Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China
Abstract
1. Introduction
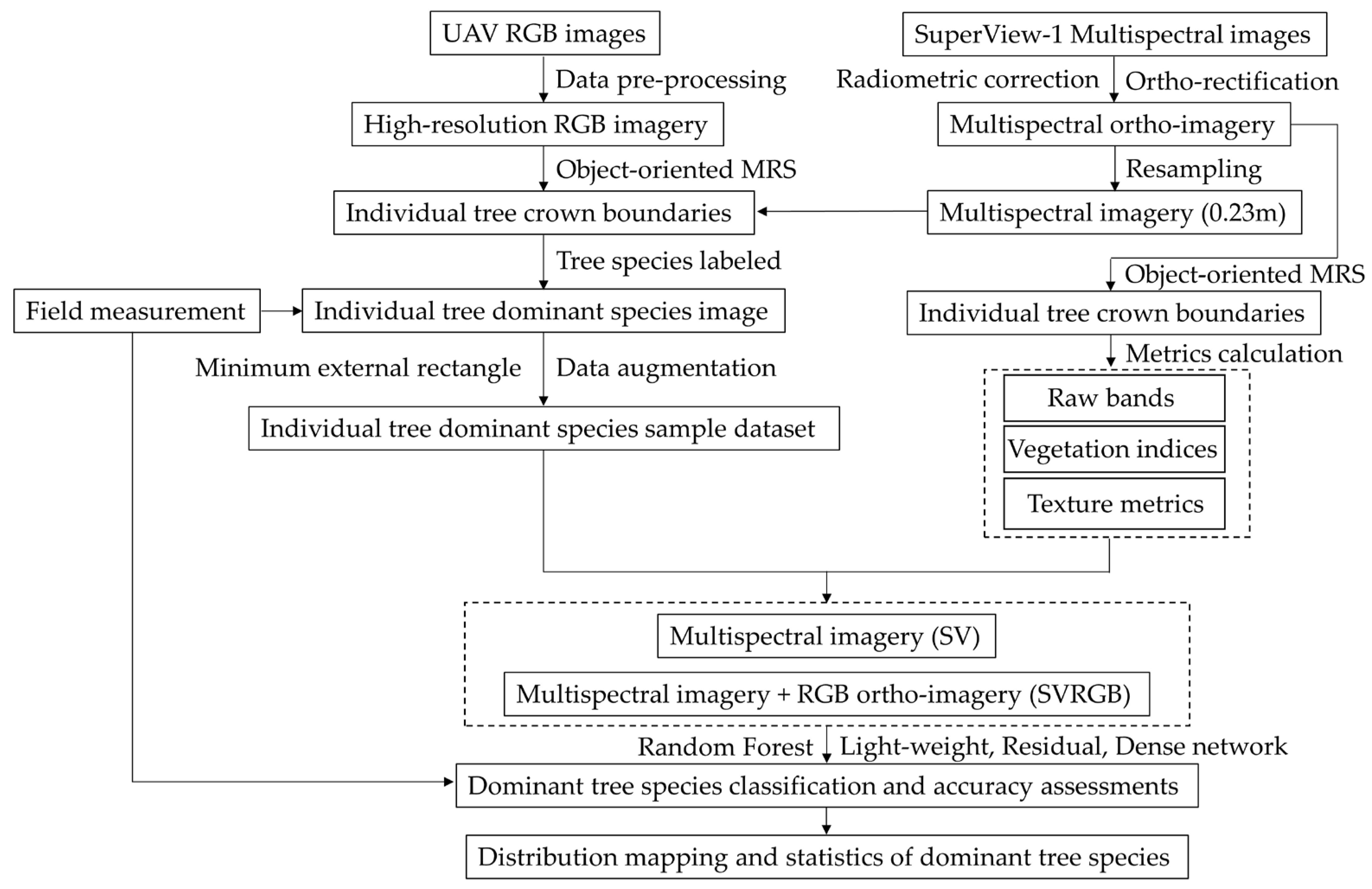
2. Materials and Methods
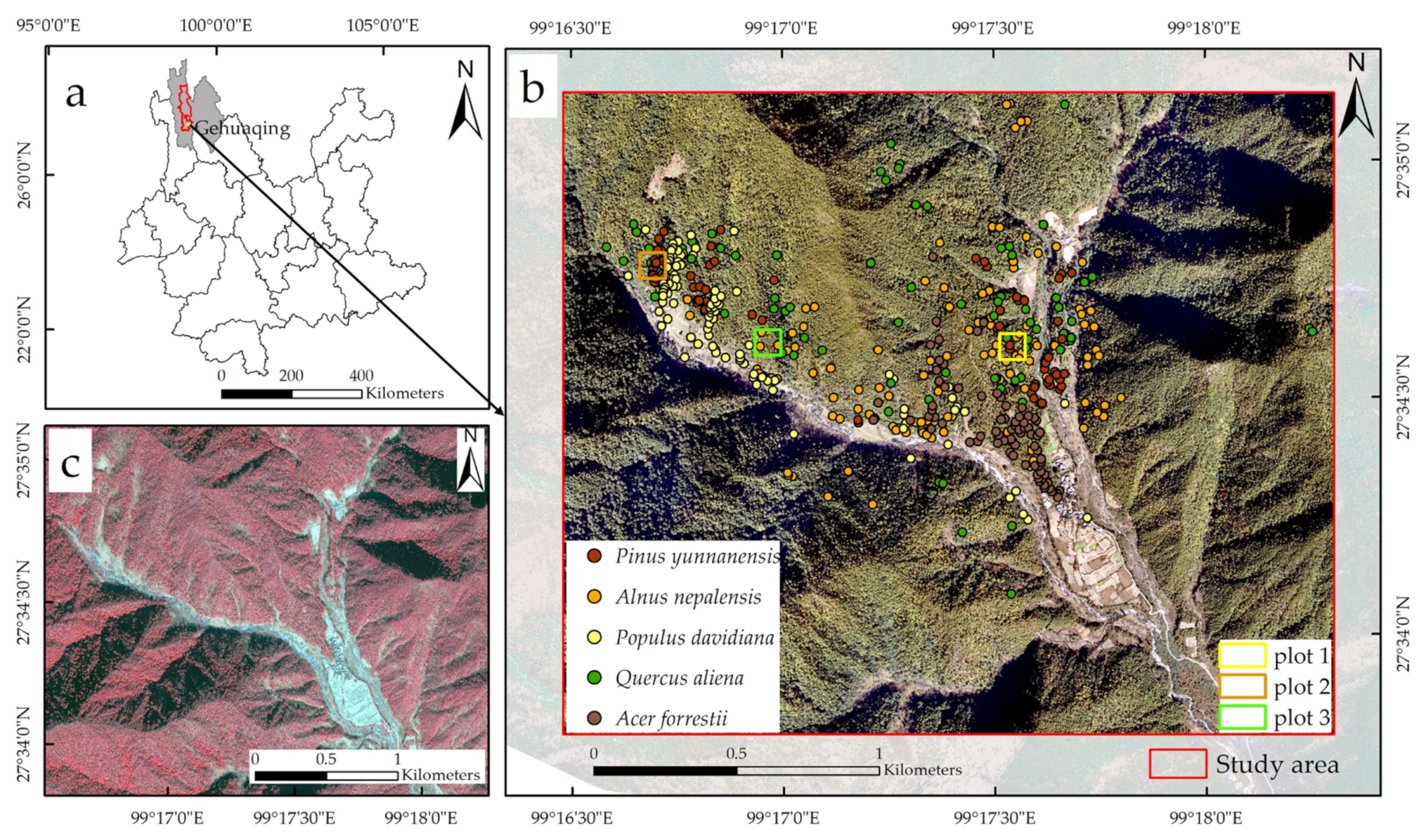
2.1. Study Area
2.2. Field Measurements
2.3. Remote Sensing Data
2.4. Data Pre-Processing
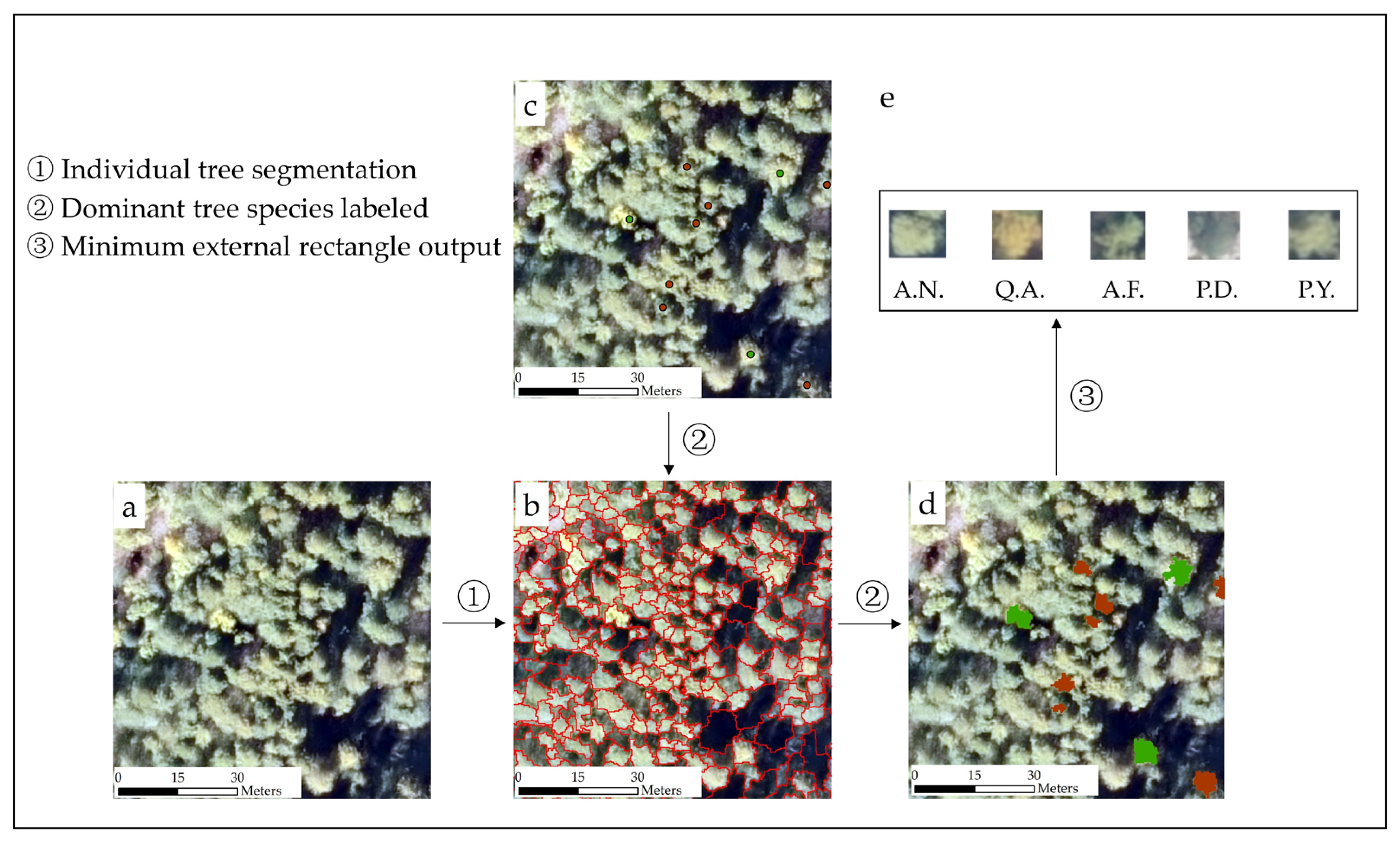
2.5. Sample Dataset of Individual Tree
2.5.1. Data Augmentation
2.5.2. Remote Sensing Sample Dataset of Individual Trees
2.6. Spectral and Texture Metrics Calculation
2.7. Random Forest and Deep Learning Network Classifier
2.7.1. Random Forest
2.7.2. Deep Learning Networks
- (1)
- Lightweight network MobileNetV2
- (2)
- Residual network ResNet34
- (3)
- Dense network DenseNet121
2.8. Validation of Individual Tree Segmentation and Tree Species Classification
2.9. Effects of the Number of Training Samples on the Performance of Dominant Tree Species Classification
3. Results
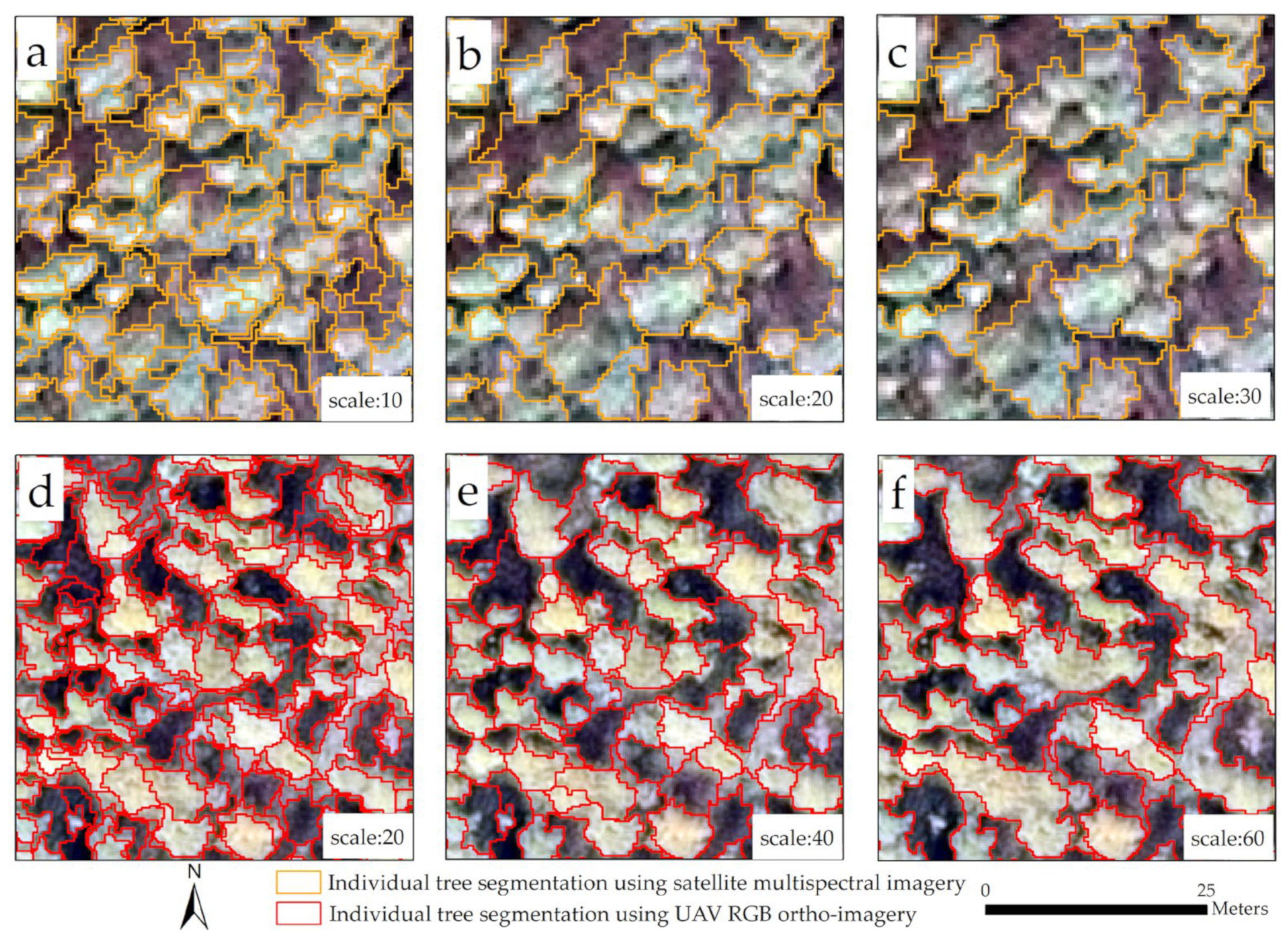
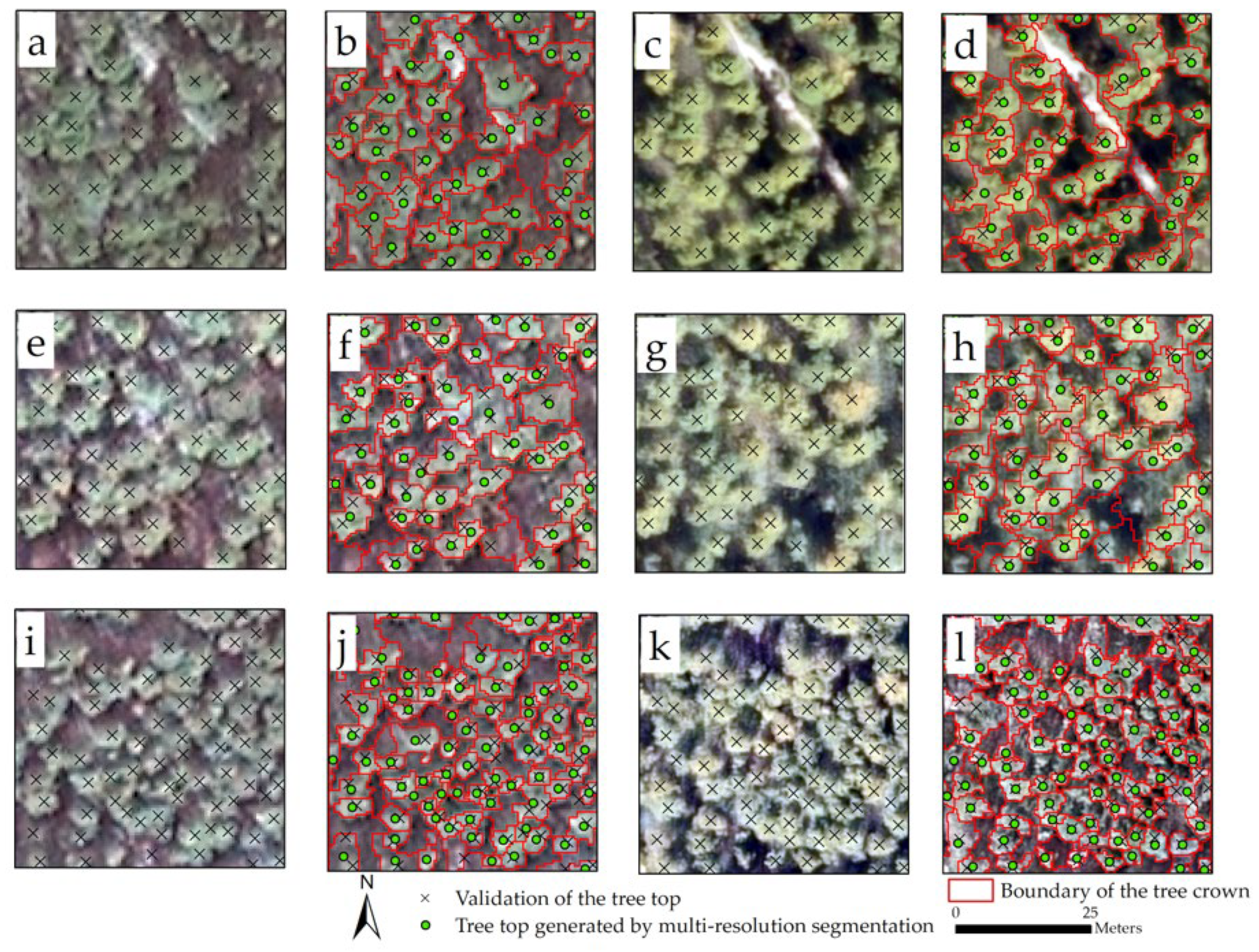
3.1. Individual Tree Segmentation
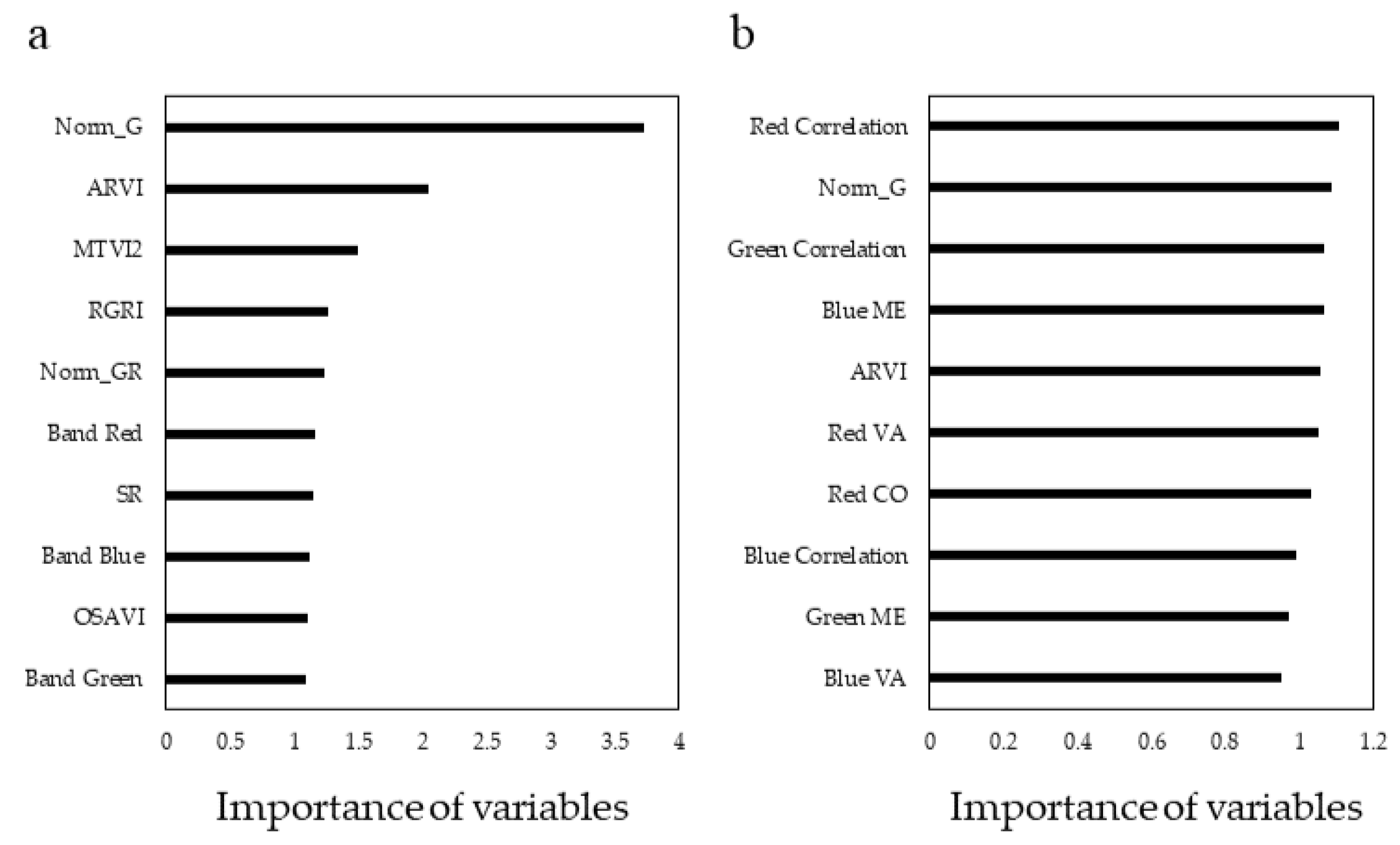
3.2. Feature Optimization and Analysis
3.3. Training of Deep Learning Networks
3.4. Accuracy Assessment
3.5. Mapping of Five Dominant Tree Species
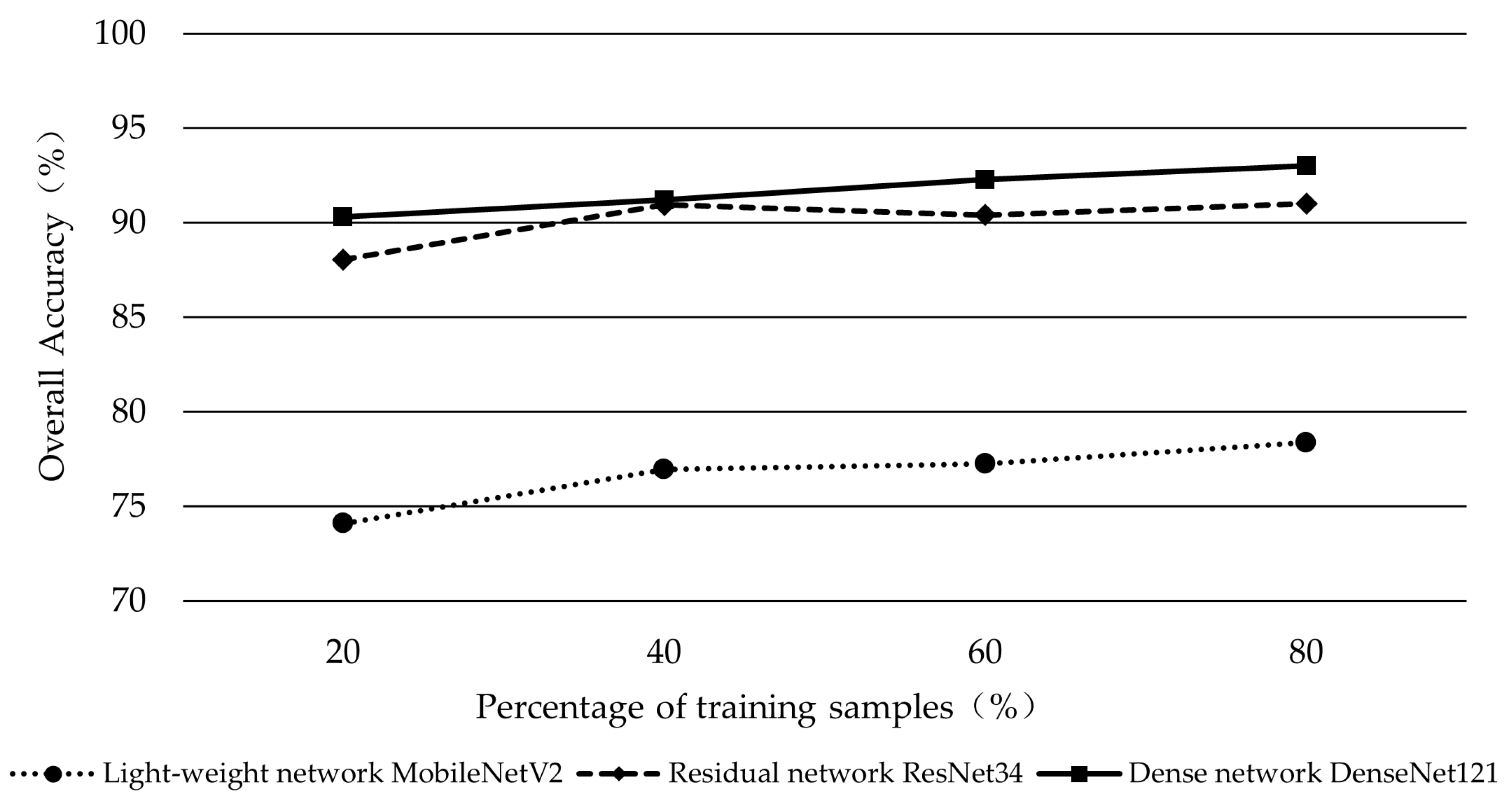
3.6. Effects of the Number of Training Samples on the Classification Performance of Three Deep Learning Networks
4. Discussion
4.1. Influence of Different Data Sources on Individual Tree Segmentation
4.2. Performance of Deep Learning Networks and Effect of Texture metrics on Tree Species Classification
4.3. Effect of Variation in the Number of Training Samples on Classification Accuracy
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, E.H.; Tian, G.L.; Chiu, C.Y. Soil Microbial Communities in Natural and Managed Cloud Montane Forests. Forests 2017, 8, 33. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Bian, Z.Q.; Ding, S.Y. Consequences of Spatial Heterogeneity of Forest Landscape on Ecosystem Water Conservation Service in the Yi River Watershed in Central China. Sustainability 2020, 12, 1170. [Google Scholar] [CrossRef]
- Zald, H.S.J.; Spies, T.A.; Harmon, M.E.; Twery, M.J. Forest Carbon Calculators: A Review for Managers, Policymakers, and Educators. J. For. 2016, 114, 134–143. [Google Scholar] [CrossRef]
- Brockerhoff, E.G.; Barbaro, L.; Castagneyrol, B.; Forrester, D.I.; Gardiner, B.; González-Olabarria, J.R.; Lyver, P.O.B.; Meurisse, N.; Oxbrough, A.; Taki, H.; et al. Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodivers. Conserv. 2017, 26, 3005–3035. [Google Scholar] [CrossRef]
