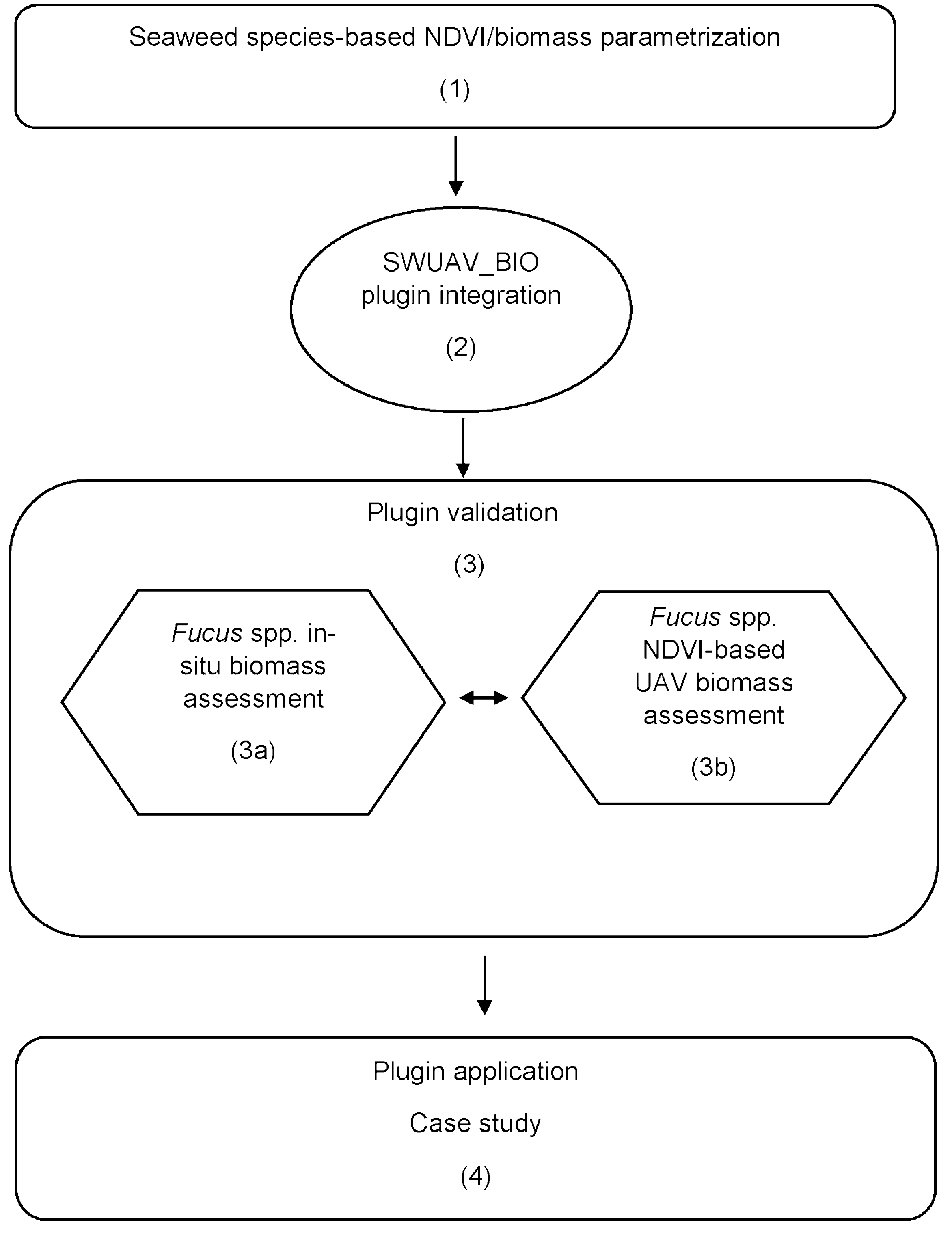
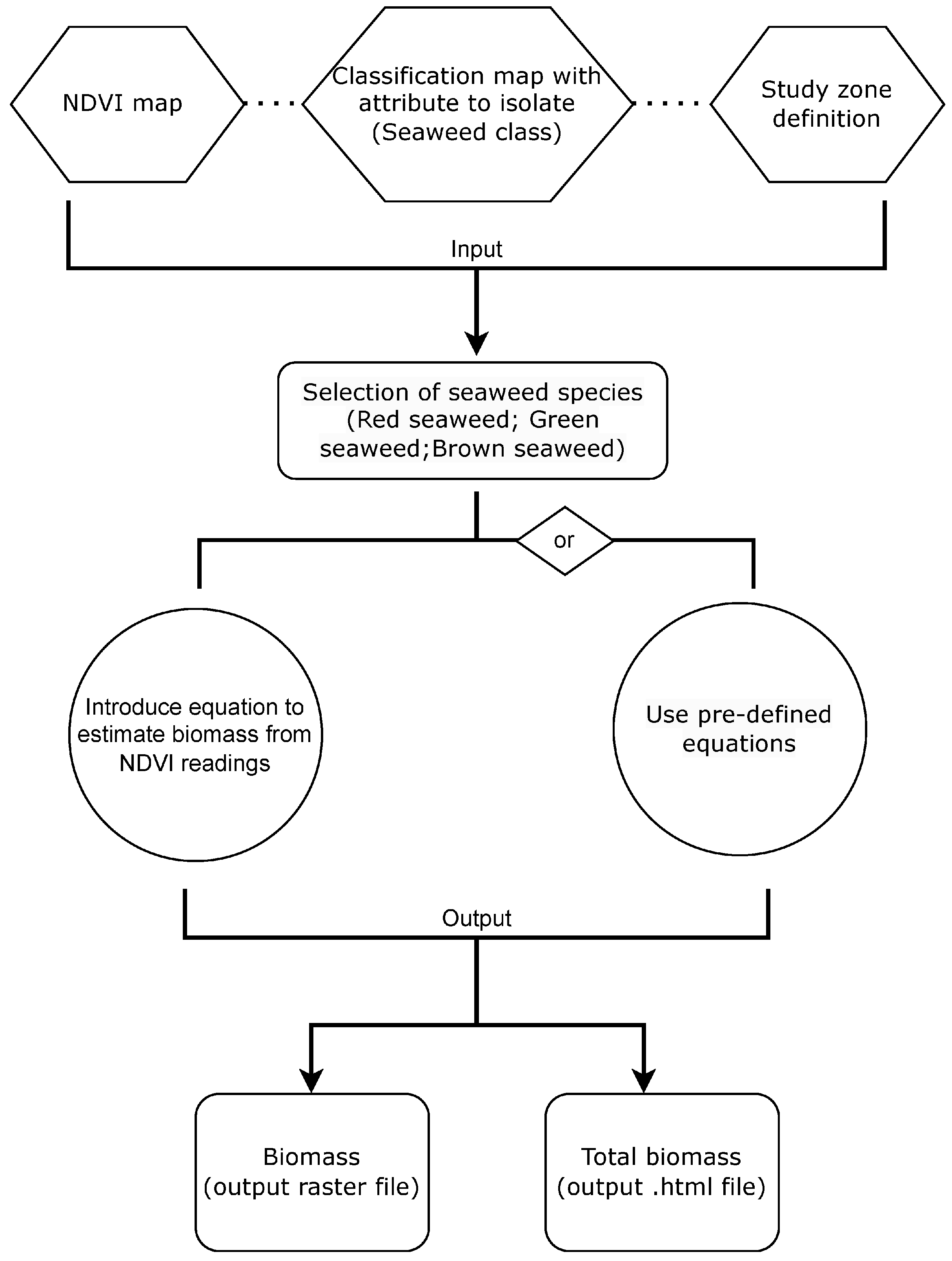
2.2. Plugin Design
2.3. Plugin Validation
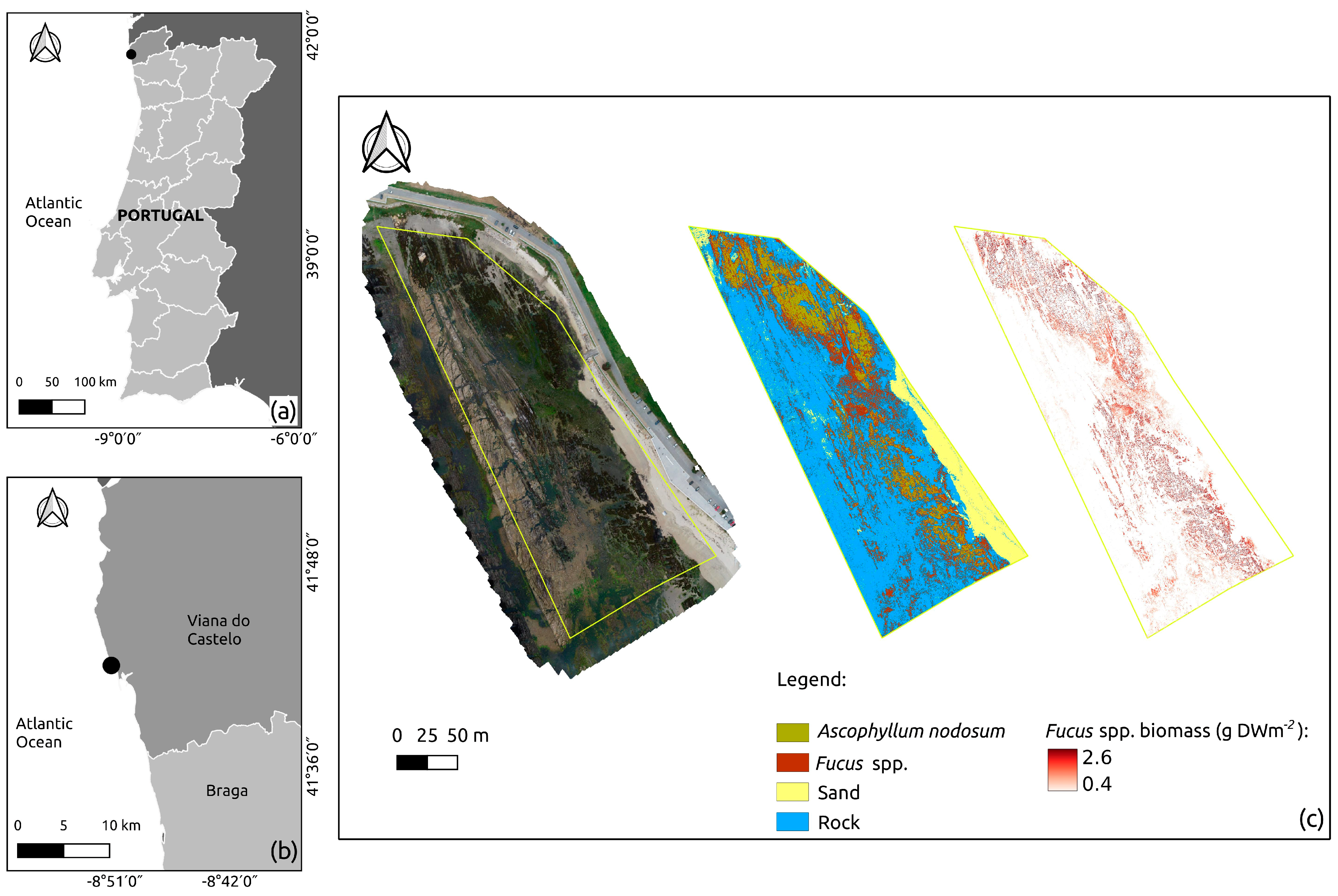
2.3.1. Study Area
2.3.2. In Situ Data Collection
2.3.3. Multispectral UAV Flight
2.4. Plugin Application—Case Study
3. Results
3.1. NDVI−Seaweed Biomass Relationship
3.2. Plugin Validation
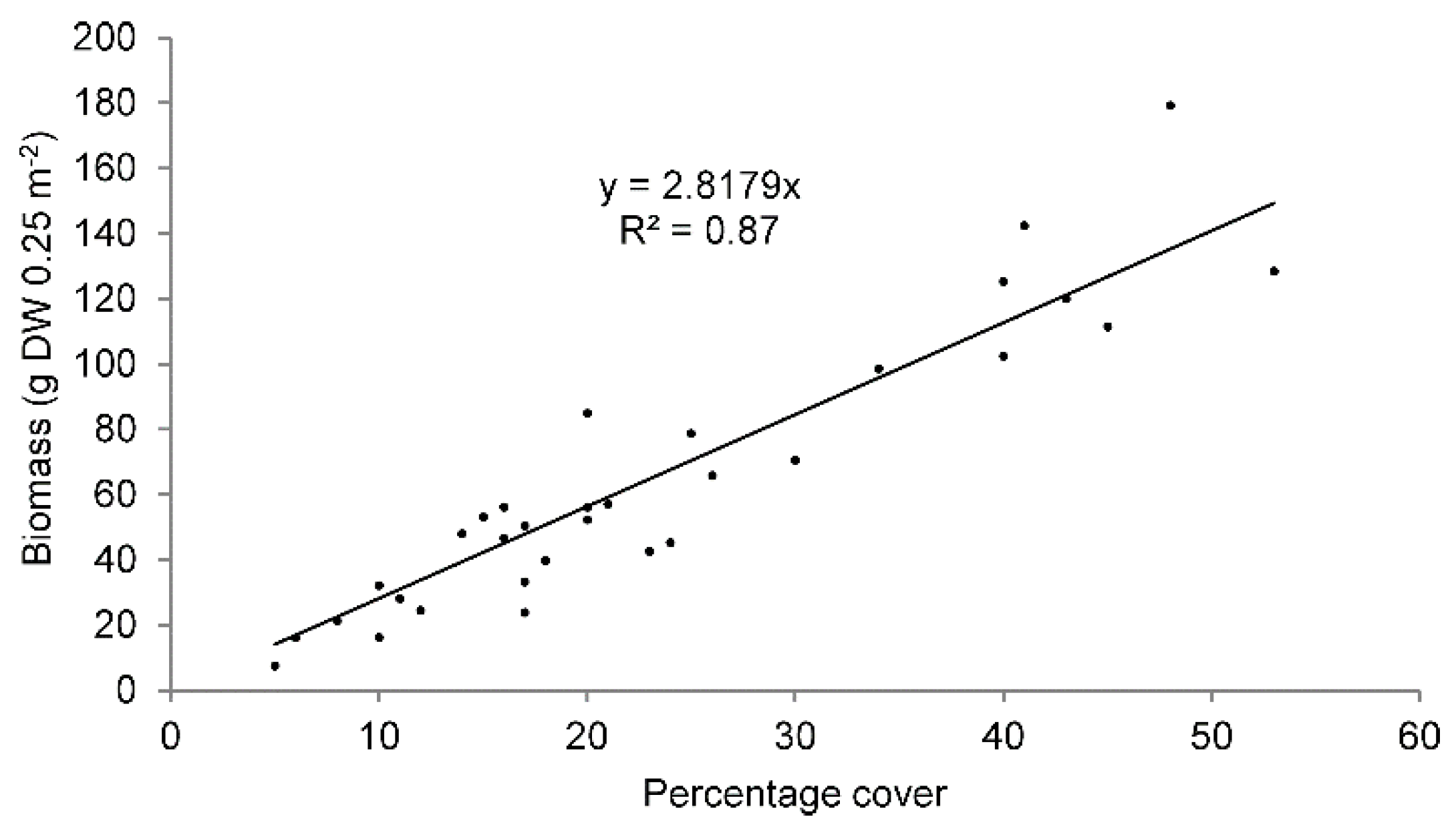
3.2.1. In Situ Data Collection
3.2.2. Multispectral UAV Flight
3.3. Plugin Application—Case Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Girão, M.; Ribeiro, I.; Ribeiro, T.; Azevedo, I.C.; Pereira, F.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Actinobacteria Isolated From Laminaria ochroleuca: A Source of New Bioactive Compounds. Front. Microbiol. 2019, 10, 683. [Google Scholar] [CrossRef]
- Hafting, J.T.; Craigie, J.S.; Stengel, D.B.; Loureiro, R.R.; Buschmann, A.H.; Yarish, C.; Edwards, M.D.; Critchley, A.T. Prospects and challenges for industrial production of seaweed bioactives. J. Phycol. 2015, 51, 821–837. [Google Scholar] [CrossRef]
- Cotas, J.; Pacheco, D.; Gonçalves, A.M.M.; Silva, P.; Carvalho, L.G.; Pereira, L. Seaweeds’ nutraceutical and biomedical potential in cancer therapy: A concise review. J. Cancer Metastasis Treat. 2021, 7, 13. [Google Scholar] [CrossRef]
- López-Hortas, L.; Flórez-Fernández, N.; Torres, M.D.; Ferreira-Anta, T.; Casas, M.P.; Balboa, E.M.; Falqué, E.; Domínguez, H. Applying Seaweed Compounds in Cosmetics, Cosmeceuticals and Nutricosmetics. Mar. Drugs 2021, 19, 552. [Google Scholar] [CrossRef] [PubMed]
- Underwood, A.J. Experimental ecology of rocky intertidal habitats: What are we learning? J. Exp. Mar. Biol. Ecol. 2000, 250, 51–76. [Google Scholar] [CrossRef] [PubMed]
- Murfitt, S.L.; Allan, B.M.; Bellgrove, A.; Rattray, A.; Young, M.A.; Ierodiaconou, D. Applications of unmanned aerial vehicles in intertidal reef monitoring. Sci. Rep. 2017, 7, 10259. [Google Scholar] [CrossRef] [PubMed]
- Borges, D.; Araujo, R.; Azevedo, I.; Pinto, I.S. Sustainable management of economically valuable seaweed stocks at the limits of their range of distribution: Ascophyllum nodosum (Phaeophyceae) and its southernmost population in Europe. J. Appl. Phycol. 2020, 32, 1365–1375. [Google Scholar] [CrossRef]
