Abstract
With the increasing occurrence of cyanobacteria blooms, it is crucial to improve our ability to monitor impacted lakes accurately, efficiently, and safely. Cyanobacteria are naturally occurring in many waters globally. Some species can release neurotoxins which cause skin irritations, gastrointestinal illness, pet/livestock fatalities, and possibly additional complications after long-term exposure. Using a DJI M300 RTK Unmanned Aerial Vehicle equipped with a MicaSense 10-band dual camera system, six New Hampshire lakes were monitored from May to September 2022. Using the image spectral data coupled with in situ water quality data, a random forest classification algorithm was used to predict water quality categories. The analysis yielded very high overall classification accuracies for cyanobacteria cell (93%), chlorophyll-a (87%), and phycocyanin concentrations (92%). The 475 nm wavelength, normalized green-blue difference index—version 4 (NGBDI_4), and normalized green-red difference index—version 4 (NGRDI_4) indices were the most important features for these classifications. Logarithmic regressions illuminated relationships between single bands/indices with water quality data but did not perform as well as the classification algorithm approach. Ultimately, the UAS multispectral data collected in this study successfully classified cyanobacteria cell, chlorophyll-a, and phycocyanin concentrations in the studied NH lakes.
1. Introduction
Globally, waterbodies are changing rapidly due to human development and changes in climate. These anthropogenic effects impact the structure of freshwater communities as well as the physical and chemical characteristics that drive many ecosystem responses [1]. Environmental monitoring is necessary to understand these occurrences and track changes over time to understand full ecosystem processes and inter-ecosystem interactions such as the effects of nutrient cycling within a changing climate [2].
As a result of changes such as increasing air temperature and altered nutrient balances, cyanobacteria blooms are occurring on many lakes and ponds in New Hampshire (USA) each summer. Cyanobacteria were one of the first organisms on Earth to produce oxygen and are naturally occurring in many of the world’s waterbodies [3]. Although recently reclassified into the prokaryotic kingdom, Monera, cyanobacteria are classified as a bacterium despite their numerous algal characteristics [4]. Excess phosphorus entering the water column stimulates the growth and proliferation of cyanobacteria leading to eutrophic conditions while also triggering public health advisories, limiting recreational use, and reducing property values. Cyanobacteria harmful algal blooms (CHABs) are a subgroup of cyanobacteria blooms which release various forms of toxins that can cause skin irritations, nausea, vomiting, diarrhea, and fevers, as well as pet and livestock fatalities [5,6,7]. Existing and emerging studies have attempted to assess any possible connections with long term exposure and anthropogenic diseases [8,9]. Traditionally, scientists have studied cyanobacteria blooms in lakes and ponds with in situ field observations in addition to the analysis of water samples in laboratories. Monitoring this biological component of aquatic ecosystems is necessary to determine the internal processes at play and overall health of the local environment [10]. Understanding the occurrence, concentration, spatial patterns, and duration of CHABs will help communities and regulating agencies further understand CHABs from which to create effective management plans.
In the past decade, the integration of unmanned aerial systems (UAS), also known as drones, to study freshwater ecosystems has increased in scope and accuracy due to technological advances of lightweight UAS and the development of very high spatial and spectral resolution cameras. With the improved development of UAS including new and more effective sensors, longer flight periods, and improved processing software, scientists and researchers have begun using this tool to study aquatic systems. Spatial resolutions of UAS now include sub-meter pixels down to a few centimeters [11]. Multispectral sensors with high spectral resolution sense electromagnetic energy with narrow band widths [11,12]. UAS are more versatile than satellites with a minimum flying height of a few centimeters off the water’s surface to 121 m (as permissible by Part 107 Federal Air Administration (FAA) regulations) while maintaining their high spatial resolutions [13,14]. Not to be confused with the imagery captured by satellites, the use of UAS in environmental studies is an emerging low cost and user-friendly application that provides the ability for rapid and frequent deployment of specific study sites, low altitude flights below the cloud deck, modification capabilities, and easy navigation of small lakes or stream networks which are difficult or impossible to assess with traditional methods or current satellite technology. These advances in UAS technology are well suited for small waterbodies and inland waters such as many of the lakes in NH.
The use of a multispectral sensor, which captures a spectral response in designated portions of the electromagnetic spectrum, enhances our ability to discriminate objects of interest both on land and water [15]. Multispectral sensors often capture imagery yielding one value in each of the blue, green, red, red edge, and NIR portions of the spectrum. This allows for increased accuracy in the detection, identification, and quantification of certain aquatic ecosystem components, particularly blue-green and green algae [16]. The MicaSense RedEdge-MX and RedEdge-MX Blue Dual Camera Imaging System was used for this study (Seattle, WA, USA). This dual camera system captures electromagnetic energy, in the form of reflectance, centered around the following 10 wavelengths: 444 (coastal blue), 475 (blue), 531 (green), 560 (green), 650 (red), 668 (red), 705 (red edge), 717 (red edge), 740 (red edge), and 842 (NIR) nm. In this study, reflectance values were extracted from the UAS-collected imagery and used to calculate derivative bands or indices to find a relationship with collected water quality data (the reference data).
Many studies have found the use of green, red, red edge, and NIR wavelengths are best for studying phytoplankton, chlorophyll-a, and submerged aquatic vegetation [11,17,18,19]. In addition, the blue portion of the spectrum has also been incorporated to study cyanobacteria [20,21]. Many others have used derivative bands made by mathematically combining multiple bands such as a normalized difference vegetation index (NDVI) and other indices to determine chlorophyll-a, algal, and/or cyanobacteria measurements [13,21,22,23,24,25,26]. This approach is rapidly evolving and less often applied to UAS applications than to satellite applications [27]. There is limited research within New England using remote sensing methods to study cyanobacteria in small lakes; a region where cyanobacteria blooms are becoming an ever increasing public and ecological issue.
Therefore, the goal of this study was to investigate an alternate method for quantifying the cell concentration of cyanobacteria in New Hampshire waterbodies using UAS multispectral data over the course of the growing season to identify areas within the waterbodies that exceed thresholds set by agencies and/or this study. To do this, each flight was paired with collected water samples. The water samples were analyzed for cyanobacteria cell concentration, chlorophyll-a, phycocyanin, and phycoerythrin to calibrate the relationship and model between the in situ sample data and the UAS imagery. These water quality parameters were selected based on the current NH State cyanobacteria monitoring procedures (cell concentration) of the New Hampshire Department of Environmental Protection (NHDES), and standard monitoring practices through pigment analyses for cyanobacteria (chlorophyll-a, phycocyanin, and phycoerythrin) [10,11,13,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. The specific objectives of this study were:
- To build a dataset of both UAS reflectance data and water quality data for cyanobacteria in NH lakes.
- To target cyanobacteria blooms from multiple lakes in New Hampshire with different dominant species of cyanobacteria over the course of the bloom’s life cycle.
- To determine the best spectral relationship for classification between very high spatial resolution multispectral UAS imagery and cyanobacteria for the sampled lakes.
2. Materials and Methods
2.1. Site Selection and Descriptions
Candidate lakes in New Hampshire were identified for this study through a series of criteria using a scoring system developed for this analysis. This scoring system consisted of data on previous bloom duration, concentrations, and dominant cyanobacteria species, lake size, public accessibility, and distance from water quality and image analysis laboratories. The candidate lakes were then located on the FAA Sectional Aeronautical Chart and removed if in a restricted airspace. Stakeholders for the remaining lakes were contacted to determine their willingness to collaborate. Silver Lake (Hollis), Keyser Pond (Henniker), and French Pond (Henniker) were then selected as the three primary lakes to include in this study. These three lakes were sampled roughly every other week from the first week in May to the first week of September 2022. Greenwood Pond (Kingston), Showell Pond (Sandown), and Tucker Pond (Salisbury) were added to the study once a cyanobacteria bloom had begun on the respective waterbody to add to the dataset during August and September of 2022 (Figure 1).
The six selected lakes span a variety of lake trophic classifications including lakes designated as oligotrophic, mesotrophic, and eutrophic (Table 1). Identified based on ambient water quality by the State of New Hampshire, oligotrophic lakes are those with low nutrient concentrations and high water clarity. Eutrophic lakes contain high levels of nutrients and degraded water clarity Mesotrophic lakes contain higher levels of nutrients than oligotrophic lakes but not as high as eutrophic lakes, and moderate levels of water clarity degradation [36].

Table 1.
Summary of New Hampshire lakes flown with the UAS and sampled for water quality measurements in 2022.
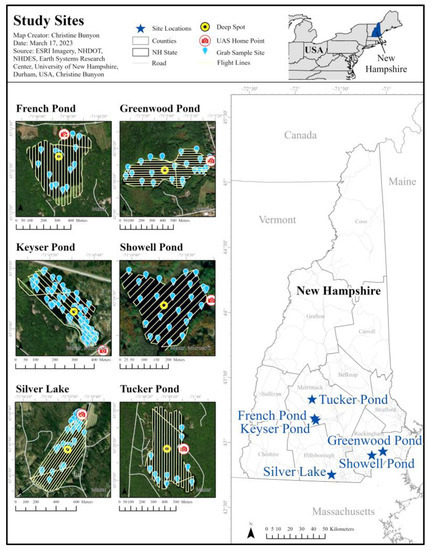
Figure 1.
Study site locations within the state of New Hampshire.
2.1.1. French Pond
French Pond spans 16 hectares in Henniker, NH, is classified as a eutrophic lake, and has a maximum depth of 12 m (Table 1) [37]. There is a state-owned boat launch along the northern shore of the lake and one campground along the eastern shore. The rest of the shoreline is primarily forested and scattered with shoreline properties within a historically agricultural watershed. French Pond has had fewer cyanobacteria advisories than Silver Lake and Keyser Pond with just two from 2017 through 2021 which lasted 22 and 60 days, respectively. However, French Pond had many advisories from 2006 through 2017 (eight in total). Species of cyanobacteria present have included Aphanizomenon, Dolichospermum, Microcystis, and Woronichinia. French Pond did not experience any blooms of cyanobacteria in 2022 (Table 1).
2.1.2. Greenwood Pond
Greenwood Pond covers 20 hectares in Kingston, NH and has a maximum depth of 4 m (Table 1) [37]. The water was accessed from the town beach on the eastern shores of the lake. Classified as a mesotrophic lake, Greenwood Pond has experienced cyanobacteria blooms since 2004 which have become more frequent since 2017. There have been four separate NHDES advisories for CHABs from 2017 through 2021. Each has lasted between 4 and 34 days (average of 14 days). Species of cyanobacteria present have included Planktothrix, Oscillatoria, and Dolichospermum. In 2022, Greenwood Pond experienced a short bloom of cyanobacteria, with the advisory lasting only 8 days from the end of July into August (Table 1). The bloom was reported to be of scattered surface accumulations of Planktothrix.