- Hartley, M.J. Rationale and methods for conserving biodiversity in plantation forests. For. Ecol. Manag. 2002, 155, 81–95. [Google Scholar] [CrossRef]
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Modzelewska, A.; Fassnacht, F.E.; Stereńczak, K. Tree species identification within an extensive forest area with diverse management regimes using airborne hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101960. [Google Scholar] [CrossRef]
- Piiroinen, R.; Fassnacht, F.E.; Heiskanen, J.; Maeda, E.; Mack, B.; Pellikka, P. Invasive tree species detection in the Eastern Arc Mountains biodiversity hotspot using one class classification. Remote Sens. Environ. 2018, 218, 119–131. [Google Scholar] [CrossRef]
- Jonsson, M.; Bengtsson, J.; Gamfeldt, L.; Moen, J.; Snäll, T. Levels of forest ecosystem services depend on specific mixtures of commercial tree species. Nat. Plants 2019, 5, 141–147. [Google Scholar] [CrossRef]
- Ali, A.; Mattsson, E. Disentangling the effects of species diversity, and intraspecific and interspecific tree size variation on aboveground biomass in dry zone homegarden agroforestry systems. Sci. Total Environ. 2017, 598, 38–48. [Google Scholar] [CrossRef]
- Fichtner, A.; Hardtle, W.; Li, Y.; Bruelheide, H.; Kunz, M.; von Oheimb, G. From competition to facilitation: How tree species respond to neighbourhood diversity. Ecol. Lett. 2017, 20, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. Remote Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Feret, J.-B.; Asner, G.P.; Sensing, R. Tree Species Discrimination in Tropical Forests Using Airborne Imaging Spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Li, J.L.; Hu, B.X.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171, 104–114. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Sam, L.; Akanksha; Martin-Torres, F.J.; Kumar, R. UAVs as remote sensing platform in glaciology: Present applications and future prospects. Remote Sens. Environ. 2016, 175, 196–204. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Fu, Y. Forest type identification by random forest classification combined with SPOT and multitemporal SAR data. J. For. Res. 2018, 29, 1407–1414. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Cho, M.A.; Malahlela, O.E.; Ramoelo, A.J. Assessing the utility WorldView-2 imagery for tree species mapping in South African subtropical humid forest and the conservation implications: Dukuduku forest patch as case study. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 349–357. [Google Scholar] [CrossRef]