- Melo, R.; Sousa-Pinto, I.; Antunes, S.C.; Costa, I.; Borges, D. Temporal and spatial variation of seaweed biomass and assemblages in Northwest Portugal. J. Sea Res. 2021, 174, 102079. [Google Scholar] [CrossRef]
- Candiago, S.; Remondino, F.; De Giglio, M.; Dubbini, M.; Gattelli, M. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña, J.M. High-Throughput 3-D Monitoring of Agricultural-Tree Plantations with Unmanned Aerial Vehicle (UAV) Technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef]
- Honrado, J.L.E.; Solpico, D.B.; Favila, C.M.; Tongson, E.; Tangonan, G.L.; Libatique, N.J.C. UAV imaging with low-cost multispectral imaging system for precision agriculture applications. In Proceedings of the 2017 IEEE Global Humanitarian Technology Conference (GHTC), San Jose, CA, USA, 19–22 October 2017; pp. 1–7. [Google Scholar]
- Soubry, I.; Patias, P.; Tsioukas, V. Monitoring vineyards with UAV and multi-sensors for the assessment of water stress and grape maturity. J. Unmanned Veh. Syst. 2017, 5, 37–50. [Google Scholar] [CrossRef]
- López-García, P.; Intrigliolo, D.S.; Moreno, M.A.; Martínez-Moreno, A.; Ortega, J.F.; Pérez-Álvarez, E.P.; Ballesteros, R. Assessment of Vineyard Water Status by Multispectral and RGB Imagery Obtained from an Unmanned Aerial Vehicle. Am. J. Enol. Vitic. 2021, 72, 285–297. [Google Scholar] [CrossRef]
- Devia, C.A.; Rojas, J.P.; Petro, E.; Martinez, C.; Mondragon, I.F.; Patino, D.; Rebolledo, M.C.; Colorado, J. High-Throughput Biomass Estimation in Rice Crops Using UAV Multispectral Imagery. J. Intell. Robot. Syst. 2019, 96, 573–589. [Google Scholar] [CrossRef]
- Borges, D.; Padua, L.; Azevedo, I.C.; Silva, J.; Sousa, J.J.; Sousa–Pinto, I.; Gonçalves, J.A. Classification of an Intertidal Reef by Machine Learning Techniques Using UAV Based RGB and Multispectral Imagery. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 64–67. [Google Scholar]
- Collin, A.; Dubois, S.; James, D.; Houet, T. Improving Intertidal Reef Mapping Using UAV Surface, Red Edge, and Near-Infrared Data. Drones 2019, 3, 67. [Google Scholar] [CrossRef]