2.1.3. Keyser Pond
Keyser Pond covers 7 hectares in Henniker, NH and has a maximum depth of 5 m (Table 1) [37]. Unlike Silver Lake, there is no state beach area, but a state operated boat launch is present along with a privately owned campground and cottage rentals which attract seasonal visitors. Keyser Pond is classified as a mesotrophic lake. Within the past five years, 2017 through 2021, Keyser Pond has had four advisories which have lasted between 22 and 112 days, with an average of 65 days. The first listed advisory was posted in 2007, but the second was not until 2015. Since then, cyanobacteria blooms at Keyser Pond have followed an increasing trend in duration. Species of cyanobacteria present have included Chrysosporum, Dolichospermum, Oscillatoria, Planktothrix, and Spirulina. In 2022, Keyser Pond experienced a full lake bloom primarily of Chrysosporum for 34 days from July into August (Table 1). This bloom drastically decreased the depth of secchi disk transparency measurements and consisted of some Planktothrix and Dolichospermum.
2.1.4. Showell Pond
Showell pond spans 8 hectares in Sandown, NH and has a maximum depth over 3 m (Table 1) [27] With permission, sampling was conducted from the lawn of a private residence along the eastern shores of the lake at the end of Showell Pond Lane. Classified as a mesotrophic lake, Showell Pond has recorded consistent blooms of cyanobacteria dating back to 2006 [38]. There were only two NHDES advisories from CHABs from 2017 through 2021. The advisories lasted 76 days in 2018 and 90 days in 2019 (an average of 83 days). Species of cyanobacteria present have included Dolichospermum, Coelosphaerium, Oscillatoria, and Aphanizomenon. In 2022, Showell Pond experienced a bloom similar to Keyser Pond. Lasting for 56 days, the dominant species were Chrysosporum, Planktothrix, and Spirulina, and turned the entire lake a greenish-brown color (Table 1).
2.1.5. Silver Lake
Silver Lake spans 16 hectares in Hollis, NH and has a maximum depth of 7 m (Table 1) [37]. The Silver Lake State Park beach area is located along the northern shores of the lake. The wind predominantly blows towards the State Park beach, bringing with it accumulations of substances on the surface of the lake. Classified as an oligotrophic lake, Silver Lake has experienced cyanobacteria blooms since 1991 [39]. Silver Lake has had a total of nine separate NHDES advisories for CHABs from 2017 through 2021. Each has lasted between 5 and 89 days (average of 37 days). Species of cyanobacteria present have included Microcystis and Dolichospermum. Woronichinia were also present in 2021 and 2022. In 2022, Silver Lake experienced two separate advisories, one from June into July, and the second from August into November. In total, these advisories were active for 140 days (Table 1). The 2022 blooms were dominated by species of Microcystis and Dolichospermum with some presence of Woronichinia. Surface scum appeared along the state park beach and swimming area.
2.1.6. Tucker Pond
Tucker Pond covers 22 hectares in Salisbury, NH and has a maximum depth of 6 m (Table 1). With permission, sampling was conducted from the dock of a private residence along the eastern shoreline on Sixth Road. Classified as a mesotrophic lake, Tucker Pond has had only three listed NHDES advisories from CHABs, all of which have occurred since 2019 [40]. These advisories lasted between 14 and 132 days (average of 68 days). Woronichinia is the dominant species present. In 2022, Tucker Pond experienced a cyanobacteria bloom lasting 99 days from August through November. Consisting of Worochinia and Microcystis, this bloom appeared as surface accumulations along the south and southwestern shorelines.
2.2. Workflow
Data analysis for this project followed two separate workflows which converge as seen in Figure 2. On each sampling day, UAS imagery was collected prior to the collection of water samples. UAS imagery was processed and spectral indices were developed and applied to the imagery. Water samples were analyzed for cyanobacteria speciation and cell count, phycocyanin and phycoerythrin concentrations, and chlorophyll-a concentration. The workflow then converges for the statistical analyses of classification and regressions to link the water quality data to the UAS imagery. The results of these analyses were evaluated for accuracy and adjusted as needed.
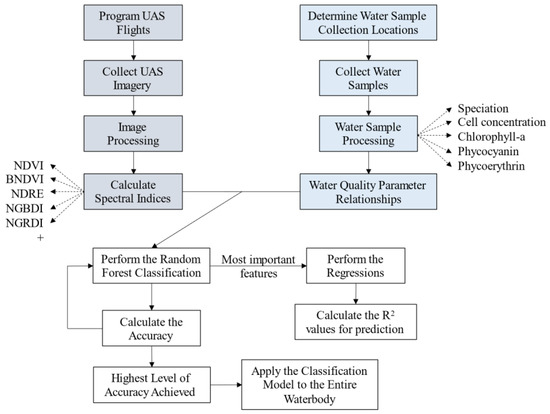
Figure 2.
Flow chart of methodology. Gray boxes are part of the UAS methodology, blue boxes are part of the water quality methodology, and white boxes are part of the data analysis.
2.3. UAS Methodology
A FAA Part 107 licensed remote pilot in command operated the UAS with at least one visual observer present each time. The necessary permit was acquired to fly the UAS from Silver Lake State Park. A DJI Matrice 300 RTK (Nanshan, Shenzhen, China) was used as the primary aerial vehicle with the MicaSense RedEdge-MX and RedEdge-MX Blue Dual Camera Imaging System (Seattle, WA, USA) (Figure 3). This dual sensor consists of two cameras which function simultaneously to capture a total of 10 bands (i.e., wavelengths) of electromagnetic energy (Table 2). The UAS flight paths were preprogrammed using the DJI Pilot software on the enterprise smart controller for the Matrice 300 RTK. Because the sensors were not in communication with the aircraft, a smart device was connected to the Wi-Fi of the sensors to set the trigger speed. Initially, each flight was flown at 100 m above ground level, with 80% forward and side overlap of images along and between flight lines. Flight lines are flown parallel to each other over the entire area of study as shown in Figure 1. At a speed of 10 m/s, the sensors captured images approximately every 1.31 s. On and following 29 June, all flights were flown at 120 m above ground level with 80% forward and side overlap, and at a speed of 10 m/s. This adjustment was implemented to aid with post-processing difficulties in the imagery from flying over water. This processing was conducted using Agisoft Metashape (St. Petersburg, Russia). Each pixel covered roughly 9 cm2 of ground area for flights flown 100 m above ground level, and 11 cm2 of ground area for flights flown at 120 m above ground level. Sky conditions varied throughout the season for the flights and included overcast and bright days. Therefore, all images were corrected for solar illumination using the downwelling light sensor 2.0 at the time of capture to ensure all image dates were directly comparable. Wind speed below 8 m/s was required to reduce the risk of flying an unstable aircraft. Total flight times varied depending on the lake size but were between 12 and 27 min, all of which required only a single set of batteries per flight.
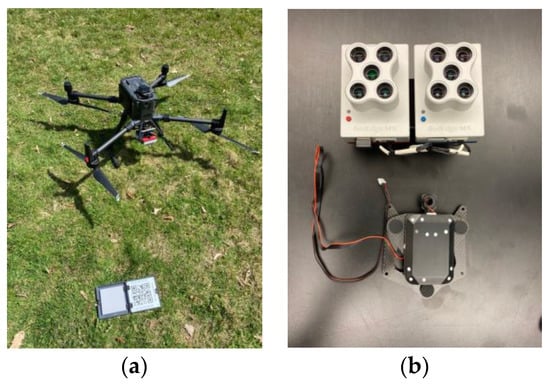
Figure 3.
DJI Matrice 300 RTK with calibration plate (a) and MicaSense RedEdge-MX Dual Camera Imaging System with downwelling light sensor 2.0 (b).

Table 2.
Wavelengths (nm) captured by the MicaSense RedEdge-MX Dual Camera Imaging System with centers and band widths (nm) identified in parenthesis.
2.3.1. Image Processing
The objective of image processing was to extract average pixel values from each band captured by the sensor for a specified area surrounding each water sampling point. Three software packages were used: Agisoft Metashape, eCognition (Munich, Germany), and ArcGIS Pro (Redlands, CA, USA). Images were loaded into Agisoft Metashape to create an orthomosaic by stitching the individual images into a single scene (i.e., a mosaic). In Agisoft Metashape, all the images were calibrated using an image of the reflectance panel captured on the day of sampling to enable direct comparison between image dates. A batch processing workflow was then conducted to (1) align the images, (2) build a dense cloud, (3) build a digital elevation model, and (4) construct the orthomosaic. The final orthomosaic was given the WGS84 geographic and UTM 19N projected coordinate systems and exported as a tiff file. Meanwhile, the GPS point of each water quality sampling location was collected using the Fulcrum® (San Francisco, CA, USA) data collection mobile application and input into ArcGIS Pro 3.0 with the same coordinate systems. A 2 m2 buffer was created around each point which was roughly equivalent to half the canoe’s length squared (approximately 600 pixels). Once a day of sampling had both an orthomosaic tiff file and a sampling location buffer shapefile, eCognition was used to extract average pixel values per band from within the buffer area for each sampling location on the imagery. A ruleset was created for the vector-based segmentation and applied using the buffer areas at the pixel level. The vector-based segmentation overlays the buffered polygon data layer on the orthomosaic. It then identifies each pixel of the orthomosaic which falls within or touches each buffered area. The data from these pixels were then extracted. Attributes for each 2 m2 sampling area included brightness, mean, and standard deviation for each of the ten bands. These attributes were then exported as a new shapefile. This new shapefile, containing only the 2 m2 sampling areas and image attribute data, was then loaded back into ArcGIS Pro 3.0 to read and export the attribute data into Microsoft Excel. This process was repeated 21 times, once for each sampling day.
2.3.2. Calculating Derivative Bands
For aquatic studies of cyanobacteria, chlorophyll-a, and/or submerged aquatic vegetation, scientists have used the NDVI, normalized difference red edge index (NDRE), BNDVI, coastal blue normalized difference vegetation index (cBNDVI), normalized green blue difference index (NGBDI), normalized green red difference index (NGRDI) (aka NDGI), surface algal bloom index (SABI), florescence line height—blue (FLH Blue), SHI index (named after the author Kun Shi), and cyanobacteria index (CI) derivative bands (Table 3). The CI identifies the presence and concentration of chlorophyll-a (when centered around 681 nm) through the spectral reflectance of the sample (Equation (1)) [13,25]. When using alternative pigments to identify the CI, such as phycocyanin, the spectral shape is centered around 655 nm [25,26]. Due to slightly different bands captured in this study compared to others which used the CI, two versions of the CI were used, both of which were slightly different than in the previously referenced studies.
For this study, 78 versions of 10 derivative bands were created (Table 3, Appendix C). These derivative bands were selected based on those used in existing aquatic-UAS studies and algebraically calculated from the extracted band averages per water sample site using Microsoft Excel. A complete list of indices used can be found in Appendix C.

Table 3.
Derivative bands used and/or created. See Appendix C for complete list of derivative bands. * See Equation (1).
Table 3.
