Advances in the Application of Small Unoccupied Aircraft Systems (sUAS) for High-Throughput Plant Phenotyping
Abstract
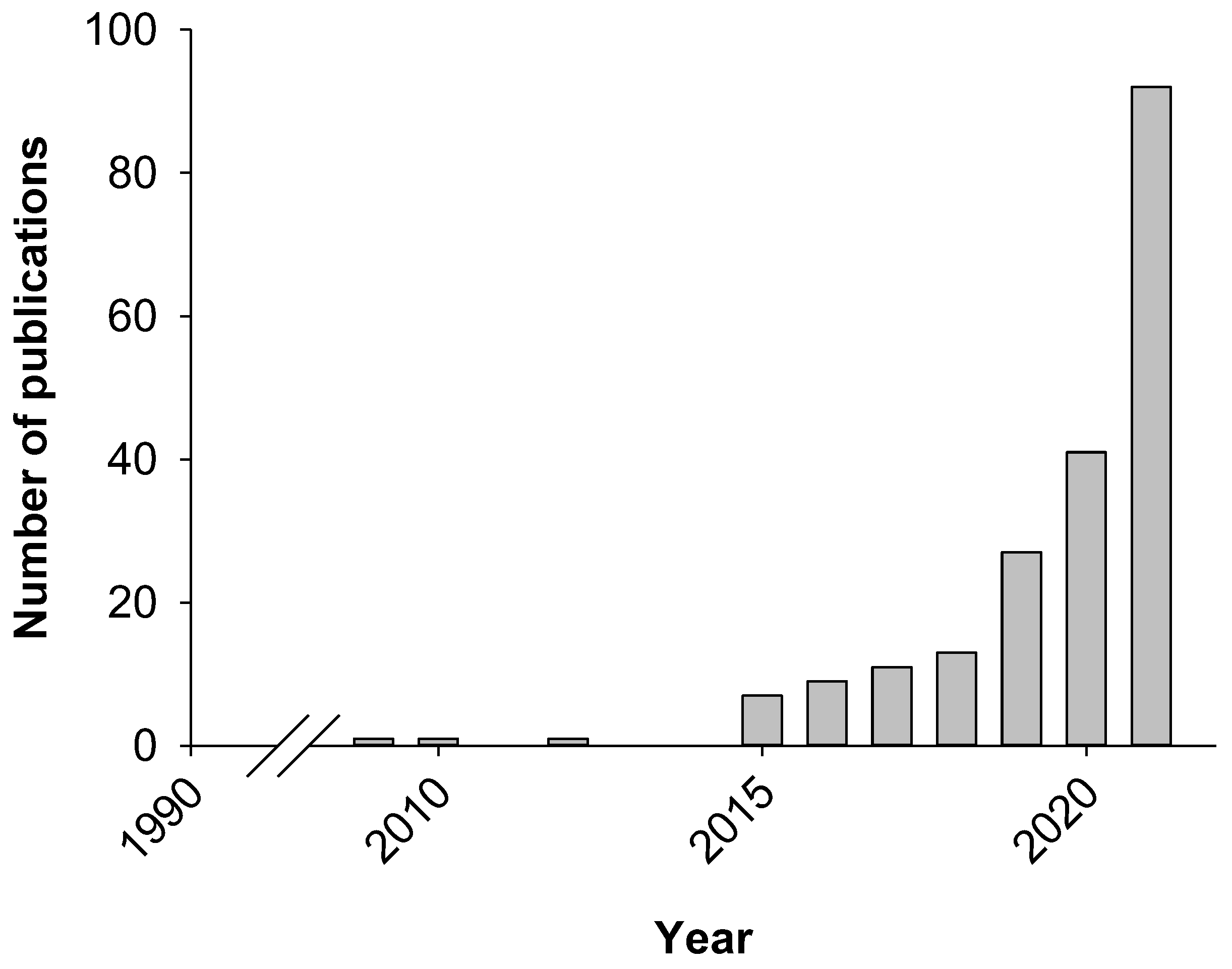
1. Introduction
Literature Survey
2. Common sUAS-Based Imaging Techniques in HTPP
2.1. Color Imaging
2.2. Thermal Imaging
2.3. Imaging Spectroscopy
2.4. Light Detection and Ranging (LiDAR) Imaging
3. sUAS-Based Image Processing and Data Analysis
4. Common sUAS-Based Trait Extraction for HTPP
4.1. Plant Growth
| Crop | Sensor | Traits | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RGB | Multi-Spectral | Hyper-Spectral | Thermal | LiDAR | Plant Height | Canopy Cover | Leaf Area Index | Biomass | Salinity Stress | Drought Stress | Nutrient Stress | NUE, WUE | Diseases | Yield | Veg. Index | Other Traits | Source | |
| Soybean | X | X | X | Borra-Serrano et al. [69] | ||||||||||||||
| X | X | X | X | X | Maimaitijiang et al. [50] | |||||||||||||
| X | X | Sagan et al. [55] | ||||||||||||||||
| Corn | X | X | X | X | X | Su et al. [122] | ||||||||||||
| X | X | X | X | X | Yang et al. [123] | |||||||||||||
| X | X | X | X | X | Han et al. [85] | |||||||||||||
| X | X | Wang et al. [135] | ||||||||||||||||
| X | X | Chivasa et al. [49] | ||||||||||||||||
| X | X | Stewart et al. [136] | ||||||||||||||||
| Cotton | X | X | X | X | Xu et al. [52] | |||||||||||||
| X | X | Thompson et al. [53] | ||||||||||||||||
| X | X | X | X | X | Xu et al. [52] | |||||||||||||
| X | X | Thorp et al. [4] | ||||||||||||||||
| Wheat | X | X | Yang et al. [86] | |||||||||||||||
| X | X | Perich et al. [111] | ||||||||||||||||
| X | Moghimi et al. [56] | |||||||||||||||||
| X | X | X | X | Gracia-Romero et al. [137] | ||||||||||||||
| X | X | Sankaran et al. [125] | ||||||||||||||||
| X | X | Camino et al. [138] | ||||||||||||||||
| Wheat | X | X | X | Gonzalez-Dugo et al. [139] | ||||||||||||||
| X | X | X | Ostos-Garrido et al. [57] | |||||||||||||||
| Sorghum | X | X | Hu et al. [124] | |||||||||||||||
| X | X | Sagan et al. [55] | ||||||||||||||||
| Barley | X | X | X | Ostos-Garrido et al. [57] | ||||||||||||||
| X | X | X | X | Kefauver et al. [110] | ||||||||||||||
| Dry bean | X | X | X | Sankaran et al. [51] | ||||||||||||||
| X | X | X | Sankaran et al. [134] | |||||||||||||||
| Rice | X | X | X | X | X | Fenghua et al. [70] | ||||||||||||
| Potato | X | X | Sugiura et al. [140] | |||||||||||||||
| Blueberry | X | X | X | Patrick and Li [121] | ||||||||||||||
| Peanut | X | X | Patrick et al. [141] | |||||||||||||||
| Citrus | X | X | X | Ampatzidis and Patel [87] | ||||||||||||||
| Tomato | X | X | X | Johansen et al. [142] | ||||||||||||||
| Sugar beet | X | X | X | Harkel et al. [106] | ||||||||||||||
| Bioenergy crop | X | X | X | Maesano et al. [102] | ||||||||||||||
4.2. Abiotic Stress Resilience and Adaptation
4.3. Nutrient- and Water-Use Efficiencies and Crop Yield
4.4. Disease Detection and Crop Resilience to Biotic Stress
4.5. Other Areas of Application
5. Comparing Crop Trait Estimation from Imaging Sensors on Terrestrial versus sUAS Platforms
5.1. Plant Height Estimation
5.2. Canopy Cover and Leaf Area Index
5.3. Biomass
5.4. Yield Estimation