- Van der Wal, D.; van Dalen, J.; Wielemaker-van den Dool, A.; Dijkstra, J.T.; Ysebaert, T. Biophysical control of intertidal benthic macroalgae revealed by high-frequency multispectral camera images. J. Sea Res. 2014, 90, 111–120. [Google Scholar] [CrossRef]
- Brodie, J.; Ash, L.V.; Tittley, I.; Yesson, C. A comparison of multispectral aerial and satellite imagery for mapping intertidal seaweed communities. Aquat. Conserv. 2018, 28, 872–881. [Google Scholar] [CrossRef]
- Taddia, Y.; Russo, P.; Lovo, S.; Pellegrinelli, A. Multispectral UAV monitoring of submerged seaweed in shallow water. Appl. Geomat. 2020, 12, 19–34. [Google Scholar] [CrossRef]
- Tait, L.; Bind, J.; Charan-Dixon, H.; Hawes, I.; Pirker, J.; Schiel, D. Unmanned Aerial Vehicles (UAVs) for Monitoring Macroalgal Biodiversity: Comparison of RGB and Multispectral Imaging Sensors for Biodiversity Assessments. Remote Sens. 2019, 11, 2332. [Google Scholar] [CrossRef]
- Rossiter, T.; Furey, T.; McCarthy, T.; Stengel, D.B. Application of multiplatform, multispectral remote sensors for mapping intertidal macroalgae: A comparative approach. Aquat. Conserv. 2020, 30, 1595–1612. [Google Scholar] [CrossRef]
- Bell, T.W.; Nidzieko, N.J.; Siegel, D.A.; Miller, R.J.; Cavanaugh, K.C.; Nelson, N.B.; Reed, D.C.; Fedorov, D.; Moran, C.; Snyder, J.N.; et al. The Utility of Satellites and Autonomous Remote Sensing Platforms for Monitoring Offshore Aquaculture Farms: A Case Study for Canopy Forming Kelps. Front. Mar. Sci. 2020, 7, 520223. [Google Scholar] [CrossRef]
- Cavanaugh, K.C.; Cavanaugh, K.C.; Bell, T.W.; Hockridge, E.G. An Automated Method for Mapping Giant Kelp Canopy Dynamics from UAV. Front. Environ. Sci. 2021, 8, 587354. [Google Scholar] [CrossRef]
- Saccomanno, V.R.; Bell, T.; Pawlak, C.; Stanley, C.K.; Cavanaugh, K.C.; Hohman, R.; Klausmeyer, K.R.; Cavanaugh, K.; Nickels, A.; Hewerdine, W.; et al. Using unoccupied aerial vehicles to map and monitor changes in emergent kelp canopy after an ecological regime shift. Remote Sens. Ecol. Conserv. 2022, 9, 62–75. [Google Scholar] [CrossRef]