Derivative bands used and/or created. See Appendix C for complete list of derivative bands. * See Equation (1).
| Derivative Band | Description | General Equation | Source |
|---|---|---|---|
| NDVI | Normalized difference vegetation index | (NIR − Red)/(NIR + Red) | [21,22,24,41] |
| NDRE | Normalized difference red edge | (NIR − RedEdge)/(NIR + RedEdge) | [23,42] |
| BNDVI | Blue normalized difference vegetation index | (NIR − Blue)/(NIR + Blue) | [21] |
| cBNDVI | Coastal blue normalized difference vegetation index | (NIR − Coastal Blue)/(NIR + Coastal Blue) | [21] |
| NGBDI | Normalized green blue difference index | (Green − Blue)/(Green + Blue) | [43] |
| NGRDI aka NDGI | Normalized difference glacier index | (Green − Red)/(Green + Red) | [24] |
| SABI | Surface algal bloom index | (NIR − Red)/(Blue + Green) | [23,44] |
| FLH Blue | Fluorescence line height | Green − (Red + (Blue − Red)) | [23,45] |
| SHI | The author’s last name is Shi | (eRed − eNIR)/(eRed + eNIR) | [23,46] |
| CI * | Cyanobacteria Index | SS = (λ − λ−)/(λ+ − λ−) | [25,26] |
Equation (1): Formula for calculating the cyanobacteria index centered for phycocyanin. SS represents the spectral shape, λ represents 655 nm, λ+ represents the adjacent bands longer than 655 (for example, 681 nm), and λ− represents the adjacent bands shorter than 655 nm (for example, 620 nm). Cyanobacteria is present when the SS is greater than 0. Equation modified from Mishra et al. (2019) and Sharp et al. (2021) who both needed to compensate for atmospheric reflectance from satellite imagery [25,26].
SS = (λ − λ−)/(λ+ − λ−)
2.4. Water Quality Sample Methodology
2.4.1. Water Sample Collection
Water sample collection immediately followed each UAS flight with as little delay as possible (beginning within minutes). Sampling sites within each waterbody were selected using a random stratified sampling approach. If bloom conditions upon arriving at a waterbody were likely present from a visual standpoint, or known according to the NHDES Cyano Mapper, sampling locations were selected to target areas which appeared to have high concentrations, while spacing them out throughout the lake.
After arriving at a sampling site, the canoe was anchored to prevent drift. Per common lake water quality monitoring practices, a reading of surface water temperature and dissolved oxygen (percent and concentration) was collected using a YSI ProODO (Yellow Springs, OH, USA) which was routinely calibrated each sampling day. If the site was also the deep spot of the waterbody, a full temperature and dissolved oxygen profile was recorded. A water clarity measurement was then collected using a secchi disk and view scope off the shady side of the canoe. Notes on date, time, sky condition, and sample depth were also entered into an electronic data sheet on the Fulcrum® mobile application. These data were collected following standard practices but were determined not to be needed as part of the final data analysis in this study. The GPS location along with photos were collected at each site. A two-meter integrated core sample was collected of lake water at each site (if the site was >3 m deep and Secchi Disk Transparency (SDT) > 2 m deep) and mixed before pouring into the sample bottles for laboratory analyses. However, this case was not common. If the site was <3 m deep or SDT < 2 m, a subsurface grab was conducted with a mixing bucket which was then mixed and poured into the sample bottles.
Five of the six lakes included in this study experienced cyanobacteria blooms in 2022 according to the NHDES Harmful Algal Bloom Monitoring Program ranging from 8 to 140 days in total (Table 1). Silver Lake was the only one to experience two bloom advisories. In total, 7, 7, and 4 UAS flights occurred, with 45, 63, and 16 water samples collected from Silver Lake, Keyser Pond, and French Pond, respectively. One UAS flight occurred and 25, 26, and 12 water quality samples were collected at Greenwood Pond, Showell Pond, and Tucker Pond, respectively. Twenty-two water quality samples were not used for the water quality to UAS spectral data analysis due to poor reflectance data (Table 4).

Table 4.
Dates of sampling in 2022 with the total number of samples collected per lake.
2.4.2. Cyanobacteria Speciation and Cell Counts
A compound light microscope and a gridded Sedgewick–Rafter slide were used for the microscopy analysis of cyanobacteria speciation and cell counts. Samples were refrigerated and analyzed within 48 h of collection. The sample was inverted three times before 1 mL was extracted and pipetted on the slide. Results per sample included the total number of cells per genus, and the total number of cells per milliliter of sample.
2.4.3. Fluorometry
Analysis of phycocyanin and phycoerythrin was conducted per guidelines set by the UNH Center for Freshwater Biology using a FluoroQuik (Amiscience Corporation, Fremont, CA, USA) (Appendix A). Prior to reading samples, the FluoroQuik was blanked with deionized water, standards were measured, and pigment concentrations were recorded. Approximately 3 mg/L of sample was added to the cuvette and placed inside the FluoroQuik. Each sample was run in triplicate to calculate a sample mean for both phycocyanin and phycoerythrin relative concentrations in µg/L.
2.4.4. Chlorophyll-a Extraction
Analysis of chlorophyll-a concentration was conducted per guidelines set forth by the UNH Water Quality Analysis Laboratory using a hot ethanol extraction and spectrophotometric analysis. According to the laboratory standard operating procedure (SOP), “this method has the benefit of extraction without grinding, avoiding toxic methanol or acetone exposure” (Appendix B). After the hot ethanol bath preparation the day prior and settling overnight, the absorbance of each sample was measured at 665 nm and 750 nm before and after the addition of 0.25% HCl. Using these values along with the total volume of the sample that was able to be filtered, total chlorophyll-a concentration in µg/L was calculated.
2.5. Statistical Analysis
Field data entry was conducted using the Fulcrum® mobile application and then input into Microsoft Excel. Statistical analyses for water quality parameter simple linear regressions were performed using R (Indianapolis, IN, USA). This step was important to identify which water quality parameters had significant relationships to cell concentration, the parameter used by the NHDES for issuing cyanobacteria advisories. The water quality parameters with significant linear relationships in our dataset were used for the random forest classification algorithm to identify areas of the waterbody that are above or below thresholds set by agencies and/or this study.
Classification of the UAS imagery and indices for each water quality parameter was performed using the random forest classification algorithm in Python (Python Software Foundation, Wilmington, DE, USA). This algorithm uses a supervised machine learning process with two parts. First, the user inputs training areas of known data to teach the algorithm. Then, based on these training areas, the computer algorithm classifies the remaining data. The accuracy of the classification is then determined from the water samples set aside independent of the training data. The parameters within the random forest Python script yielded 500 “trees” and were repeated 20 times to produce an average overall classification accuracy.
Each viable water quality parameter, cell concentration, chlorophyll-a, and phycocyanin, were simplified (i.e., recoded) into classes. For recreational waters, the NHDES currently issues cyanobacteria advisories when cell concentrations exceed 70,000 cells/mL or if over 50% of the cells within an algal bloom are cyanobacteria. Cell concentration classes followed the State of New Hampshire 70,000 cells/mL threshold, with “Low” being samples less than the threshold, and “High” being samples equal to or greater than the threshold. Classes for chlorophyll-a were selected based on the NHDES designated chlorophyll-a concentrations for trophic classes and the World Health Organization (WHO) chlorophyll-a thresholds for potential exposure to cyanotoxins [36,47]. Mesotrophic and oligotrophic lakes are defined by having chlorophyll-a concentrations less than 11 μg/L (in addition to other factors), while eutrophic and nuisance statuses are applied to lakes with greater than 11 μg/L of chlorophyll-a [36]. Chlorophyll-a concentrations of 10 μg/L or less indicate low to not low risk of exposure, while concentrations at or above 10 μg/L indicate moderate to high risk of exposure to algal toxins [37]. Therefore, chlorophyll-a concentrations less than 10 μg/L were identified as “Low”, and concentrations greater than or equal to 10 μg/L were identified as “High” for this study. Lastly, classes for phycocyanin were created following guidelines set in Almuhtaram et al., 2018. Because a significant correlation was found between phycocyanin and cell concentration, the equation of the line of best fit (y = 0.0005x + 19.822) was used with 70,000 input as the x value to determine the threshold for “High” and “Low” phycocyanin concentrations as 54.822 μg/L. Each water quality parameter was divided into only two classes to maintain the necessary 60 samples per class for accurate classification using an error matrix. An error matrix is the standard methodology for recording thematic accuracy in remote sensing [48].
For the random forest classification analysis, the reference data (water quality samples) and the UAS data were divided randomly into 50% for training the algorithm and 50% for validation or assessing the accuracy of the result. In other words, the algorithm is fed the entire dataset of paired water sample results (Highs or Lows for one water quality parameter at a time), with the UAS reflectance and indices (UAS data) per sample as the input to the model. The random forest classification algorithm then randomly subsets 50% of the data to train the model. It learns, based on the UAS data, the resulting water quality classification is either High or Low for each sample in the training subset. The remaining 50% of the dataset is then used for validation. The algorithm then takes the input UAS data and assigns water quality classifications based on what it learned with the training subset (to produce the output of the model). The accuracy of the model (i.e., properly assigning samples as either High or Low compared to the known water quality results) is determined using an error matrix [49].
Other variations of training/validation (45% and 55%) were also selected by adjusting the validation size in the python script from 0.5 to 0.45 then 0.55 but produced marginally less accurate results. The alternate validation sizes were tested per standard best practices when conducting the random forest classification algorithm to produce the highest possible accuracies [50]. Additionally, samples with poor reflectance data were initially left in but then removed. Poor reflectance samples were determined if a sample site’s 2 m buffer contained pixels affected by orthomosaic holes, sun glint, emergent vegetation, or shoreline. These samples were identified and removed when the band standard deviation for pixels within the 2 m area was over 100 DN. Lastly, the random forest algorithm was run with all 78 UAS parameters included, with the 10% least important features removed, and with only the top 10% most important features included. The condition that produced the highest overall accuracies for each water quality parameter were when the sample set was divided into 50% training and 50% validation, with poor reflectance samples removed, and with only using the top 10% most important features. The random forest algorithm returned high overall accuracies with a smaller standard deviation for each water quality parameter. Accuracy results are presented as error matrices from which overall, user’s, and producer’s accuracies were calculated [49]. Simple linear and logarithmic regressions for the most important UAS bands and indices (found through the random forest algorithm) to each water quality parameter were conducted in Microsoft Excel. Determining which features were most important was based on the average feature importance score produced through the random forest classification and computing the average of the 20 iterations per water quality parameter. This analysis indicates which UAS features contribute most to the classification of a sample being “predicted” from those used to “train” the model. Simple regressions were calculated simply because this is the approach many similar studies apply [10,21,22,23,24,26,31,32], but produced less desirable results than from the random forest classification algorithm.
Lastly, to visually illustrate the random forest classification algorithm, the model was applied to every pixel of a waterbody to classify the water quality throughout the lake. UAS data from every pixel over a lake, one day at a time, were extracted (reflectance values per band) or calculated (indices) for the top 10% most important features identified from the random forest classification algorithm (a total of 8 features). The lake pixels were then classified as either “High” or “Low” for each of the three water quality parameters separately.