- Johansen, K.; Phinn, S. Mapping indicators of riparian vegetation health using IKONOS and Landsat-7 ETM+ image data in Australian tropical savannas. In Proceedings of the IGARSS 2004, 2004 IEEE International Geoscience and Remote Sensing Symposium, Anchorage, AK, USA, 20–24 September 2004; Volume 1553, pp. 1559–1562. [Google Scholar]
- Mallinis, G.; Koutsias, N.; Tsakiri-Strati, M.; Karteris, M. Object-based classification using Quickbird imagery for delineating forest vegetation polygons in a Mediterranean test site. ISPRS J. Photogramm. Remote Sens. 2008, 63, 237–250. [Google Scholar] [CrossRef]
- Deur, M.; Gasparovic, M.; Balenović, I. Tree Species Classification in Mixed Deciduous Forests Using Very High Spatial Resolution Satellite Imagery and Machine Learning Methods. Remote Sens. 2020, 12, 3926. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.O.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, J.B.; Lian, J.Y.; Fan, Z.J.; Ouyang, X.J.; Ye, W.H. Seeing the forest from drones: Testing the potential of lightweight drones as a tool for long-term forest monitoring. Biol. Conserv. 2016, 198, 60–69. [Google Scholar] [CrossRef]
- Reis, B.P.; Martins, S.V.; Fernandes, E.I.; Sarcinelli, T.S.; Gleriani, J.M.; Leite, H.G.; Halassy, M. Forest restoration monitoring through digital processing of high resolution images. Ecol. Eng. 2019, 127, 178–186. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Nuijten, R.; Coops, N.; Goodbody, T.; Pelletier, G. Examining the Multi-Seasonal Consistency of Individual Tree Segmentation on Deciduous Stands Using Digital Aerial Photogrammetry (DAP) and Unmanned Aerial Systems (UAS). Remote Sens. 2019, 11, 739. [Google Scholar] [CrossRef]
- Chen, L.; Jia, J.; Wang, H.J. An overview of applying high resolution remote sensing to natural resources survey. Remote Sens. Nat. Resour. 2019, 31, 1–7. [Google Scholar]
- Marques, P.; Pádua, L.; Adão, T.; Hruška, J.; Peres, E.; Sousa, A.; Sousa, J.J. UAV-Based Automatic Detection and Monitoring of Chestnut Trees. Remote Sens. 2019, 11, 855. [Google Scholar] [CrossRef]
- Ryherd, S.; Woodcock, C. Combining Spectral and Texture Data in the Segmentation of Remotely Sensed Images. Photogramm. Eng. Remote Sens. 1996, 62, 181–194. [Google Scholar]
- Blaschke, T. Object-based contextual image classification built on image segmentation. In Proceedings of the IEEE Workshop on Advances in Techniques for Analysis of Remotely Sensed Data, Greenbelt, MD, USA, 27–28 October 2003; pp. 113–119. [Google Scholar]