- Conser, E.; Shanks, A.L. Density of benthic macroalgae in the intertidal zone varies with surf zone hydrodynamics. Phycologia 2019, 58, 254–259. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Wang, K.; Zhang, S.; Li, J. Estimation of Seaweed Biomass Based on Multispectral UAV in the Intertidal Zone of Gouqi Island. Remote Sens. 2022, 14, 2143. [Google Scholar] [CrossRef]
- Praeger, C.; Vucko, M.J.; McKinna, L.; de Nys, R.; Cole, A. Estimating the biomass density of macroalgae in land-based cultivation systems using spectral reflectance imagery. Algal. Res. 2020, 50, 102009. [Google Scholar] [CrossRef]
- Che, S.; Du, G.; Wang, N.; He, K.; Mo, Z.; Sun, B.; Chen, Y.; Cao, Y.; Wang, J.; Mao, Y. Biomass estimation of cultivated red algae Pyropia using unmanned aerial platform based multispectral imaging. Plant Methods 2021, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Hu, C.; Ming-Xia, H.E. Remote estimation of biomass of Ulva prolifera macroalgae in the Yellow Sea. Remote Sens. Environ. 2017, 192, 217–227. [Google Scholar] [CrossRef]
- Karki, S.; Bermejo, R.; Wilkes, R.; Monagail, M.M.; Daly, E.; Healy, M.; Hanafin, J.; McKinstry, A.; Mellander, P.-E.; Fenton, O.; et al. Mapping Spatial Distribution and Biomass of Intertidal Ulva Blooms Using Machine Learning and Earth Observation. Front. Mar. Sci. 2021, 8, 633128. [Google Scholar] [CrossRef]
- Kriegler, F.J.; Malila, W.A.; Nalepka, R.F.; Richardson, W. Preprocessing Transformations and Their Effects on Multispectral Recognition. In Proceedings of the Remote Sensing of Environment, VI, Ann Arbor, MI, USA, 1 January 1969; p. 97. [Google Scholar]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with Erts. In NASA Special Publication; NASA: Washington, DC, USA, 1974; Volume 351, p. 309. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Jensen, J.R. Introductory Digital Image Processing: A Remote Sensing Perspective; Prentice Hall, Inc.: Hoboken, NJ, USA, 1986. [Google Scholar]
- Tucker, C.J.; Sellers, P.J. Satellite remote sensing of primary production. Int. J. Remote Sens. 1986, 7, 1395–1416. [Google Scholar] [CrossRef]
- Chao Rodríguez, Y.; Domínguez Gómez, J.A.; Sánchez-Carnero, N.; Rodríguez-Pérez, D. A comparison of spectral macroalgae taxa separability methods using an extensive spectral library. Algal. Res. 2017, 26, 463–473. [Google Scholar] [CrossRef]