3. Results
3.1. Water Quality Parameter Relationships
Table 5 shows the correlations between the four water quality parameters. A significant linear relationship exists between many of the parameters but is not consistent across all lakes. Overall, there is an emerging trend between cell concentration and phycocyanin, cell concentration and chlorophyll-a, and phycocyanin and chlorophyll-a. Although significant results were found between cell concentration, chlorophyll-a, and phycocyanin, phycoerythrin did not follow suit except for Silver Lake (Table 5). Because there were only four water samples collected at Silver Lake exceeding the state threshold (cell concentration), these results were not deemed suitable to use phycoerythrin as a surrogate measurement for cell concentration.

Table 5.
Simple linear regression outputs between water quality parameters (R2 values). Bold, italicized, and blue values indicate significant results (p < 0.01). Cells = cell concentration (cells/mL), Chl-a = chlorophyll-a concentration (μg/L), PC = phycocyanin concentration (μg/L), PE = phycoerythrin (μg/L). Datapoints for cell concentrations over 1 million (n = 3) were removed for this analysis. For example, Cells/PC is the simple linear regression between cell concentrations and phycocyanin concentrations.
3.2. Random Forest Classification Algorithm
Building a relationship between data collected by the UAS and the water quality data was conducted using the random forest (RF) classification algorithm. The results for each water quality parameter are shown in Table 6. The average accuracies for the three water quality parameters were 92.9%, 87.4%, and 91.7% with standard deviations of 3% which shows the random forest classification algorithm produced both accurate and precise results to determine if samples were above or below state thresholds (properly classified within the High or Low classes).

Table 6.
Overall classification results and standard deviations from the random forest (RF) classification algorithm for each water quality parameter. Results were generated using the top 10% most important features identified from the RF algorithm, with samples that had poor/obstructed reflectance values removed. Average overall accuracies were calculated from 20 repetitions.
3.3. Simple Regressions
The R2 value from a simple regression explains how much variation in the water quality parameter value can be explained by the UAS parameter. Higher R2 values were generated from logarithmic regressions than linear and are shown in Table 7. The UAS parameter legend for index equations can be found in Appendix C. Table 7 shows the results from the simple logarithmic regressions for the top 10% of the most important features (UAS bands and indices) of the 78 features originally used for each classification to the selected water quality parameters.

Table 7.
Simple logarithmic regression results for the top 10% most important features of the UAS parameters from the random forest classification algorithm for each water quality parameter. * Indicates p < 0.01, ** indicates p < 0.001. The scatter plots for the highest three R2 values per water quality parameter are provided in Appendix D.
3.4. Application of Results to Keyser Pond
The most complete dataset portraying before, during, and after conditions of a cyanobacteria bloom in 2022 was for Keyser Pond. Here, 15 samples were collected from the 12 May 2022 through 15 June 2022 prior to the onset of the bloom and subsequent advisory. Once the cyanobacteria bloom had established, 36 samples were collected from 7 July 2022 through 27 July 2022. Twelve more samples were collected on 16 August 2022 after the bloom had subsided. The cyanobacteria bloom caused the water to turn a greenish brown color for the duration of the bloom. Figure 4 illustrates this trend through the water quality data over the course of the field season. Ideally, this pattern was desired for each lake for this study to capture UAS data from blooms with varying concentrations, but only occurred at Keyser Pond.
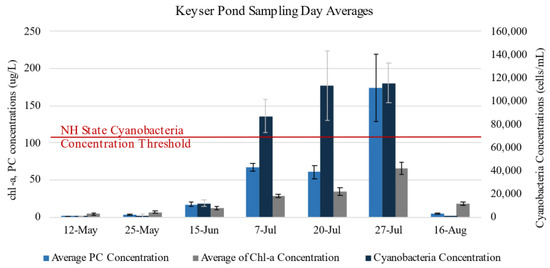
Figure 4.
Sampling day averages for cyanobacteria concentration (cells/mL), chlorophyll-a (chl-a) concentration (μg/L), and phycocyanin (PC) concentration (μg/L) for Keyser Pond over the course of the sampling season. Samples collected from floating clumps detached from the benthic mat bloom were not included here (n = 2, cell concentrations were over 1 and 4 million cells/mL).
A visual application of the random forest classification algorithm applied to Keyser Pond before, during, and after the 2022 cyanobacteria advisory is provided in Figure 5 (15 June through 16 August). Each pixel is classified as either “High” (above the NHDES threshold for cyanobacteria, ≥70,000 cells/mL) or “Low” (below the NHDES threshold for cyanobacteria < 70,000 cells/mL). Technical difficulties were experienced with the UAS’ internal GPS on 20 July which resulted in an incomplete orthomosaic as the flight was terminated prematurely for safety. Shadows from trees along the north-eastern shore also affected the model’s results and can be seen on 20 and 27 July.
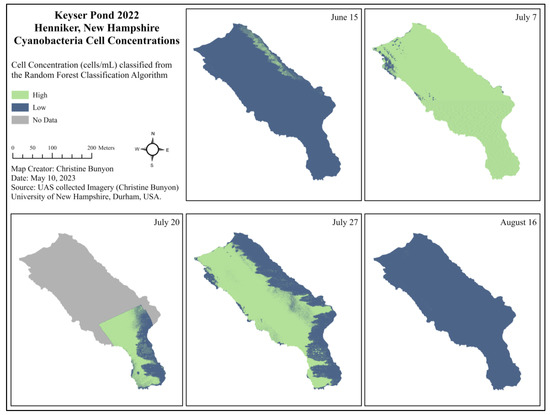
Figure 5.
Cell concentrations (cells/mL) per pixel classified from the random forest classification algorithm as High or Low to represent areas of Keyser Pond exceeding the NHDES threshold for cyanobacteria (70,000 cells/mL) before (15 June), during (7 July, 20 July, and 27 July), and after (16 August) the 2022 NHDES cyanobacteria bloom advisory.
4. Discussion
Cyanobacteria blooms have been a growing concern for many lake stakeholders in New England, particularly in New Hampshire. Monitoring cyanobacteria blooms has become an increased need in many lake communities. Contrary to traditional water quality sampling, monitoring cyanobacteria blooms using a UAS allows the user to assess the entire waterbody, reduces sample analysis and processing times, and increases sampler safety. The use of a very high spatial resolution multispectral camera flown on a DJI Matrice 300 RTK was investigated to capture reflectance values centered around ten different wavelengths of light of lakes known to experience cyanobacteria blooms.
4.1. Explanation and Interpretation of Results
Through building paired datasets of both water quality and UAS spectral data, multivariate classification and regression analyses were conducted. Most importantly, discerning if a sample was above or below the New Hampshire state threshold for cyanobacteria cell concentration yielded an overall accuracy of 93%, a chlorophyll-a concentration above or below 10 μg/L had an overall accuracy of 87%, and a phycocyanin concentration above or below 54.8% was 92% accurate overall. Therefore, this process could help lake stakeholders make informed management decisions regarding closures of certain use areas of the waterbody throughout the bloom season. Looking at the random forest algorithm results, a potential explanation for the lower overall accuracy for chlorophyll-a could possibly be attributed to the method in which chlorophyll-a was extracted. During this laboratory procedure, water samples were filtered through a 47 mm filter with pore sizes of 0.7 µm (Appendix B). This was the only analysis to include a filtration step. Although unlikely, it is possible that some dissolved chlorophyll-a, less than 0.7 µm, was able to flow through the filter rather than be trapped by it, thus not being measured in the hot ethanol and fluorescence portion of the analysis. It is also possible that chlorophyll-a was captured on filters from organisms other than cyanobacteria, including green algae or plant cells as chlorophyll-a is not exclusive to cyanobacteria.
Multivariate regressions proved difficult with this dataset due to both structural multicollinearity and data multicollinearity in the spectral data. Simple regression was the preferred method for similar studies. Variation in cell concentrations, chlorophyll-a concentrations, and phycocyanin concentrations can be poorly explained by individual spectral features (Table 7). Simple regressions were calculated for the top ten percent of the most important features (determined from the average feature important scores from the random forest algorithm). As shown in Table 7, the reflectance data from the Blue 475 wavelength, NGBDI_4, and NGRDI_4 indices were the three most important features for classifying cell concentrations into “High” and “Low” categories based on the UAS or spectral data. The Blue 475 band is part of the NGBDI_4 equation, which also contains the green 560 band (Appendix C). The Green 560 band is also found in the NGRDI_4 equation along with the red 668 band. However, the Green 560 band alone was not found to be within the top 10% of the most important features. The NGRDI_4, Green 531, and FLHblue_2 features were the most important features for classifying chlorophyll-a concentrations into high and low categories based on the spectral data. The FLHblue_2 equation uses the Blue 444, Green 531, and Red 650, 668 bands. Lastly, the Blue 475, NGBDI_4, and CI_2 features were most important for classifying phycocyanin concentrations into high and low categories based on spectral data. The CI_2 contains data from the red 650 and 668 bands, in addition to the red edge at 705 nm. Four of the total seventy-eight features were found to be in the top 10% most important features for all three water quality parameters: Blue 475, NGRDI_4, CI_2, and Green_531 (Table 7).
Identifying which spectral features were most important for studying cyanobacteria concentrations with surrogate water quality parameters can guide emerging studies. Based on these findings, future studies should use sensors capable of capturing imagery from or near 444 nm, 475 nm, 531 nm, 560 nm, 650 nm, 668 nm, and 705 nm; and especially, those in bold. The MicaSense RedEdge-MX Dual Camera Imaging System senses wavelengths comparable to the Landsat 8 and Sentinel 2 satellites [51,52]. The important similarities of the MicaSense sensor to the Sentinel 2 (S2) satellite include the 475 band (490 S2), 560 band (560 S2), and 668 (664 S2).
The collection and processing of the water quality samples took over 4.5 times longer than the collection and processing of the UAS data. The three most time-consuming components were determining cell and chlorophyll-a concentrations, and physically collecting each water quality sample via canoe. Other studies have evaluated the applicability and reliability of using phycocyanin as indicators for cyanobacteria rather than relying on the time intensive cell counting for cell concentration [10,29,30,53,54]. This study showed significant relationships between cell and phycocyanin concentrations at French Pond, Keyser Pond, Silver Lake, Tucker Pond, and for the entire dataset. The ability to use fluorometry to measure phycocyanin rather than the time intensive method of counting cells to determine cell concentrations or filtering and analyzing samples to measure chlorophyll-a concentrations would drastically improve the speed at which analyses could be made and results shared with communities. In the time it took to collect all the water samples alone, the entire UAS methodology could have been conducted and completed (Figure 6). This time comparison does not include tasks shared by both processes which include time traveled to each lake, communication with lake residents and stakeholders, or data analysis.