6. Suggested Directions for Future Research
7. Discussion
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent Advances of Hyperspectral Imaging Technology and Applications in Agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.A.T.; Edmeades, G.O. Breeding and Cereal Yield Progress. Crop. Sci. 2010, 50, S-85–S-98. [Google Scholar] [CrossRef]
- Thorp, K.R.; Thompson, A.L.; Harders, S.J.; French, A.N.; Ward, R.W. High-Throughput Phenotyping of Crop Water Use Efficiency via Multispectral Drone Imagery and a Daily Soil Water Balance Model. Remote Sens. 2018, 10, 1682. [Google Scholar] [CrossRef]
- Ayankojo, I.T.; Thorp, K.R.; Morgan, K.; Kothari, K.; Ale, S. Assessing the Impacts of Future Climate on Cotton Production in the Arizona Low Desert. Trans. ASABE 2020, 63, 1087–1098. [Google Scholar] [CrossRef]
- Ayankojo, I.T.; Morgan, K.T. Increasing Air Temperatures and Its Effects on Growth and Productivity of Tomato in South Florida. Plants 2020, 9, 1245. [Google Scholar] [CrossRef]
- Ali, M.H.; Talukder, M.S.U. Increasing water productivity in crop production—A synthesis. Agric. Water Manag. 2008, 95, 1201–1213. [Google Scholar] [CrossRef]
- Brauman, K.A.; Siebert, S.; Foley, J.A. Improvements in crop water productivity increase water sustainability and food security—A global analysis. Environ. Res. Lett. 2013, 8, 024030. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- De Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; De Lacerda, C.F.; Silva, J.V.; Da Costa, P.H.A.; Gomes-Filho, E. Effects of salt stress on plant growth, stomatal response and solute accumulation of different maize genotypes. Braz. J. Plant Physiol. 2004, 16, 31–38. [Google Scholar] [CrossRef]
- Thu, T.T.P.; Yasui, H.; Yamakawa, T. Effects of salt stress on plant growth characteristics and mineral content in diverse rice genotypes. Soil Sci. Plant Nutr. 2017, 63, 264–273. [Google Scholar] [CrossRef]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C.; et al. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 3890. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Byrd, S.A.; Rowland, D.L.; Bennett, J.; Zotarelli, L.; Wright, D.; Alva, A.; Nordgaard, J. Reductions in a Commercial Potato Irrigation Schedule during Tuber Bulking in Florida: Physiological, Yield, and Quality Effects. J. Crop Improv. 2014, 28, 660–679. [Google Scholar] [CrossRef]
- Rowland, D.L.; Faircloth, W.H.; Payton, P.; Tissue, D.T.; Ferrell, J.A.; Sorensen, R.B.; Butts, C.L. Primed acclimation of cultivated peanut (Arachis hypogaea L.) through the use of deficit irrigation timed to crop developmental periods. Agric. Water Manag. 2012, 113, 85–95. [Google Scholar] [CrossRef]
- Vincent, C.; Rowland, D.; Schaffer, B.; Bassil, E.; Racette, K.; Zurweller, B. Primed acclimation: A physiological process offers a strategy for more resilient and irrigation-efficient crop production. Plant. Sci. 2019, 203, 29–40. [Google Scholar] [CrossRef]