- Congedo, L. Semi-Automatic Classification Plugin: A Python tool for the download and processing of remote sensing images in QGIS. J. Open Source Softw. 2021, 6, 3172. [Google Scholar] [CrossRef]
- Arekhi, M.; Goksel, C.; Balik Sanli, F.; Senel, G. Comparative Evaluation of the Spectral and Spatial Consistency of Sentinel-2 and Landsat-8 OLI Data for Igneada Longos Forest. ISPRS Int. J. Geo.-Inf. 2019, 8, 56. [Google Scholar] [CrossRef]
- Oddi, L.; Cremonese, E.; Ascari, L.; Filippa, G.; Galvagno, M.; Serafino, D.; Cella, U.M.d. Using UAV Imagery to Detect and Map Woody Species Encroachment in a Subalpine Grassland: Advantages and Limits. Remote Sens. 2021, 13, 1239. [Google Scholar] [CrossRef]
- Myers, D.J.; Schweik, C.M.; Wicks, R.; Bowlick, F.; Carullo, M. Developing a land cover classification of salt marshes using UAS time-series imagery and an open source workflow. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, XLII-4/W8, 155–162. [Google Scholar] [CrossRef]
- Smyth, T.A.G.; Wilson, R.; Rooney, P.; Yates, K.L. Extent, accuracy and repeatability of bare sand and vegetation cover in dunes mapped from aerial imagery is highly variable. Aeolian Res. 2022, 56, 100799. [Google Scholar] [CrossRef]
- Barillé, L.; Robin, M.; Harin, N.; Bargain, A.; Launeau, P. Increase in seagrass distribution at Bourgneuf Bay (France) detected by spatial remote sensing. Aquat. Bot. 2010, 92, 185–194. [Google Scholar] [CrossRef]
- QGIS.org. QGIS 3.16. Geographic Information System Installation Guide. QGIS Association. Available online: https://github.com/qgis/QGIS/blob/master/INSTALL.md (accessed on 31 March 2020).
- GDAL/OGR, c. GDAL/OGR Geospatial Data Abstraction software Library. Open Source Geospatial Foundation. Available online: https://gdal.org/ (accessed on 31 March 2020).
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- GRASS Development Team. Geographic Resources Analysis Support System (GRASS) Software, Version 7.8. Open Source Geospatial Foundation. Available online: https://grass.osgeo.org/ (accessed on 31 March 2020).
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; Available online: https://www.python.org/ (accessed on 31 March 2020).