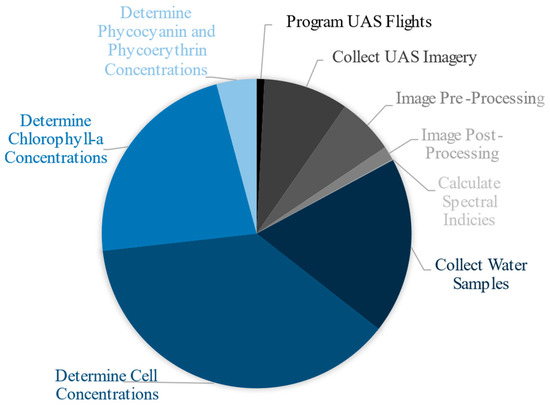
Figure 6.
Approximate comparison of the amount of time each task took to complete. Blue sections represent those associated with the collection and processing of water quality parameters. Grayscale sections represent those associated with the collection and processing of UAS parameters. Traditional water quality tasks took roughly 310 h to complete while UAS tasks took roughly 65 h to complete.
4.2. Limitations
Five of the six lakes included in this study experienced cyanobacteria blooms in 2022 according to the NHDES Harmful Algal Bloom Monitoring Program (Table 1). However, samples showing bloom forming conditions were only collected at Keyser Pond, Showell Pond, and Silver Lake. There were no collected samples indicating bloom conditions from French Pond, Greenwood Pond, or Tucker Pond as follows:
- French Pond did not have any cyanobacteria blooms during the 2022 field season; thus no “High” cell concentration samples were collected.
- The cyanobacteria bloom present at Greenwood Pond in 2022 was very rapid. Once sampling was conducted, the cyanobacteria bloom had subsided.
- At Tucker Pond, a pixilated surface bloom of Worochinia and Microcystis was present in small groupings along the southern and southwestern shores. Notes from lake residents indicated the bloom only appeared to span roughly 15 feet into the lake. On the day of sampling, the concentrations of cyanobacteria were found to be below the state of New Hampshire’s advisory threshold of 70,000 cells/mL.
- On par with the Microcystis and Worochinia bloom in Tucker Pond, Silver Lake’s bloom of primarily Microcystis and Dolichospermum proved difficult to capture. The ribbon of high cyanobacteria concentrations was isolated to the northern shores along the state park beach area, extending roughly 20 feet out into the lake at most. As a result, only a small fraction of the total samples collected throughout the lake surpassed the state threshold. During the peak of the first advisory on 6/29/22, collected samples ranged from 1150 to over 3.4 million cells/mL as the bloom was unevenly distributed throughout the lake.
Samples above the state threshold were collected at Keyser Pond, Showell Pond, and Silver Lake in 2022. Keyser Pond and Showell Pond contained very homogenous blooms of primarily Chyrosoporium with some Planktothrix in 2022 which turned the entire waterbodies a greenish-brown color. These blooms decreased water clarity measured with a secchi disk and view scope to less than 1 m. Cell concentrations ranged from 70,256 to 182,640 cells/mL at Keyser Pond, and from 320,400 to 650,250 cells/mL at Showell Pond for the samples collected during the advisories. Additionally, two samples were collected at Keyser Pond from clumps of Planktothrix that had broken off the benthic mat and floated to the surface after the advisory had lifted and ambient water cyanobacteria cell concentrations had fallen beneath the state threshold. These two samples, collected on 8/16/22, contained cell concentrations over 1 and 4 million cells/mL.
In addition to lake-specific analyses, this study identified the need for a more detailed analysis to be completed at a species-specific level. Different species of cyanobacteria alter lakes in varied ways. As seen in Keyser and Showell Ponds, blooms of Chrysosporum turned the entire waterbodies a greenish-brown color. This color alteration extended from the surface down into the water column. Planktothrix appeared in specks within the water column and as free-floating clumps that had detached from the benthic substrate. Microcystis was predominately found at Silver Lake in surface scum isolated to a single section of the waterbody. Often forming early in the morning and mixing with the water column as the wind and solar angle increased. This scum repeatedly came and dissipated quickly which caused the advisory to last for months and be illusive to the UAS.
Additional limitations arose in the image collection and processing phases of the UAS methodology. Due to the battery life of the UAS and not having a motorized boat, smaller lakes were targeted for this study. Lakes without islands or hidden coves were selected to maintain line of sight by the UAS pilot in command and to make canoeing to sites easier. Image processing in Agisoft Metashape proved difficult for waterbodies. Traditionally, tie points are used to properly align overlapping photos. However, the software struggles to identify tie points over a homogenous water’s surface, thus creating holes (Figure 7). The solution to this problem was to fly the UAS higher to include more of the lake edge in more photos. Although not a perfect solution, this worked well enough for the purposes of this study. This limitation would be a hindering factor for wide lakes or those that are very large. Any in-lake features such as islands, floating docks, moored boats, etc. would help to build tie points over this homogenous surface. This challenge was also stated by many other scientists [17,24,31,32,33,55,56,57]. Due to this issue, ten water quality sampling points were not included in the UAS spectral data to water quality parameter analyses because they occurred in the reflectance data “holes.” Another limitation to using UAS for environmental monitoring is caused by the weather. The DJI Matrice 300 RTK could only safely fly on days where the wind speed (including gusts) was less than 8 MPH or 3.6 m/s, and there was no chance of rain in the immediate forecast. The wind proved to be more difficult but was generally at its lowest earlier in the morning. However, this timing was beneficial since it occurred when the sun angle and glint were at their lowest even though some edges of the waterbodies were within shadows from shoreline vegetation on sunny days.
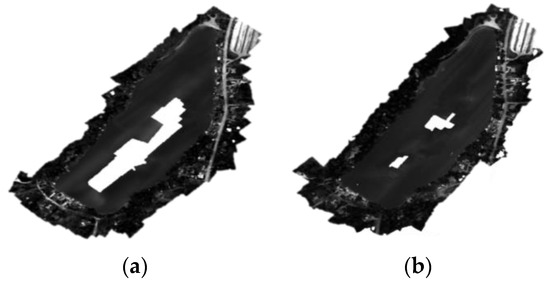
Figure 7.
Examples of black and white orthomosaic outputs for Silver Lake created from images flown at 100 m (a) and 120 m (b). More points in the point cloud were generated from the higher flying height which resulted in a more complete orthomosaic of Silver Lake.
4.3. Relation to Similar Studies
Contrary to other studies, this study involved multiple visits to various lakes with different dominant species of cyanobacteria from May to September in 2022. The revisitation allowed sampling to be conducted during various stages of the bloom cycle, ambient weather conditions, and seasonal changes in other water quality parameters including total suspended solids and emergent/submerged aquatic vegetation cover (not measured). It is difficult to draw usable conclusions from one flight over one lake on one day with only a handful of collected water samples for analysis without this replication.
However, many scientists have attempted to find correlations between chlorophyll-a concentrations and spectral indices collected by a UAS equipped sensor with less rigorous field studies. These studies include correlations to general “algae” [22,24,32], cyanobacteria [26], and a toxin associated with some species of cyanobacteria, microcystin [23]. A variety of sensors including a Parrot Sequoia multispectral sensor, a Canon ELPH 110HS, and a modified digital camera were used [21,24,32]. Two studies used the MicaSense RedEdge sensor [22,23]. A variety of indices were built for the chlorophyll-a regressions. R2 values ranged from 0.004 to 0.88 depending on the index used. The most common index was a NDVI or modified blue NDVI (BNDVI). The NDVI_3 and BNDVI_3 regressions to chlorophyll-a in this study produced R2 values of 0.50 and 0.66 respectively but were not of the most important for the random forest classification. These indices produced R2 values of 0.15 and 0.16 [23], 0.51 [32], 0.70 [22], 0.77 to 0.87 [21] and 0.88 [24]. These regressions were also represented in various forms including linear, logarithmic, and polynomial.
The logarithmic R2 values found in this study (Table 7) are comparable to those found in Sharp et al., 2021 [26]. However, the indices which showed the best correlations for the lakes studied in New Hampshire were not similar to those used in these referenced studies. The NGRDI did not produce a significant linear relationship with chlorophyll-a in Kim et al., 2021, though it produced one of the highest R2 values for chlorophyll-a concentrations of the most important features for classification in this study [24]. The difference might be attributed to one being a linear and the other a logarithmic line of best fit, or due to different wavelengths of light used in the equations per the sensor’s capabilities although they are both designed to be NGRDI = (green − red)/(green + red). García-Fernández et al., 2021 used the NGBDI to assess the quality of grape plants for wine production using a UAS equipped RGB sensor [43]. Although not an aquatic study, the NGBDI was used to assess alterations to growth due to water stress. This index proved to be very important for determining the presence of cyanobacteria associated parameters likely because it uses data from the blue and green portions of the electromagnetic spectrum (Table 7).
The use of phycocyanin concentration for assessing cyanobacteria blooms is growing in momentum [27]. This study serves as an additional source for verifying the cyanobacteria cell concentration to phycocyanin concentration relationship in addition to Almuhtaram et al., 2018 and Bertone et al., 2018 [53,54]. Few papers have discussed connecting phycocyanin concentration to UAS spectral features [26,34,35]. Sharp et al., 2021 studied a single cyanobacteria bloom in California, USA, over the summer of 2019 [26]. Sharp and colleagues included four visits to the lake for measurements of chlorophyll-a and phycocyanin paired with the overpass of the Sentinel-3a satellite. One visit corresponded with a small UAS (sUAS) mission over 12 sites in one portion of the lake when the dominant taxa were Dolichospermum, Gleotrichia, and Microcystis, but a CI was not recreated due to the limited wavelengths of the UAS sensor used. The sUAS was only used to map chlorophyll-a concentrations throughout the study area using a band ratio relationship from a previously published paper designed for a different waterbody. Pyo et al., 2018 studied the relationship between phycocyanin and chlorophyll-a to hyperspectral imagery using modeled simulations [35]. They produced R2 values between 0.55 and 0.75 for their two and three band ratios when plotted against estimated phycocyanin, and between 0.25 and 0.56 for chlorophyll-a. J. M. Ahn et al., 2021 also used a UAS mounted hyperspectral sensor to form a correlation with phycocyanin concentrations (R2 = 0.85) using a “generic algorithm” [34].
Using a multivariate classification algorithm approach rather than only simple regressions allowed the overall success of this study to drastically increase. Although less commonly conducted compared to simple regressions, models for remotely sensed data to water quality data using machine learning algorithms produce high accuracies across the board in many recently published studies. Surface sediment classification using an object-based classification method from UAS multispectral data in tidal flats produced an overall classification accuracy of 72.8% [58]. Scientists mapping percent cover of emergent vegetation in freshwater waterbodies of California (USA) used the random forest classification algorithm to discern overall accuracies of 82% [59]. In addition, researchers studying two lakes in China classified general water quality into three classes based on designated uses. Using a convolutional neural network with four convolutional layers, overall accuracies reached 92.5% within their study [60].
The argument can be made from a public safety standpoint for recreational waters, that knowing if cyanobacteria is classified as above or below regulatory thresholds is the primary goal. Only then would distinguishing between cell concentrations be useful, i.e., 25,000 cells/mL and 45,000 cells/mL. In other words, due to the high accuracy a classification approach produces, stakeholders can know if the waterbody is safe for the designated use. With the random forest algorithm method, this study found very high overall and user’s accuracies from 87.4 to 92.9% for the three water quality parameters with UAS multispectral spectral data.