- Elisabeth, A.A. The Importance of Scale in Object-based Mapping of Vegetation Parameters with Hyperspectral Imagery. Photogramm. Eng. Remote Sens. 2007, 73, 905–912. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Zhong, T.; Zhao, W.; Sun, Q.; Ba, S.; Zhang, Z.; et al. Tree species classification using UAS-based digital aerial photogrammetry point clouds and multispectral imageries in subtropical natural forests. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. Isprs J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. Isprs J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Natesan, S.; Armenakis, C.; Vepakomma, U. Resnet-Based Tree Species Classification Using Uav Images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W13, 475–481. [Google Scholar] [CrossRef]
- Guo, X.; Li, H.; Jing, L.; Wang, P. Individual Tree Species Classification Based on Convolutional Neural Networks and Multitemporal High-Resolution Remote Sensing Images. Sensors 2022, 22, 3157. [Google Scholar] [CrossRef]
- Yan, S.J.; Jing, L.H.; Wang, H. A New Individual Tree Species Recognition Method Based on a Convolutional Neural Network and High-Spatial Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 479. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Rood, S.B.; Peddle, D.R. See the forest and the trees: Effective machine and deep learning algorithms for wood filtering and tree species classification from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 1–16. [Google Scholar] [CrossRef]
- Onishi, M.; Ise, T. Explainable identification and mapping of trees using UAV RGB image and deep learning. Sci. Rep. 2021, 11, 903. [Google Scholar] [CrossRef]
- Zhang, C.; Xia, K.; Feng, H.; Yang, Y.; Du, X. Tree species classification using deep learning and RGB optical images obtained by an unmanned aerial vehicle. J. For. Res. 2020, 32, 1879–1888. [Google Scholar] [CrossRef]
- Li, H.; Jing, L.; Tang, Y.; Ding, H. An Improved Pansharpening Method for Misaligned Panchromatic and Multispectral Data. Sensors 2018, 18, 557. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.; Choi, K.; Cho, W.; Chang, B.; Ko, D.W. Efficient dead pine tree detecting method in the Forest damaged by pine wood nematode (Bursaphelenchus xylophilus) through utilizing unmanned aerial vehicles and deep learning-based object detection techniques. For. Sci. Technol. 2022, 18, 36–43. [Google Scholar] [CrossRef]
- Long, Y.; Gong, Y.; Xiao, Z.; Liu, Q.J. Accurate Object Localization in Remote Sensing Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2486–2498. [Google Scholar] [CrossRef]
- Cheng, G.; Zhou, P.; Han, J. Learning Rotation-Invariant Convolutional Neural Networks for Object Detection in VHR Optical Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 7405–7415. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, X.; Fu, K.; Wang, C.; Wang, H. Object Detection in High-Resolution Remote Sensing Images Using Rotation Invariant Parts Based Model. IEEE Geosci. Remote Sens. Lett. 2014, 11, 74–78. [Google Scholar] [CrossRef]