- Liu, S.; Li, X.; Larsen, D.H.; Zhu, X.; Song, F.; Liu, F. Drought Priming at Vegetative Growth Stage Enhances Nitrogen-Use Efficiency Under Post-Anthesis Drought and Heat Stress in Wheat. J. Agron. Crop Sci. 2017, 203, 29–40. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Zaman-Allah, M.; Vergara, O.; Araus, J.L.; Tarekegne, A.; Magorokosho, C.; Zarco-Tejada, P.J.; Hornero, A.; Alba, A.H.; Das, B.; Craufurd, P.; et al. Unmanned aerial platform-based multi-spectral imaging for field phenotyping of maize. Plant Methods 2015, 11, 35. [Google Scholar] [CrossRef]
- Rebetzke, G.; Fischer, R.; Deery, D.; Jimenez-Berni, J.; Smith, D. Review: High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal Remote Sensing Buggies and Potential Applications for Field-Based Phenotyping. Agronomy 2014, 4, 349–379. [Google Scholar] [CrossRef]
- Feng, L.; Chen, S.; Zhang, C.; Zhang, Y.; He, Y. A comprehensive review on recent applications of unmanned aerial vehicle remote sensing with various sensors for high-throughput plant phenotyping. Comput. Electron. Agric. 2021, 182, 106033. [Google Scholar] [CrossRef]
- Klukas, C.; Chen, D.; Pape, J.M. Integrated analysis platform: An open-source information system for high-throughput plant phenotyping. Plant. Physiol. 2014, 165, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Knecht, A.C.; Campbell, M.T.; Caprez, A.; Swanson, D.R.; Walia, H. Image Harvest: An open-source platform for high-throughput plant image processing and analysis. J. Exp. Bot. 2016, 67, 3587–3599. [Google Scholar] [CrossRef]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High Throughput In vivo Analysis of Plant Leaf Chemical Properties Using Hyperspectral Imaging. Front. Plant Sci. 2017, 8, 1348. [Google Scholar] [CrossRef] [PubMed]
- Golzarian, M.R.; Frick, R.A.; Rajendran, K.; Berger, B.; Roy, S.; Tester, M.; Lun, D.S. Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 2011, 7, 20. [Google Scholar] [CrossRef]
- Fahlgren, N.; Feldman, M.; Gehan, M.A.; Wilson, M.S.; Shyu, C.; Bryant, D.W.; Hill, S.T.; McEntee, C.J.; Warnasooriya, S.N.; Kumar, I. A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 2015, 8, 1520–1535. [Google Scholar] [CrossRef]
- Campbell, M.T.; Knecht, A.C.; Berger, B.; Brien, C.J.; Wang, D.; Walia, H. Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 2015, 168, 1476–1489. [Google Scholar] [CrossRef]
- Thorp, K.R.; Dierig, D.A. Color image segmentation approach to monitor flowering in lesquerella. Ind. Crop. Prod. 2011, 34, 1150–1159. [Google Scholar] [CrossRef]
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Kluka, C. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis w open. Plant Cell 2014, 26, 4636–4655. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef]
- Neilson, E.H.; Edwards, A.M.; Blomstedt, C.K.; Berger, B.; Møller, B.L.; Gleadow, R.M. Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. J. Exp. Bot. 2015, 66, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Arvidsson, S.; Pérez-Rodríguez, P.; Mueller-Roeber, B. A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol. 2011, 191, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Aguirrezabal, L.; Chenu, K.; Cookson, S.J.; Dauzat, M.; Hamard, P.; Thioux, J.; Rolland, G.; Bouchier-Combaud, S.; Lebaudy, A.; et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006, 169, 623–635. [Google Scholar] [CrossRef]