4.4. Recommendations for Future Research
The integration of using UASs to study and monitor cyanobacteria and harmful algal blooms is an emerging discipline [61]. Because of its infancy, there is lots of room for expansion and refinement. This study would be strengthened with a larger dataset, and one across multiple years. With a larger dataset, including more UAS reflectance data of cyanobacteria blooms with additional reference (water quality) data validation, one can then subdivide the dataset by lake, trophic class, or dominant cyanobacteria species and re-run all algorithms to create lake or species-specific trends. With a larger dataset, the number of classes used for classification could be increased to further refine assessments. It would also be interesting to build a dataset from lakes without homogenous blooms similar to Tucker Pond or Silver Lake. Once the dataset is large enough, images from a flight over the lake could then be processed to identify the status of the lake at a classified pixel level. If repeated, this could aid in determining where blooms begin on an individual lake, and where they reside. This information could be used to complement data gathered for remediation efforts such as alum treatments, physical removal, or aeration. Lastly, these findings could be applied to lakes large enough for studies incorporating imagery from satellites—particularly the Sentinel 2 satellite. This approach would only improve as satellite technology increases in spatial resolution.
5. Conclusions
The use of a 10-band multispectral sensor flown on a UAS to detect cyanobacteria blooms in NH waterbodies was found to be a viable approach to monitor the rising water quality issue of cyanobacteria blooms. Identifying cyanobacteria blooms through imagery allows the user to assess the entire waterbody, including coves, littoral areas, and open water areas which might be inaccessible by land or boat. The use of imagery also promotes sampler safety through decreased time spent at each waterbody and the elimination of the need to contact the water for sampling. Additionally, this study identified significant correlations between cyanobacteria cell concentration to chlorophyll-a concentration, and cyanobacteria cell concentration to phycocyanin concentrations. These relationships identified the validity of using secondary parameters to measure cyanobacteria concentration. In conjunction with the water quality data, the NGBDI_4, NGRDI_4, 475 nm, 560 nm, and 668 nm were found to be the most important indices and bands for identifying the presence and classification of cyanobacteria, chlorophyll-a, and phycocyanin through the UAS classification approach. These important features illuminate the ability for this methodology to be applied to data collected by the Sentinel 2 satellite for larger freshwater waterbodies. Lastly, the UAS approach took significantly less time to complete than the traditional water quality sampling and analysis approach, therefore opening the possibility for these results to be applied in larger area, state, or region-wide studies.
Author Contributions
Conceptualization, C.L.B.; methodology, C.L.B., B.T.F., A.M. and R.G.C.; software, B.T.F. and C.L.B.; validation, B.T.F. and A.M.; formal analysis, C.L.B.; resources, C.L.B., R.G.C. and A.M.; data curation, C.L.B. and B.T.F.; writing—original draft preparation, C.L.B.; writing—review and editing, C.L.B. and R.G.C.; visualization, C.L.B.; supervision, R.G.C.; project administration, R.G.C.; funding acquisition, R.G.C. All authors have read and agreed to the published version of the manuscript.
Funding
Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2975. This work was supported by the USDA National Institute of Food and Agriculture McIntire-Stennis Project 1026105.
Acknowledgments
Thank you to the NH Lay Lakes Monitoring Program and the UNH WQAL for lending laboratory supplies. Thank you to the NHDES for providing historical cyanobacteria bloom data for New Hampshire, and the citizens at each studied lake. The Fulcrum® corporation provided a one-year free trial of the data collection app for this study. Much appreciation goes to FB Environmental for their support and field equipment.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A
- Cyanobacteria Fluorometric Detection of Whole Lake Water—New Hampshire Lay Lakes Monitoring Program
For cyanomonitoring protocol, freeze all samples before reading, thaw quickly in a water bath, and read within 20 min of reaching target temperature (21–24 °C). It works well to thaw samples in small batches of 8–12 depending on the efficiency of reading. Be careful not to over warm the samples. The goal is to bring them up to temp but not over. If you over-warm samples, place them in a cool water bath to bring them back down to between 21 and 24 °C.
General tips: always place the cuvette in the fluorometer in the same orientation and ensure that there are no air bubbles or drips on the side of the cuvette.
- Materials List
Kim Wipes, Waste bucket, DI Water Bottle, Infrared thermometer, bucket for thawing samples. Cyanofluor: Meter, 4-sided glass cuvette, calibration standard cuvette. Fluoroquick (model number FQ-PC-PE-RATIO-C, serial number DL072501): Methacrylate Plastic cuvette, charging cable.
- FluoroQuik SOP
General Note: The FlouroQuik should be blanked immediately prior to analyzing lake water samples and both low and high standards should be “read” before proceeding to lake water samples. The following SOP is applicable to both dual channel instruments: the chlorophyll/phycocyanin model and the phycocyanin/phycoeurethrin model. The flouroQuik requires plastic cuvettes and the instrument measures pigments as micrograms per liter (μg/L)
- Rinse a cuvette three times with DI water.
- Fill the rinsed cuvette with DI water and place the cuvette in the FlouroQuik chamber.
- Press “measure”.
- Press “blank”.
- Press “measure” and record both pigment concentrations (e.g., chlorophyll and phycocyanin or phycocyanin and phycoeurythrin).
- Press “return”.
- Rinse the cuvette with low pigment standard.
- Fill the cuvette with low pigment standard.
- Place the cuvette in the FlouroQuik chamber.
- Press “measure”.
- Press “sample”.
- Record both pigment concentrations.
- Press “return”.
- Rinse the cuvette three times with DI water.
- Rinse the cuvette with high pigment standard.
- Fill the cuvette with high pigment standard.
- Place the cuvette in the FlouroQuik chamber.
- Press “measure”.
- Press “sample”.
- Record both pigment concentrations.
- Press “return”.
- Rinse the cuvette three times with DI water.
- Rinse the cuvette with a lake water sample.
- Fill the cuvette with the lake water sample.
- Place the cuvette in the FlouroQuik chamber.
- Press “measure”.
- Press “sample”.
- Record both pigment concentrations.
- Repeat steps 20 through 27 until all lake water samples have been analyzed.
- Analyze a low pigment concentration and a DI blank as the last two samples and record the pigment concentrations.
Appendix B
- UNH Water Quality Analysis Laboratory Standard Operating Procedure
- Chlorophyll-a—Hot Ethanol Extraction and Spectrophotometric Determination
- Introduction
This extraction is applied to samples on filters. This method has the benefit of extraction without grinding and avoiding toxic methanol or acetone exposure [62]. Samples are analyzed with spectrophotometric analysis at 665 and 750 nm using a 1 cm spectrophotometer cuvette (method in Standard Methods (10200 H) with different extinction coefficient for ethanol and conversion for chlorophyll per unit area).
- Sample Preparation
- Filter a known volume of culture onto a Whatman GF/F filter (47 mm). Swirl your culture before sampling to ensure homogeneous sampling. Keep vacuum level low (<10 in Hg) to prevent cell breakage and low filtration efficiency.
- Rinse funnel walls with small amount of DI.
- Remove the funnel. Fold the filter in quarters and place in aluminum foil. Label foil and freeze samples until analysis.
- Hot Ethanol Extraction (Day 1)
- Turn on the water bath 30–60 min before starting. Set point to 79 °C. Make sure the water is above the right inner port.
- Keep samples in dark or low light at all times. If the sample is kept in the dark, only 1.3% of the chlorophyll degrades with the 5 min. hot extraction and 24-h storage.
- Use test tubes with screw tops that have the tube numbers scribed on the side with a diamond pencil (ethanol extracts numbers written in sharpie).
- Place the filter (Whatman GFF, 47 mm) or scraped material in the tube. Add 10 mL (pipette) of 95% ethanol. Make sure the filter is totally submerged in the ethanol.
- Mark the location of the meniscus on the side of the tube and place the cap on loosely.
- Heat the tube in a water bath at 79 °C for 5 min, then mix and cool for 24 h in the dark (at room temperature and can be sealed tightly).
- Turn off the hot water bath.
- After extraction, use 95% ethanol to bring up to mark on side of tube if ethanol has evaporated, then mix.
- Clear sample by centrifugation, filtration, or settling. We normally use settling.
- Spec Analysis (Day 2)
- Analyze sample with spectrophotometric analysis at 665 and 750 nm using 1 cm cuvette (Standard Methods). The 7G on spec cuvette faces you.
- Run the blank.
- Measure blank (fill the cuvette with 3 mL of ethanol only) every time you change from 665 nm to 750 nm. Record absorbance. Click set blank.
- Run a sample.
- Take the test tube with the filter in it. Tighten the cap and then invert it to mix.
- Pipette 3 mL of sample into the cuvette.
- Read the sample at 665.
- If adsorption is over 1.5 absorbance units, dilute sample. Try 1:10 dilution first into cuvette: 300 µL sample and 2.7 mL of ethanol. This was only needed for one sample which came from a benthic mat of cyanobacteria that had broken off and floated to the surface.
- Record value.
- Set nm type 750, enter.
- Measure blank again.
- Measure the sample again but at 750.
- Record the value.
- Add 0.1 mL of 0.25 N HCl into 3 mL of sample in spec cuvette of extractant after the first reading and let sit for 3 min to phaeophytinize all chl before reading (amount of acid is very important). Measure at 665 and 750 nm again with a blank in between, record absorbance.
- Rinse 3x with DI and repeat to Step 3.
- Calculations
Calculations are made as follows:
where subscript 0 = absorption at the designated wavelength before acid addition and subscript a = absorption at the designated wavelength after acid addition. v = volume of extractant used (in liters, 0.01 for this study), A = volume of original sample that was filtered in mL, and l = path length of cell (1000 for this study).
Chlorophyll-a (μg/L) = ((28.78((6650-7500) − (665a-750a))v)/(A/l))1000
Appendix C

Table A1.