- Ju, C.-H.; Tian, Y.-C.; Yao, X.; Cao, W.-X.; Zhu, Y.; Hannaway, D. Estimating Leaf Chlorophyll Content Using Red Edge Parameters. Pedosphere 2010, 20, 633–644. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanré, D. Strategy for direct and indirect methods for correcting the aerosol effect on remote sensing: From AVHRR to EOS-MODIS. Remote Sens. Environ. 1996, 55, 65–79. [Google Scholar] [CrossRef]
- Verstraete, M.M.; Pinty, B. Designing optimal spectral indexes for remote sensing applications. IEEE Trans. Geosci. Remote Sens. 1996, 34, 1254–1265. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Peñuelas, J.; Gamon, J.A.; Griffin, K.L.; Field, C.B. Assessing community type, plant biomass, pigment composition, and photosynthetic efficiency of aquatic vegetation from spectral reflectance. Remote Sens. Environ. 1993, 46, 110–118. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.K.; Abkar, A.A. Leaf Area Index derivation from hyperspectral vegetation indicesand the red edge position. Int. J. Remote Sens. 2009, 30, 6199–6218. [Google Scholar] [CrossRef]
- Fraser, R.H.; Sluijs, J.v.d.; Hall, R.J.J. Calibrating Satellite-Based Indices of Burn Severity from UAV-Derived Metrics of a Burned Boreal Forest in NWT, Canada. Remote Sens. 2017, 9, 279. [Google Scholar] [CrossRef]
- Yuan, J.; Niu, Z.; Fu, W. Model simulation for sensitivity of hyperspectral indices to LAI, leaf chlorophyll, and internal structure parameter. In Proceedings of the Geoinformatics 2007: Remotely Sensed Data and Information, Nanjing, China, 8 August 2007. [Google Scholar] [CrossRef]
- Gamon, J.; Surfus, J. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zhu, R.; Zeng, D.; Kosorok, M.R.J. Reinforcement Learning Trees. J. Am. Stat. Assoc. 2015, 110, 1770. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.L.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2016; pp. 2261–2269. [Google Scholar]
- Benz, U.C.; Hofmann, P.; Willhauck, G.; Lingenfelder, I.; Heynen, M. Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS J. Photogramm. Remote Sens. 2004, 58, 239–258. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation. In Advances in Information Retrieval, Proceedings of the 27th European Conference on IR Research, ECIR 2005, Santiago de Compostela, Spain,21–23 March 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 345–359. [Google Scholar]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In AI 2006: Advances in Artificial Intelligence, Proceedings of the 19th Australian Joint Conference on Artificial Intelligence, Hobart, Australia, 4–8 December 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1015–1021. [Google Scholar]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a WorldView-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef]