- PyQt, 2012. PyQt Reference Guide. Available online: https://doc.qt.io/qtforpython/ (accessed on 31 March 2020).
- QGIS.org, 2020. QGIS 3.16. Geographic Information System API Documentation. QGIS Association. Available online: https://api.qgis.org/api/ (accessed on 31 March 2020).
- Hoang, T.C.; O’Leary, M.J.; Fotedar, R.K. Remote-Sensed Mapping of Sargassum spp. Distribution around Rottnest Island, Western Australia, Using High-Spatial Resolution WorldView-2 Satellite Data. J. Coast. Res. 2016, 32, 1310–1321, 1312. [Google Scholar]
- Hoang, T.; Garcia, R.; O’Leary, M.; Fotedar, R. Identification and Mapping of Marine Submerged Aquatic Vegetation in Shallow Coastal Waters with WorldView-2 Satellite Data. J. Coast. Res. 2016, 75, 1287–1291, 1285. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Richards, J.A. Remote Sensing Digital Image Analysis: An Introduction; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data: Principles and Practices; Press, C., Ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar] [CrossRef]
- Tucker, C.J. Asymptotic nature of grass canopy spectral reflectance. Appl. Opt. 1977, 16, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Huete, A.R.; Ni, W.; Miura, T. Optical-biophysical relationships of vegetation spectra without background contamination. Remote Sens. Environ. 2000, 74, 609–620. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, H.; Niu, Y.; Han, W. Mapping Maize Water Stress Based on UAV Multispectral Remote Sensing. Remote Sens. 2019, 11, 605. [Google Scholar] [CrossRef]
- Gutierrez, M.; Reynolds, M.P.; Klatt, A.R. Association of water spectral indices with plant and soil water relations in contrasting wheat genotypes. J. Exp. Bot. 2010, 61, 3291–3303. [Google Scholar] [CrossRef]
- Fyfe, S.K. Spatial and temporal variation in spectral reflectance: Are seagrass species spectrally distinct? Limnol. Oceanogr. 2003, 48, 464–479. [Google Scholar] [CrossRef]
- Vahtmäe, E.; Kotta, J.; Orav-Kotta, H.; Kotta, I.; Pärnoja, M.; Kutser, T. Predicting macroalgal pigments (chlorophyll a, chlorophyll b, chlorophyll a + b, carotenoids) in various environmental conditions using high-resolution hyperspectral spectroradiometers. Int. J. Remote Sens. 2018, 39, 5716–5738. [Google Scholar] [CrossRef]