Index equation versions. Refer to Table 3 for citations and descriptions. * Indicates this parameter is found in the top feature important scores in Table 7.
| Index and General Equation | Specific Equation | ID |
|---|---|---|
| CI | (650 − 560)/(668 − 560) | CI_1 * |
| (668 − 650)/(705 − 650) | CI_2 * | |
| NDVI = (NIR − R)/(NIR + R) | (842 − 650)/(842 + 650) | NDVI_1 |
| (842 − 650)/(842 + 668) | NDVI_2 | |
| (842 − 668)/(842 + 650) | NDVI_3 | |
| (842 − 668)/(842 + 668) | NDVI_4 | |
| cBNDVI = (NIR − cB)/(NIR + cB) | (842 − 444)/(842 + 444) | CBNDVI |
| BNDVI = (NIR − B)/(NIR + B) | (842 − 475)/(842 + 475) | BNDVI_1 |
| (842 − 444)/(842 + 475) | BNDVI_2 | |
| (842 − 475)/(842 + 444) | BNDVI_3 | |
| NDRE = (NIR − RE)/(NIR + RE) | (842 − 705)/(842 + 705) | NDRE_1 |
| (842 − 717)/(842 + 717) | NDRE_2 | |
| (842 − 740)/(842 + 740) | NDRE_3 | |
| NGBDI = (G − B)/(G + B) | (531 − 444)/(531 + 444) | NGBDI_1 |
| (531 − 475)/(531 + 475) | NGBDI_2 | |
| (560 − 444)/(560 + 444) | NGBDI_3 | |
| (560 − 475)/(560 + 475) | NGBDI_4 * | |
| NGRDI = (G − R)/(G + R) | (531 − 650)/(531 + 650) | NGRDI_1 |
| (531 − 668)/(531 + 668) | NGRDI_2 | |
| (560 − 650)/(560 + 650) | NGRDI_3 * | |
| (560 − 668)/(560 + 668) | NGRDI_4 * | |
| SABI = (NIR − R)/(B + G) | (842 − 650)/(444 + 531) | SABI_1 |
| (842 − 650)/(444 + 560) | SABI_2 | |
| (842 − 650)/(475 + 531) | SABI_3 | |
| (842 − 650)/(475 + 560) | SABI_4 | |
| (842 − 668)/(444 + 531) | SABI_5 | |
| (842 − 668)/(444 + 560) | SABI_6 | |
| (842 − 668)/(475 + 531) | SABI_7 | |
| (842 − 668)/(475 + 560) | SABI_8 | |
| KIVU = (B − R)/G | (444 − 650)/531 | KIVU_1 |
| (444 − 650)/560 | KIVU_2 | |
| (444 − 668)/531 | KIVU_3 | |
| (444 − 668)/560 | KIVU_4 | |
| (475 − 650)/531 | KIVU_5 | |
| (475 − 650)/560 | KIVU_6 | |
| (475 − 668)/531 | KIVU_7 | |
| (475 − 668)/560 | KIVU_8 | |
| FLH Blue = G − (R + (B − R)) | 531 − (650 + (444 − 650)) | FLHblue_1 * |
| 531 − (650 + (444 − 668)) | FLHblue_2 * | |
| 531 − (650 + (475 − 650)) | FLHblue_3 | |
| 531 − (650 + (475 − 668)) | FLHblue_4 | |
| 531 − (668 + (444 − 650)) | FLHblue_5 | |
| 531 − (668 + (444 − 668)) | FLHblue_6 | |
| 531 − (668 + (475 − 650)) | FLHblue_7 | |
| 531 − (668 + (475 − 668)) | FLHblue_8 | |
| 560 − (650 + (444 − 650)) | FLHblue_9 | |
| 560 − (650 + (444 − 668)) | FLHblue_10 | |
| 560 − (650 + (475 − 650)) | FLHblue_11 | |
| 560 − (650 + (475 − 668)) | FLHblue_12 | |
| 560 − (668 + (444 − 650)) | FLHblue_13 | |
| 560 − (668 + (444 − 668)) | FLHblue_14 | |
| 560 − (668 + (475 − 650)) | FLHblue_15 | |
| 560 − (668 + (475 − 668)) | FLHblue_16 | |
| MODIS normalized spectral index [46] from [23] = (eRed − eNIR)/(eRed + eNIR) | (650 − 842)/(650 + 842) | MODIS_NSI_1 |
| (650 − 842)/(668 + 842) | MODIS_NSI_2 | |
| (668 − 842)/(650 + 842) | MODIS_NSI_3 | |
| (668 − 842)/(668 + 842) | MODIS_NSI_4 |
Appendix D
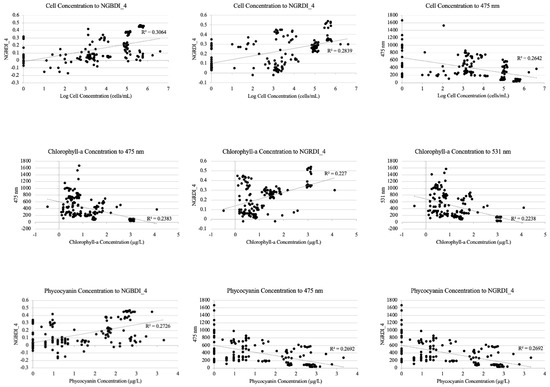
Figure A1.
Scatter plots for the three highest R2 values per water quality parameter from the simple regressions provided in Table 7.
References
- USEPA. National Lakes Assessment: The Third Collaborative Survey of Lakes in the United States (EPA 841-R-22-002). U.S. Environmental Protection Agency, Office of Water and Office of Research Development 2022. Available online: https://nationallakesassessment.epa.gov/webreport (accessed on 18 March 2023).
- Spyrakos, E.; O’Donnell, R.; Hunter, P.D.; Miller, C.; Scott, M.; Simis, S.G.H.; Neil, C.; Barbosa, C.C.F.; Binding, C.E.; Bradt, S.; et al. Optical types of inland and coastal waters. Limnol. Oceanogr. 2017, 63, 846–870. [Google Scholar] [CrossRef]
- Cole, J.J.; Prairie, Y.T.; Caraco, N.F.; McDowell, W.H.; Tranvik, L.J.; Striegl, R.G.; Duarte, C.M.; Kortelainen, P.; Downing, J.A.; Middelburg, J.J.; et al. Plumbing the global carbon cycle: Integrating inland waters into the terrestrial carbon budget. Ecosystems 2007, 10, 172–185. [Google Scholar] [CrossRef]
- Palinska, K.A.; Surosz, W. Taxonomy of cyanobacteria: A contribution to consensus approach. Hydrobiologia 2014, 740, 1–11. [Google Scholar] [CrossRef]
- Carey, C.C.; Ewing, H.A.; Cottingham, K.L.; Weathers, K.C.; Thomas, R.Q.; Haney, J.F. Occurrence and toxicity of the cyanobacterium Gloeotrichia echinulata in low-nutrient lakes in the northeastern United States. Aquat. Ecol. 2012, 46, 395–409. [Google Scholar] [CrossRef]
- Cole, J.J. The Carbon Cycle. In Fundamentals of Ecosystem Science; Elsevier: Amsterdam, The Netherlands, 2013; pp. 109–135. [Google Scholar] [CrossRef]
- NHDES. Cyanobacteria Advisory; New Hampshire Department of Environmental Services: Concord, NH, USA, 2022.
- Caller, T.A.; Doolin, J.W.; Haney, J.F.; Murby, A.J.; West, K.G.; Farrar, H.E.; Ball, A.; Harris, B.T.; Stommel, E.W. A cluster of amyotrophic lateral sclerosis in New Hampshire: A possible role for toxic cyanobacteria blooms. Amyotroph. Lateral Scler. 2009, 10, 101–108. [Google Scholar] [CrossRef]
- Codd, G.A.; Testai, E.; Funari, E.; Svirčev, Z. Cyanobacteria, Cyanotoxins, and Human Health. In Water Treatment for Purification from Cyanobacteria and Cyanotoxins; Wiley Online Library: Hoboken, NJ, USA, 2020; Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118928677.ch2 (accessed on 24 February 2023).
- Rolim, S.B.A.; Veettil, B.K.; Vieiro, A.P.; Kessler, A.B.; Gonzatti, C. Remote sensing for mapping algal blooms in freshwater lakes: A review. Environ. Sci. Pollut. Res. 2023, 30, 19602–19616. [Google Scholar] [CrossRef]
- Rhee, D.S.; Kim, Y.D.; Kang, B.; Kim, D. Applications of unmanned aerial vehicles in fluvial remote sensing: An overview of recent achievements. KSCE J. Civ. Eng. 2017, 22, 588–602. [Google Scholar] [CrossRef]
- GISGeography. Spatial Resolution vs Spectral Resolution—GIS Geography. Available online: https://gisgeography.com/spatial-resolution-vs-spectral-resolution/ (accessed on 14 January 2023).
- Becker, R.H.; Sayers, M.; Dehm, D.; Shuchman, R.; Quintero, K.; Bosse, K.; Sawtell, R. Unmanned aerial system based spectroradiometer for monitoring harmful algal blooms: A new paradigm in water quality monitoring. J. Great Lakes Res. 2019, 45, 444–453. [Google Scholar] [CrossRef]
- Lyu, P.; Malang, Y.; Liu, H.H.; Lai, J.; Liu, J.; Jiang, B.; Qu, M.; Anderson, S.; Lefebvre, D.D.; Wang, Y. Autonomous cyanobacterial harmful algal blooms monitoring using multirotor UAS. Int. J. Remote Sens. 2017, 38, 2818–2843. [Google Scholar] [CrossRef]
- Jensen, J.R. Introductory Digital Image Processing: A Remote Sensing Perspective, 4th ed.; Pearson Education Inc.: Saddle River, NJ, USA, 2018. [Google Scholar]
- Lillesand, T.; Kiefer, R.W.; Chipman, J. Remote Sensing and Image Interpretation, 7th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; Available online: https://www.wiley.com/en-us/Remote+Sensing+and+Image+Interpretation%2C+7th+Edition-p-9781118343289 (accessed on 3 December 2021).