- Wang, X.; Tan, L.; Fan, J. Performance Evaluation of Mangrove Species Classification Based on Multi-Source Remote Sensing Data Using Extremely Randomized Trees in Fucheng Town, Leizhou City, Guangdong Province. Remote Sens. 2023, 15, 1386. [Google Scholar] [CrossRef]
- Lisein, J.; Pierrot-Deseilligny, M.; Bonnet, S.; Lejeune, P. A Photogrammetric Workflow for the Creation of a Forest Canopy Height Model from Small Unmanned Aerial System Imagery. Forests 2013, 4, 922–944. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X. Individual Tree Parameters Estimation for Plantation Forests Based on UAV Oblique Photography. IEEE Access 2020, 8, 96184–96198. [Google Scholar] [CrossRef]
- Zhong, H.; Lin, W.S.; Liu, H.R.; Ma, N.; Liu, K.K.; Cao, R.Z.; Wang, T.T.; Ren, Z.Z. Identification of tree species based on the fusion of UAV hyperspectral image and LiDAR data in a coniferous and broad-leaved mixed forest in Northeast China. Front. Plant Sci. 2022, 13, 964769. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Knox, R.G.; Clark, D.B.; Blair, J.B. Sensitivity of large-footprint lidar to canopy structure and biomass in a neotropical rainforest. Remote Sens. Environ. 2002, 81, 378–392. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Maimaitijiang, M. Urban tree species classification using UAV-based multi-sensor data fusion and machine learning. GISci. Remote Sens. 2021, 58, 1–26. [Google Scholar] [CrossRef]
- Matsuki, T.; Yokoya, N.; Iwasaki, A. Hyperspectral Tree Species Classification of Japanese Complex Mixed Forest with the Aid of Lidar Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2177–2187. [Google Scholar] [CrossRef]
- Gini, R.; Sona, G.; Ronchetti, G.; Passoni, D.; Pinto, L. Improving Tree Species Classification Using UAS Multispectral Images and Texture Measures. ISPRS Int. J. Geo-Inf. 2018, 7, 315. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef]




| Scientific Name | N | DBH (cm) | Height (m) | Crown Width (m) | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Alnus nepalensis (A.N.) | 118 | 27.87 | 21.01 | 13.10 | 6.63 | 5.32 | 1.91 |
| Quercus aliena (Q.A.) | 80 | 55.72 | 19.37 | 23.41 | 6.07 | 6.76 | 2.30 |
| Populus davidiana (P.D.) | 97 | 30.96 | 23.07 | 11.34 | 5.56 | 5.44 | 2.22 |