- Hartmann, A.; Czauderna, T.; Hoffmann, R.; Stein, N.; Schreiber, F. HTPheno: An image analysis pipeline for high-throughput plant phenotyping. BMC Bioinform. 2011, 12, 148. [Google Scholar] [CrossRef]
- Friedli, M.; Kirchgessner, N.; Grieder, C.; Liebisch, F.; Mannale, M.; Walter, A. Terrestrial 3D laser scanning to track the increase in canopy height of both monocot and dicot crop species under field conditions. Plant Methods 2016, 12, 9. [Google Scholar] [CrossRef]
- Raesch, A.R.; Muller, O.; Pieruschka, R.; Rascher, U. Field Observations with Laser-Induced Fluorescence Transient (LIFT) Method in Barley and Sugar Beet. Agriculture 2014, 4, 159–169. [Google Scholar] [CrossRef]
- Bai, G.; Ge, Y.; Hussain, W.; Baenziger, P.S.; Graef, G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 2016, 128, 181–192. [Google Scholar] [CrossRef]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-Based High-Throughput Phenotyping for Maize Plant Using 3D LiDAR Point Cloud Generated With a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef] [PubMed]
- Sunil, N.; Rekha, B.; Yathish, K.R.; Sehkar, J.C.; Ramesh, P.; Vadez, V. LeasyScan-an efficient phenotyping platform for identification of pre-breeding genetic stocks in maize. Maize J. 2018, 7, 16–22. [Google Scholar]
- Vadez, V.; Kholová, J.; Hummel, G.; Zhokhavets, U.; Gupta, S.K.; Hash, C.T. LeasyScan: A novel concept combining 3D imaging and lysimetry for high-throughput phenotyping of traits controlling plant water budget. J. Exp. Bot. 2015, 66, 5581–5593. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yang, C. A review on plant high-throughput phenotyping traits using UAV-based sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Meyering, B.; Albrecht, U. Citrus rootstock evaluation utilizing UAV-based remote sensing and artificial intelligence. Comput. Electron. Agric. 2019, 164, 104900. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Costa, L. Agroview: Cloud-based application to process, analyze and visualize UAV-collected data for precision agriculture applications utilizing artificial intelligence. Comput. Electron. Agric. 2020, 174, 105457. [Google Scholar] [CrossRef]
- Li, D.; Quan, C.; Song, Z.; Li, X.; Yu, G.; Li, C.; Mohammad, A. High-Throughput Plant Phenotyping Platform (HT3P) as a Novel Tool for Estimating Agronomic Traits From the Lab to the Field. Front. Bioeng. Biotechnol. 2021, 8, 623705. [Google Scholar] [CrossRef]
- Chivasa, W.; Mutanga, O.; Biradar, C. UAV-based multispectral phenotyping for disease resistance to accelerate crop improvement under changing climate conditions. Remote Sens. 2020, 12, 2445. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Ghulam, A.; Sidike, P.; Hartling, S.; Maimaitiyiming, M.; Peterson, K.; Shavers, E.; Fishman, J.; Peterson, J.; Kadam, S.; et al. Unmanned Aerial System (UAS)-based phenotyping of soybean using multi-sensor data fusion and extreme learning machine. ISPRS J. Photogramm. Remote Sens. 2017, 134, 43–58. [Google Scholar] [CrossRef]