- Flynn, K.F.; Chapra, S.C. Remote Sensing of Submerged Aquatic Vegetation in a Shallow Non-Turbid River Using an Unmanned Aerial Vehicle. Remote Sens. 2014, 6, 12815–12836. [Google Scholar] [CrossRef]
- Silva, T.S.F.; Costa, M.P.F.; Melack, J.M.; Novo, E.M.L.M. Remote sensing of aquatic vegetation: Theory and applications. Environ. Monit. Assess. 2007, 140, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Visser, F.; Wallis, C.; Sinnott, A.M. Optical remote sensing of submerged aquatic vegetation: Opportunities for shallow clearwater streams. Limnologica 2013, 43, 388–398. [Google Scholar] [CrossRef]
- Su, T.-C.; Chou, H.-T. Application of Multispectral Sensors Carried on Unmanned Aerial Vehicle (UAV) to Trophic State Mapping of Small Reservoirs: A Case Study of Tain-Pu Reservoir in Kinmen, Taiwan. Remote Sens. 2015, 7, 10078–10097. [Google Scholar] [CrossRef]
- Van der Merwe, D.; Price, K.P. Harmful Algal Bloom Characterization at Ultra-High Spatial and Temporal Resolution Using Small Unmanned Aircraft Systems. Toxins 2015, 7, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.; Kang, G.; Kim, D.; Lee, S. A study on the evaluation of water-bloom using image processing. Environ. Sci. Pollut. Res. 2018, 25, 36775–36780. [Google Scholar] [CrossRef]
- Douglas Greene, S.B.; LeFevre, G.H.; Markfort, C.D. Improving the spatial and temporal monitoring of cyanotoxins in Iowa lakes using a multiscale and multi-modal monitoring approach. Sci. Total. Environ. 2020, 760, 143327. [Google Scholar] [CrossRef]
- Kim, E.-J.; Nam, S.-H.; Koo, J.-W.; Hwang, T.-M. Hybrid Approach of Unmanned Aerial Vehicle and Unmanned Surface Vehicle for Assessment of Chlorophyll-a Imagery Using Spectral Indices in Stream, South Korea. Water 2021, 13, 1930. [Google Scholar] [CrossRef]
- Mishra, S.; Stumpf, R.P.; Schaeffer, B.A.; Werdell, P.J.; Loftin, K.A.; Meredith, A. Measurement of Cyanobacterial Bloom Magnitude using Satellite Remote Sensing. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Sharp, S.L.; Forrest, A.L.; Bouma-Gregson, K.; Jin, Y.; Cortés, A.; Schladow, S.G. Quantifying Scales of Spatial Variability of Cyanobacteria in a Large, Eutrophic Lake Using Multiplatform Remote Sensing Tools. Front. Environ. Sci. 2021, 9, 612934. [Google Scholar] [CrossRef]
- Ogashawara, I. Determination of Phycocyanin from Space—A Bibliometric Analysis. Remote Sens. 2020, 12, 567. [Google Scholar] [CrossRef]
- Ogashawara, I.; Mishra, D.R.; Mishra, S.; Curtarelli, M.P.; Stech, J.L. A Performance Review of Reflectance Based Algorithms for Predicting Phycocyanin Concentrations in Inland Waters. Remote Sens. 2013, 5, 4774–4798. [Google Scholar] [CrossRef]
- Leland, N.J.; Haney, J.F. Alternative Methods for Analysis of Cyanobacterial Populations in Drinking Water Supplies: Fluorometric and Toxicological Applications Using Phycocyanin. J. Water Resour. Prot. 2018, 10, 740–761. [Google Scholar] [CrossRef]
- Murby, A.L. Assessing Spatial Distributions of Cyanobacteria and Microcystins in NH Lakes with Implications for Lake Monitoring; University of New Hampshire: Durham, NH, USA, 2009. [Google Scholar]
- Aguirre-Gómez, R.; Salmerón-García, O.; Gómez-Rodríguez, G.; Peralta-Higuera, A. Use of unmanned aerial vehicles and remote sensors in urban lakes studies in Mexico. Int. J. Remote Sens. 2016, 38, 2771–2779. [Google Scholar] [CrossRef]
- Guimarães, T.T.; Veronez, M.R.; Koste, E.C.; Gonzaga, L.; Bordin, F.; Inocencio, L.C.; Larocca, A.P.C.; De Oliveira, M.Z.; Vitti, D.C.; Mauad, F.F. An Alternative Method of Spatial Autocorrelation for Chlorophyll Detection in Water Bodies Using Remote Sensing. Sustainability 2017, 9, 416. [Google Scholar] [CrossRef]
- Qu, M.; Anderson, S.; Lyu, P.; Malang, Y.; Lai, J.; Liu, J.; Jiang, B.; Xie, F.; Liu, H.H.; Lefebvre, D.D.; et al. Effective aerial monitoring of cyanobacterial harmful algal blooms is dependent on understanding cellular migration. Harmful Algae 2019, 87, 101620. [Google Scholar] [CrossRef]
- Ahn, J.M.; Kim, B.; Jong, J.; Nam, G.; Park, L.J.; Park, S.; Kang, T.; Lee, J.-K.; Kim, J. Predicting Cyanobacterial Blooms Using Hyperspectral Images in a Regulated River. Sensors 2021, 21, 530. [Google Scholar] [CrossRef]
- Pyo, J.C.; Ligaray, M.; Kwon, Y.S.; Ahn, M.-H.; Kim, K.; Lee, H.; Kang, T.; Cho, S.B.; Park, Y.; Cho, K.H. High-Spatial Resolution Monitoring of Phycocyanin and Chlorophyll-a Using Airborne Hyperspectral Imagery. Remote Sens. 2018, 10, 1180. [Google Scholar] [CrossRef]
- NHDES. Sources of Information and Explanation of Lake Trophic Data. New Hampshire Department of Environmental Services. 2019. Available online: https://www.des.nh.gov/sites/g/files/ehbemt341/files/documents/2020-01/laketrophic-explain-current.pdf (accessed on 24 February 2023).
- NHFGD. (n.d.). Depth Maps of Selected NH Lakes and Ponds|Maps|New Hampshire Fish and Game Department. New Hampshire Fish and Game. Available online: https://www.wildlife.state.nh.us/maps/bathymetry.html (accessed on 24 February 2023).
- Nelson, K.; Neils, D. New Hampshire Lake Trend Report: Status and Trends of Water Quality Indicators. NHDES. 2020. Available online: https://www4.des.state.nh.us/OneStopPub/TrophicSurveys/r-wd-20-08.pdf (accessed on 24 February 2023).
- Nye, T.L. Microcystins in Water, Gastropods and Bivalves from Silver Lake, New Hampshire. Master’s Thesis, University of New Hampshire, Durham, NH, USA, 1997. [Google Scholar]
- Steiner, S.; Nelson, K. New Hampshire Volunteer Lake Assessment Program: 2014 Lakes Region Regional Report. NHDES. Available online: https://www.des.nh.gov/sites/g/files/ehbemt341/files/documents/2020-01/r-wd-16-08.pdf (accessed on 24 February 2023).
- Mishra, S.; Mishra, D.R. Normalized difference chlorophyll index: A novel model for remote estimation of chlorophyll-a concentration in turbid productive waters. Remote Sens. Environ. 2012, 117, 394–406. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident Detection of Crop Water Stress, Nitrogen Status and Canopy Density Using Ground-Based Multispectral Data. 2000. Available online: https://www.researchgate.net/publication/43256762_Coincident_detection_of_crop_water_stress_nitrogen_status_and_canopy_density_using_ground_based_multispectral_data (accessed on 24 February 2023).
- García-Fernández, M.; Sanz-Ablanedo, E.; Rodríguez-Pérez, J.R. High-Resolution Drone-Acquired RGB Imagery to Estimate Spatial Grape Quality Variability. Agronomy 2021, 11, 655. [Google Scholar] [CrossRef]
- Alawadi, F. Detection of surface algal blooms using the newly developed algorithm surface algal bloom index (SABI). In Proceedings of the SPIE 7825, Remote Sensing of the Ocean, Sea Ice, and Large Water Regions, Toulouse, France, 20–23 September 2010. [Google Scholar] [CrossRef]
- Kabbara, N.; Benkhelil, J.; Awad, M.; Barale, V. Monitoring water quality in the coastal area of Tripoli (Lebanon) using high-resolution satellite data. ISPRS J. Photogramm. Remote Sens. 2008, 63, 488–495. [Google Scholar] [CrossRef]
- Shi, K.; Zhang, Y.; Xu, H.; Zhu, G.; Qin, B.; Huang, C.; Liu, X.; Zhou, Y.; Lv, H. Long-Term Satellite Observations of Microcystin Concentrations in Lake Taihu during Cyanobacterial Bloom Periods. Environ. Sci. Technol. 2015, 49, 6448–6456. [Google Scholar] [CrossRef] [PubMed]
- USEPA. National Lakes Assessment: A Collaborative Survey of the Nation’s Lakes (EPA 841-R-09-001). 2009. Available online: https://www.epa.gov/sites/default/files/2013-11/documents/nla_newlowres_fullrpt.pdf (accessed on 24 February 2023).
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data Principles and Practices, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Story, M.; Congalton, R.G. Accuracy Assessment: A User’s Perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Fraser, B.T.; Congalton, R.G. Monitoring Fine-Scale Forest Health Using Unmanned Aerial Systems (UAS) Multispectral Models. Remote Sens. 2021, 13, 4873. [Google Scholar] [CrossRef]
- U.S. Geological Survey. (n.d.). What Are the Band Designations for the Landsat Satellites? Available online: https://www.usgs.gov/faqs/what-are-band-designations-landsat-satellites (accessed on 1 April 2023).
- The European Space Agency. (n.d.). Spatial Resolution. Sentinel Online. Available online: https://sentinels.copernicus.eu/web/sentinel/user-guides/sentinel-2-msi/resolutions/spatial (accessed on 1 April 2023).
- Almuhtaram, H.; Cui, Y.; Zamyadi, A.; Hofmann, R. Cyanotoxins and Cyanobacteria Cell Accumulations in Drinking Water Treatment Plants with a Low Risk of Bloom Formation at the Source. Toxins 2018, 10, 430. [Google Scholar] [CrossRef]
- Bertone, E.; Burford, M.A.; Hamilton, D.P. Fluorescence probes for real-time remote cyanobacteria monitoring: A review of challenges and opportunities. Water Res. 2018, 141, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, T.T.; Veronez, M.R.; Koste, E.C.; Souza, E.M.; Brum, D.; Gonzaga, L.; Mauad, F.F. Evaluation of Regression Analysis and Neural Networks to Predict Total Suspended Solids in Water Bodies from Unmanned Aerial Vehicle Images. Sustainability 2019, 11, 2580. [Google Scholar] [CrossRef]
- Veronez, M.R.; Kupssinskü, L.S.; Guimarães, T.T.; Koste, E.C.; Da Silva, J.M.; De Souza, L.V.; Oliverio, W.F.M.; Jardim, R.S.; Koch, I.; De Souza, J.G.; et al. Proposal of a Method to Determine the Correlation between Total Suspended Solids and Dissolved Organic Matter in Water Bodies from Spectral Imaging and Artificial Neural Networks. Sensors 2018, 18, 159. [Google Scholar] [CrossRef]
- Wu, D.; Li, R.; Zhang, F.; Liu, J. A review on drone-based harmful algae blooms monitoring. Environ. Monit. Assess. 2019, 191, 211. [Google Scholar] [CrossRef]
- Kim, K.-L.; Kim, B.-J.; Lee, Y.-K.; Ryu, J.-H. Generation of a Large-Scale Surface Sediment Classification Map Using Unmanned Aerial Vehicle (UAV) Data: A Case Study at the Hwang-do Tidal Flat, Korea. Remote Sens. 2019, 11, 229. [Google Scholar] [CrossRef]
- Kislik, C.; Genzoli, L.; Lyons, A.; Kelly, M. Application of UAV Imagery to Detect and Quantify Submerged Filamentous Algae and Rooted Macrophytes in a Non-Wadeable River. Remote Sens. 2020, 12, 3332. [Google Scholar] [CrossRef]
- Pu, F.; Ding, C.; Chao, Z.; Yu, Y.; Xu, X. Water-Quality Classification of Inland Lakes Using Landsat8 Images by Convolutional Neural Networks. Remote Sens. 2019, 11, 1674. [Google Scholar] [CrossRef]
- Fraser, B.T.; Bunyon, C.L.; Reny, S.; Lopez, I.S.; Congalton, R.G. Analysis of Unmanned Aerial System (UAS) Sensor Data for Natural Resource Applications: A Review. Geographies 2022, 2, 303–340. [Google Scholar] [CrossRef]
- Sartory, D.P.; Grobbelaar, J.U. Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 1984, 114, 177–187. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).