- Bell, T.W.; Cavanaugh, K.C.; Siegel, D.A. Remote monitoring of giant kelp biomass and physiological condition: An evaluation of the potential for the Hyperspectral Infrared Imager (HyspIRI) mission. Remote Sens. Environ. 2015, 167, 218–228. [Google Scholar] [CrossRef]
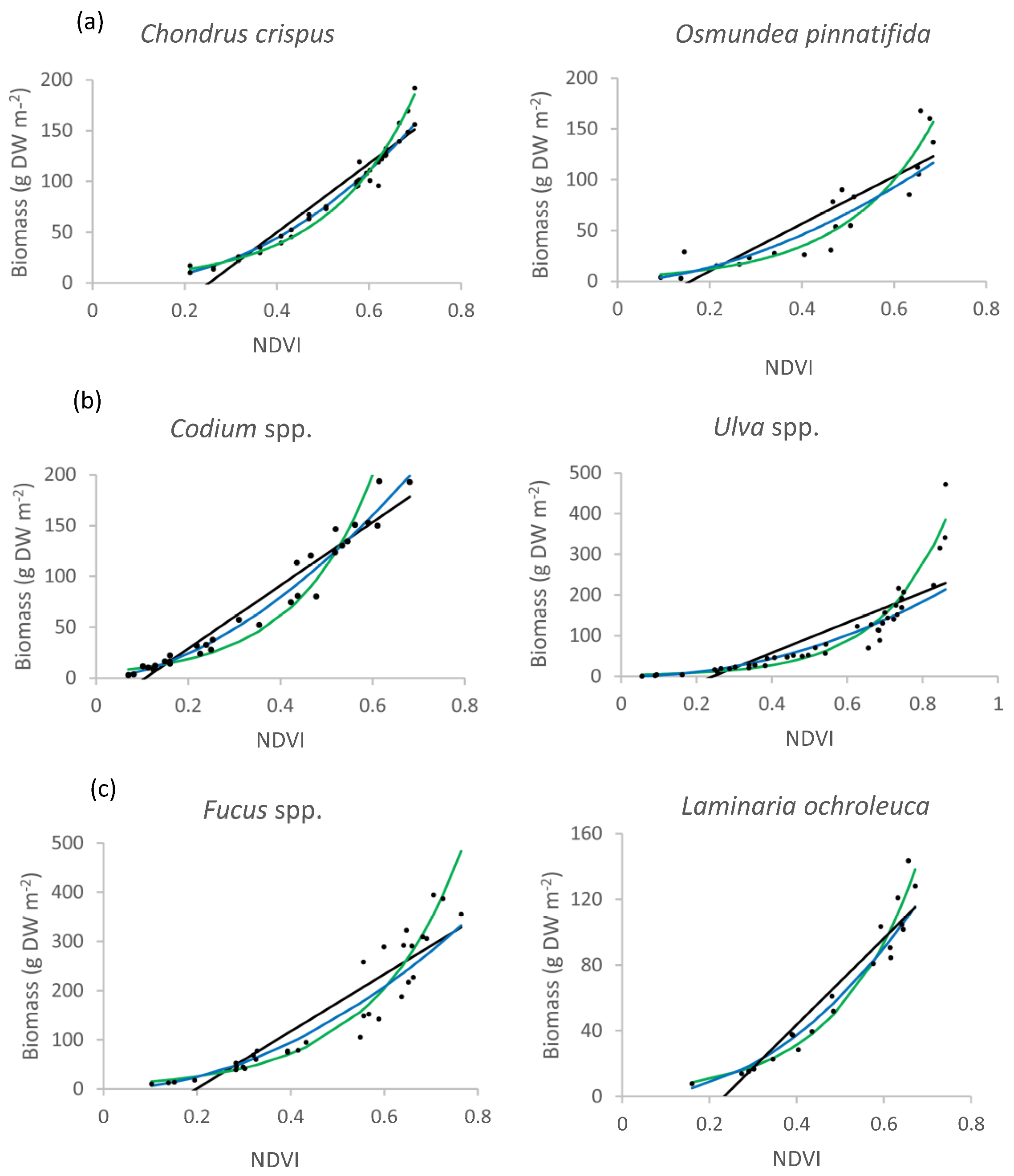

| Species | Model | Equation | R2 | RMSE |
|---|---|---|---|---|
| Chondrus crispus | Linear | y = 338.79 x − 85.673 | 0.89 | 17.16 |
| Exponential | y = 4.4908 e5.3261 x | 0.97 | 8.84 | |
| Power | y = 351.36 × 2.2640 | 0.95 | 12.88 | |
| Osmundea pinnatifida | Linear | y = 233.16 x − 36.700 | 0.78 | 23.22 |
| Exponential | y = 4.1340 e5.3085 x | 0.84 | 18.87 | |
| Power | y = 225.08 x1.7403 | 0.84 | 21.82 | |
| Codium spp. | Linear | y = 310.55 x − 33.123 | 0.95 | 14.07 |
| Exponential | y = 5.6390 e5.9438 x | 0.92 | 29.04 | |
| Power | y = 397.32 x1.7299 | 0.98 | 10.77 | |
| Ulva spp. | Linear | y = 370.68 x − 90.415 | 0.70 | 55.55 |
| Exponential | y = 2.7744 e5.7304 x | 0.93 | 28.14 | |
| Power | y = 291.15 x2.0691 | 0.95 | 51.60 | |
| Fucus spp. | Linear | y = 580.28 x − 114.880 | 0.84 | 48.76 |
| Exponential | y = 8.8671 e5.2320x | 0.95 | 41.75 | |
| Power | y = 560.91 x1.9453 | 0.95 | 43.86 | |
| Laminaria ochroleuca | Linear | y = 262.87 x − 61.477 | 0.90 | 13.26 |
| Exponential | y = 3.5349 e5.4575x | 0.97 | 9.56 | |
| Power | y = 275.52 x2.1824 | 0.96 | 10.80 |
| Percentage Cover (%) | Estimated Biomass (g DW 0.25 m−2) | Relative Error (%) |
|---|---|---|
| 26 | 73 | 20 |
| 35 | 99 | 15 |
| 51 | 144 | 10 |
| Thematic Map Classes | Reference Data Classes | Total | |||
|---|---|---|---|---|---|
| Mixed Algae | Mussels and Rock | Rock, Barnacles and Limpets | Sand | ||
| Mixed algae | 0.04 | 0.03 | 0.02 | 0.00 | 0.09 |
| Mussels and Rock | 0.03 | 0.04 | 0.04 | 0.00 | 0.11 |
| Rock, Barnacles and Limpets | 0.05 | 0.06 | 0.21 | 0.00 | 0.32 |
| Sand | 0.00 | 0.00 | 0.00 | 0.48 | 0.48 |
| Total | 0.12 | 0.13 | 0.26 | 0.48 | 1.00 |
| Classes | Omission Errors | Commission Errors | Precision | Recall | F1 Score |
|---|---|---|---|---|---|