- Sankaran, S.; Quirós, J.J.; Miklas, P.N. Unmanned aerial system and satellite-based high resolution imagery for high-throughput phenotyping in dry bean. Comput. Electron. Agric. 2019, 165, 104965. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Paterson, A.H. Multispectral imaging and unmanned aerial systems for cotton plant phenotyping. PLoS ONE 2019, 14, e0205083. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.; Thorp, K.; Conley, M.; Elshikha, D.; French, A.; Andrade-Sanchez, P.; Pauli, D. Comparing Nadir and Multi-Angle View Sensor Technologies for Measuring in-Field Plant Height of Upland Cotton. Remote Sens. 2019, 11, 700. [Google Scholar] [CrossRef]
- Kawamura, K.; Asai, H.; Yasuda, T.; Khanthavong, P.; Soisouvanh, P.; Phongchanmixay, S. Field phenotyping of plant height in an upland rice field in Laos using low-cost small unmanned aerial vehicles (UAVs). Plant Prod. Sci. 2020, 23, 452–465. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitijiang, M.; Sidike, P.; Eblimit, K.; Peterson, K.; Hartling, S.; Esposito, F.; Khanal, K.; Newcomb, M.; Pauli, D.; et al. UAV-Based High Resolution Thermal Imaging for Vegetation Monitoring, and Plant Phenotyping Using ICI 8640 P, FLIR Vue Pro R 640, and thermoMap Cameras. Remote Sens. 2019, 11, 330. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Anderson, J.A. Aerial hyperspectral imagery and deep neural networks for high-throughput yield phenotyping in wheat. Comput. Electron. Agric. 2020, 172, 105299. [Google Scholar] [CrossRef]
- Ostos-Garrido Francisco, J.; Huang, Y.; Li, Z.; Peña, J.M.; De Castro, A.I.; Torres-Sánchez, J.; Pistón, F. High-Throughput Phenotyping of Bioethanol Potential in Cereals Using UAV-Based Multi-Spectral Imagery. Front. Plant Sci. 2019, 10, 948. [Google Scholar] [CrossRef]
- Koh, L.P.; Wich, S.A. Dawn of Drone Ecology: Low-Cost Autonomous Aerial Vehicles for Conservation. Trop. Conserv. Sci. 2012, 5, 121–132. [Google Scholar] [CrossRef]
- Marino, S.; Alvino, A. Detection of Spatial and Temporal Variability of Wheat Cultivars by High-Resolution Vegetation Indices. Agronomy 2019, 9, 226. [Google Scholar] [CrossRef]
- Atzberger, C. Advances in Remote Sensing of Agriculture: Context Description, Existing Operational Monitoring Systems and Major Information Needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef]
- Hassler, S.C.; Baysal-Gurel, F. Unmanned Aircraft System (UAS) Technology and Applications in Agriculture. Agronomy 2019, 9, 618. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.G.; Ghonge, M.; Jawandhiya, P.M. Review of Unmanned Aircraft System (UAS). SSRN Electron. J. 2013, 2, 1646–1658. [Google Scholar] [CrossRef]
- Lelong, C.; Burger, P.; Jubelin, G.; Roux, B.; Labbé, S.; Baret, F. Assessment of Unmanned Aerial Vehicles Imagery for Quantitative Monitoring of Wheat Crop in Small Plots. Sensors 2008, 8, 3557–3585. [Google Scholar] [CrossRef] [PubMed]
- Hunt, E.R.; Doraiswamy, P.C.; McMurtrey, J.E.; Daughtry, C.S.T.; Perry, E.M.; Akhmedov, B. A visible band index for remote sensing leaf chlorophyll content at the Canopy scale. Int. J. Appl. Earth Obs. Geoinf. 2012, 21, 103–112. [Google Scholar] [CrossRef]