| Acer forrestii (A.F.) | 80 | 37.90 | 18.65 | 14.59 | 4.93 | 6.97 | 2.20 |
| Pinus yunnanensis (P.Y.) | 75 | 38.91 | 17.95 | 22.23 | 8.04 | 5.31 | 1.58 |
| Class | Training | Validation | Total |
|---|---|---|---|
| A.N. | 472 | 118 | 590 |
| Q.A. | 320 | 80 | 400 |
| P.D. | 388 | 97 | 485 |
| A.F. | 320 | 80 | 400 |
| P.Y. | 300 | 75 | 375 |
| Total | 1800 | 450 | 2250 |
| Metrics | Equation | Reference |
|---|---|---|
| Difference Vegetation Index (DVI) | ρnir − ρred | [49] |
| Atmospherically Resistant Vegetation Index (ARVI) | (ρnir − ρrb)/(ρnir + ρrb), ρrb = ρred – γ × (ρblue − ρred), γ = 0.5 | [50] |
| Green Normalized Difference Vegetation Index (GNDVI) | (ρnir − ρgreen)/(ρnir + ρgreen) | [51] |
| Modified triangular vegetation index 2 (MTVI2) | [1.5 × (1.2 × (ρnir − ρgreen) − 2.5 × (ρred −ρgreen)]/[(2 × ρnir+1)2 – (6 × ρnir – 5 × ρred0.5) – 0.5]0.5 | [52] |
| Normalized Difference Vegetation Index (NDVI) | (ρnir – ρred)/(ρnir + ρred) | [52] |
| Simple Ration Vegetation Index (SR) | ρnir/ρred | [53] |
| Soil Adjusted Vegetation Index (SAVI) | 1.5 × (ρnir − ρred)/(ρnir + ρred + 0.5) | [52] |
| Ratio Vegetation Index (RVI) | ρred/ρnir | [54] |
| Normalized Greenness (Norm G) | ρgreen/(ρred + ρgreen+ ρblue) | [55] |
| Normalized Green-Red Ratio (Norm GR) | (ρgreen – ρred)/(ρgreen + ρred) | [55] |
| Optimized Soil Adjusted Vegetation Index (OSAVI) | (ρnir – ρred)/(ρnir + ρred + 0.16) | [56] |
| Red Green Ratio Index (RGRI) | ρred/ρgreen | [57] |
| Metrics | Equation |
|---|---|
| Correlation (CR) | CR = |
| Contrast (CO) | CO = |
| Dissimilarity (DI) | DI = |
| Entropy (EN) | EN = (−) |
| Homogeneity (HO) | HO = |
| Mean (ME) | ME = |
| Variance (VA) | VA = ) |
| Density | Nt | No | Nc | r (%) | p (%) | F1 (%) |
|---|---|---|---|---|---|---|
| Satellite multispectral imagery | ||||||
| Low | 28 | 13 | 6 | 68.3 | 82.4 | 74.7 |
| Middle | 31 | 21 | 4 | 59.6 | 88.6 | 71.3 |
| High | 49 | 29 | 8 | 62.8 | 85.9 | 72.6 |
| UAV high-resolution RGB imagery | ||||||
| Low | 27 | 14 | 3 | 65.9 | 90.0 | 76.1 |
| Middle | 38 | 14 | 6 | 73.1 | 86.4 | 79.2 |
| High | 52 | 26 | 8 | 66.7 | 86.7 | 75.4 |
| Data Source | Nt | No | Nc | r (%) | p (%) | F1 (%) |
|---|---|---|---|---|---|---|
| Satellite multispectral imagery | 273 | 177 | 91 | 60.7 | 75.0 | 67.1 |
| UAV high-resolution RGB imagery | 318 | 132 | 111 | 70.7 | 74.1 | 72.4 |
| Class | A.N. | Q.A. | A.F. | P.D. | P.Y. |
|---|---|---|---|---|---|
| Spectral metrics | |||||
| A.N. | 23 | 2 | 6 | 3 | 5 |
| Q.A. | 1 | 7 | 3 | 3 | 0 |
| A.F. | 2 | 0 | 6 | 2 | 0 |
| P.D. | 1 | 0 | 2 | 10 | 0 |
| P.Y. | 2 | 1 | 1 | 2 | 8 |