| Mixed algae | 0.68 | 0.55 | 0.45 | 0.32 | 0.37 |
| Mussels and Rock | 0.67 | 0.61 | 0.39 | 0.33 | 0.35 |
| Rock, Barnacles and Limpets | 0.21 | 0.35 | 0.65 | 0.79 | 0.71 |
| Sand | 0.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| Thematic Map Classes | Reference Data Classes | Total | |||
|---|---|---|---|---|---|
| Ascophyllum nodosum | Fucus spp. | Sand | Rock | ||
| Ascophyllum nodosum | 0.04 | 0.03 | 0.02 | 0.00 | 0.09 |
| Fucus spp. | 0.03 | 0.04 | 0.04 | 0.00 | 0.11 |
| Sand | 0.05 | 0.06 | 0.21 | 0.00 | 0.32 |
| Rock | 0.00 | 0.00 | 0.00 | 0.48 | 0.48 |
| Total | 0.12 | 0.13 | 0.26 | 0.48 | 1.00 |
| Classes | Omission Errors | Commission Errors | Precision | Recall | F1 Score |
|---|---|---|---|---|---|
| Ascophyllum nodosum | 0.43 | 0.42 | 0.58 | 0.57 | 0.58 |
| Fucus spp. | 0.39 | 0.72 | 0.28 | 0.61 | 0.39 |
| Sand | 0.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| Rock | 0.16 | 0.00 | 1.00 | 0.84 | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borges, D.; Duarte, L.; Costa, I.; Bio, A.; Silva, J.; Sousa-Pinto, I.; Gonçalves, J.A. New Methodology for Intertidal Seaweed Biomass Estimation Using Multispectral Data Obtained with Unoccupied Aerial Vehicles. Remote Sens. 2023, 15, 3359. https://doi.org/10.3390/rs15133359
Borges D, Duarte L, Costa I, Bio A, Silva J, Sousa-Pinto I, Gonçalves JA. New Methodology for Intertidal Seaweed Biomass Estimation Using Multispectral Data Obtained with Unoccupied Aerial Vehicles. Remote Sensing. 2023; 15(13):3359. https://doi.org/10.3390/rs15133359
Chicago/Turabian StyleBorges, Débora, Lia Duarte, Isabel Costa, Ana Bio, Joelen Silva, Isabel Sousa-Pinto, and José Alberto Gonçalves. 2023. "New Methodology for Intertidal Seaweed Biomass Estimation Using Multispectral Data Obtained with Unoccupied Aerial Vehicles" Remote Sensing 15, no. 13: 3359. https://doi.org/10.3390/rs15133359
APA StyleBorges, D., Duarte, L., Costa, I., Bio, A., Silva, J., Sousa-Pinto, I., & Gonçalves, J. A. (2023). New Methodology for Intertidal Seaweed Biomass Estimation Using Multispectral Data Obtained with Unoccupied Aerial Vehicles. Remote Sensing, 15(13), 3359. https://doi.org/10.3390/rs15133359