- Jing, R.; Gong, Z.; Zhao, W.; Pu, R.; Deng, L. Above-bottom biomass retrieval of aquatic plants with regression models and SfM data acquired by a UAV platform—A case study in Wild Duck Lake Wetland, Beijing, China. ISPRS J. Photogramm. Remote Sens. 2017, 134, 122–134. [Google Scholar] [CrossRef]
- Aldao, E.; González-de Santos, L.M.; González-Jorge, H. LiDAR Based Detect and Avoid System for UAV Navigation in UAM Corridors. Drones 2022, 6, 185. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Borra-Serrano, I.; De Swaef, T.; Quataert, P.; Aper, J.; Saleem, A.; Saeys, W.; Saeys, W.; Roldán-Ruiz, I.; Lootens, P. Closing the Phenotyping Gap: High Resolution UAV Time Series for Soybean Growth Analysis Provides Objective Data from Field Trials. Remote Sens. 2020, 12, 1644. [Google Scholar] [CrossRef]
- Fenghua, Y.; Tongyu, X.; Wen, D.; Hang, M.; Guosheng, Z.; Chunling, C. Radiative transfer models (RTMs) for field phenotyping inversion of rice based on UAV hyperspectral remote sensing. Int. J. Agric. Biol. Eng. 2017, 10, 150–157. [Google Scholar] [CrossRef]
- Andrade-Sanchez, P.; Gore, M.A.; Heun, J.T.; Thorp, K.R.; Carmo-Silva, A.E.; French, A.N.; Salvucci, M.E.; White, J.W. Development and evaluation of a field-based high-throughput phenotyping platform. Funct. Plant Biol. 2014, 41, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, N. Imaging technologies for plant high-throughput phenotyping: A review. Front. Agric. Sci. Eng. 2018, 5, 406–419. [Google Scholar] [CrossRef]
- Ludovisi, R.; Tauro, F.; Salvati, R.; Khoury, S.; Mugnozza, G.S.; Harfouche, A. Uav-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front. Plant Sci. 2017, 8, 1681. [Google Scholar] [CrossRef]
- Nilsson, H. Remote Sensing and Image Analysis in Plant Pathology. Annu. Rev. Phytopathol. 1995, 33, 489–528. [Google Scholar] [CrossRef] [PubMed]
- Chaerle, L.; Van Der Straeten, D. Imaging techniques and the early detection of plant stress. Trends Plant Sci. 2000, 5, 495–501. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitiyiming, M.; Fishman, J. Effects of Ambient Ozone on Soybean Biophysical Variables and Mineral Nutrient Accumulation. Remote Sens. 2018, 10, 562. [Google Scholar] [CrossRef]
- Urban, J.; Ingwers, M.W.; McGuire, M.A.; Teskey, R.O. Increase in leaf temperature opens stomata and decouples net photosynthesis from stomatal conductance in Pinus taeda and Populus deltoides x nigra. J. Exp. Bot. 2017, 68, 1757–1767. [Google Scholar] [CrossRef]
- Jones, H.G. Use of thermography for quantitative studies of spatial and temporal variation of stomatal conductance over leaf surfaces. Plant Cell Environ. 1999, 22, 1043–1055. [Google Scholar] [CrossRef]
- Jones, H.G. Plants and Microclimate: A Quantitative Approach to Environmental Plant Physiology, 3rd ed.; Cambridge University Press: Cambridge, UK, 2013; ISBN 9780521279598. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. N. Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Ferrio, J.P.; Bertran, E.; Nachit, M.M.; Català, J.; Araus, J.L. Estimation of grain yield by near-infrared reflectance spectroscopy in durum wheat. In Euphytica; Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; Volume 137, pp. 373–380. [Google Scholar] [CrossRef]