| Overall Accuracy | 60.00% | Kappa Accuracy | 47.80% | ||
| Spectral and texture metrics | |||||
| A.N. | 24 | 1 | 4 | 2 | 5 |
| Q.A. | 2 | 7 | 1 | 1 | 0 |
| A.F. | 1 | 1 | 8 | 2 | 0 |
| P.D. | 1 | 0 | 3 | 12 | 1 |
| P.Y. | 1 | 1 | 2 | 3 | 7 |
| Overall Accuracy | 64.44% | Kappa Accuracy | 53.61% | ||
| Class | A.N. | Q.A. | A.F. | P.D. | P.Y. |
|---|---|---|---|---|---|
| Light-weight network MobileNetV2 | |||||
| A.N. | 20 | 2 | 2 | 2 | 1 |
| Q.A. | 3 | 7 | 1 | 3 | 3 |
| A.F. | 2 | 0 | 13 | 0 | 0 |
| P.D. | 4 | 1 | 1 | 15 | 0 |
| P.Y. | 0 | 0 | 1 | 0 | 9 |
| Over Accuracy | 71.11% | Kappa Accuracy | 63.01% | ||
| Residual network ResNet34 | |||||
| A.N. | 24 | 1 | 1 | 2 | 0 |
| Q.A. | 1 | 8 | 0 | 2 | 0 |
| A.F. | 3 | 0 | 13 | 0 | 3 |
| P.D. | 0 | 1 | 0 | 16 | 0 |
| P.Y. | 1 | 0 | 4 | 0 | 10 |
| Over Accuracy | 78.89% | Kappa Accuracy | 72.86% | ||
| Dense network DenseNet121 | |||||
| A.N. | 24 | 2 | 3 | 1 | 0 |
| Q.A. | 1 | 7 | 0 | 2 | 0 |
| A.F. | 4 | 0 | 14 | 0 | 2 |
| P.D. | 0 | 1 | 0 | 17 | 0 |
| P.Y. | 0 | 0 | 1 | 0 | 11 |
| Over Accuracy | 81.11% | Kappa Accuracy | 75.53% | ||
| Class | A.N. | Q.A. | A.F. | P.D. | P.Y. |
|---|---|---|---|---|---|
| Light-weight network MobileNetV2 | |||||
| A.N. | 25 | 0 | 2 | 1 | 1 |
| Q.A. | 1 | 8 | 1 | 1 | 0 |
| A.F. | 0 | 1 | 12 | 0 | 1 |
| P.D. | 1 | 0 | 1 | 18 | 0 |
| P.Y. | 2 | 1 | 2 | 0 | 11 |
| Over Accuracy | 82.22% | Kappa Accuracy | 77.09% | ||
| Residual network ResNet34 | |||||
| A.N. | 27 | 1 | 0 | 1 | 0 |
| Q.A. | 0 | 8 | 0 | 0 | 0 |
| A.F. | 2 | 1 | 16 | 0 | 1 |
| P.D. | 0 | 0 | 0 | 19 | 0 |
| P.Y. | 0 | 0 | 2 | 0 | 12 |
| Over Accuracy | 91.11% | Kappa Accuracy | 88.49% | ||
| Dense network DenseNet121 | |||||
| A.N. | 28 | 2 | 0 | 0 | 0 |
| Q.A. | 1 | 8 | 0 | 0 | 0 |
| A.F. | 0 | 0 | 17 | 0 | 1 |
| P.D. | 0 | 0 | 0 | 20 | 0 |
| P.Y. | 0 | 0 | 1 | 0 | 12 |
| Over Accuracy | 94.44% | Kappa Accuracy | 92.79% | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Shen, X.; Cao, L. Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China. Remote Sens. 2023, 15, 2697. https://doi.org/10.3390/rs15102697
Chen X, Shen X, Cao L. Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China. Remote Sensing. 2023; 15(10):2697. https://doi.org/10.3390/rs15102697
Chicago/Turabian StyleChen, Xianggang, Xin Shen, and Lin Cao. 2023. "Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China" Remote Sensing 15, no. 10: 2697. https://doi.org/10.3390/rs15102697
APA StyleChen, X., Shen, X., & Cao, L. (2023). Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China. Remote Sensing, 15(10), 2697. https://doi.org/10.3390/rs15102697