- Berger, B.; Parent, B.; Tester, M. High-throughput shoot imaging to study drought responses. J. Exp. Bot. 2010, 61, 3519–3528. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Thorp, K.R.; Gore, M.A.; Andrade-Sanchez, P.; Carmo-Silva, A.E.; Welch, S.M.; White, J.W.; French, A.N. Proximal hyperspectral sensing and data analysis approaches for field-based plant phenomics. Comput. Electron. Agric. 2015, 118, 225–236. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Yang, H.; Xu, B.; Li, Z.; Yang, X. Clustering Field-Based Maize Phenotyping of Plant-Height Growth and Canopy Spectral Dynamics Using a UAV Remote-Sensing Approach. Front. Plant Sci. 2018, 9, 1638. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Hassan, M.A.; Xu, K.; Zheng, C.; Rasheed, A.; Zhang, Y.; Jin, X.; Xia, X.; Xiao, Y.; He, Z. Assessment of Water and Nitrogen Use Efficiencies Through UAV-Based Multispectral Phenotyping in Winter Wheat. Front. Plant Sci. 2020, 11, 927. [Google Scholar] [CrossRef] [PubMed]
- Ampatzidis, Y.; Partel, V. UAV-Based High Throughput Phenotyping in Citrus Utilizing Multispectral Imaging and Artificial Intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef]
- Thiel, M.; Rath, T.; Ruckelshausen, A. Plant moisture measurement in field trials based on NIR spectral imaging a feasibility study. In Proceedings of the 2nd International CIGR Workshop on Image Analysis in Agriculture, Budapest, Hungary, 6–27 August 2010; pp. 16–29. [Google Scholar]
- Serrano, L.; González-Flor, C.; Gorchs, G. Assessment of grape yield and composition using the reflectance based Water Index in Mediterranean rainfed vineyards. Remote Sens. Environ. 2012, 118, 249–258. [Google Scholar] [CrossRef]
- Yi, Q.; Bao, A.; Wang, Q.; Zhao, J. Estimation of leaf water content in cotton by means of hyperspectral indices. Comput. Electron. Agric. 2013, 90, 144–151. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Joshi, S.; Thoday-Kennedy, E.; Pasam, R.K.; Tibbits, J.; Hayden, M.; Spangenberg, G.; Kant, S. High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J. Exp. Bot. 2020, 71, 4604–4615. [Google Scholar] [CrossRef]
- Mohd Asaari, M.S.; Mishra, P.; Mertens, S.; Dhondt, S.; Inzé, D.; Wuyts, N.; Scheunders, P. Close-range hyperspectral image analysis for the early detection of stress responses in individual plants in a high-throughput phenotyping platform. ISPRS J. Photogramm. Remote Sens. 2018, 138, 121–138. [Google Scholar] [CrossRef]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Sytar, O.; Brestic, M.; Zivcak, M.; Olsovska, K.; Kovar, M.; Shao, H.; He, X. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. Sci. Total Environ. 2017, 578, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, N.; Ecarnot, M.; Rabatel, G.; Roumet, P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in Wheat. Field Crop. Res. 2011, 122, 25–31. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Huang, Y.; Yang, T.; Li, J.; Zhu, K.; Zhang, J.; Yang, B.; Shao, C.; Peng, J. Optimization of multi-source UAV RS agro-monitoring schemes designed for field-scale crop phenotyping. Precis. Agric. 2021, 22, 1768–1802. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.M.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Dupuis, J.; Kuhlmann, H.; Oerke, E.C.; Plümer, L. Generation and application of hyperspectral 3D plant models: Methods and challenges. Mach. Vis. Appl. 2016, 27, 611–624. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogr. Earth Environ. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A. In-Field High-Throughput Phenotyping of Cotton Plant Height Using LiDAR. Remote Sens. 2017, 9, 377. [Google Scholar] [CrossRef]
- Lin, Y. LiDAR: An important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput. Electron. Agric. 2015, 119, 61–73. [Google Scholar] [CrossRef]
- Maesano, M.; Khoury, S.; Nakhle, F.; Firrincieli, A.; Gay, A.; Tauro, F.; Harfouche, A. UAV-Based LiDAR for High-Throughput Determination of Plant Height and Above-Ground Biomass of the Bioenergy Grass Arundo donax. Remote Sens. 2020, 12, 3464. [Google Scholar] [CrossRef]
- Hollaus, M.; Wagner, W.; Eberhöfer, C.; Karel, W. Accuracy of large-scale canopy heights derived from LiDAR data under operational constraints in a complex alpine environment. ISPRS J. Photogramm. Remoten Sens. 2006, 60, 323–338. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; Rex, F.E.; de Almeida, D.R.A.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Zambrano, A.M.A.; da Cunha Neto, E.M.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef]
- Picos, J.; Bastos, G.; Míguez, D.; Alonso, L.; Armesto, J. Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR. Remote Sens. 2020, 12, 885. [Google Scholar] [CrossRef]
- ten Harkel, J.; Bartholomeus, H.; Kooistra, L. Biomass and crop height estimation of different crops using UAV-based LiDAR. Remote Sens. 2020, 12, 17. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Peng, J.; Huang, Y.; Li, J.; Zhang, J.; Yang, B.; Liao, X. Estimating Maize Above-Ground Biomass Using 3D Point Clouds of Multi-Source Unmanned Aerial Vehicle Data at Multi-Spatial Scales. Remote Sens. 2019, 11, 2678. [Google Scholar] [CrossRef]
- Wang, C.; Nie, S.; Xi, X.; Luo, S.; Sun, X. Estimating the Biomass of Maize with Hyperspectral and LiDAR Data. Remote Sens. 2016, 9, 11. [Google Scholar] [CrossRef]
- Wang, X.; Thorp, K.R.; White, J.W.; French, A.N.; Poland, J.A. Approaches for geospatial processing of field-based high-throughput plant phenomics data from ground vehicle platforms. Trans. ASABE 2016, 59, 1053–1067. [Google Scholar] [CrossRef]
- Kefauver, S.C.; Vicente, R.; Vergara-Díaz, O.; Fernandez-Gallego, J.A.; Kerfal, S.; Lopez, A.; Melichar, J.P.E.; Molins, M.D.S.; Araus, J.L. Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley. Front. Plant Sci. 2017, 8, 1733. [Google Scholar] [CrossRef]
- Perich, G.; Hund, A.; Anderegg, J.; Roth, L.; Boer, M.P.; Walter, A.; Liebisch, F.; Aasen, H. Assessment of Multi-Image Unmanned Aerial Vehicle Based High-Throughput Field Phenotyping of Canopy Temperature. Front. Plant Sci. 2020, 11, 150. [Google Scholar] [CrossRef]
- Agisoft LLC. Agisoft Metashape User Manual; Agisoft Metashape: Sankt Petersburg, Russia, 2020; p. 160. [Google Scholar]
- Pix4D S.A. User Manual Pix4Dmapper v. 4.1; Pix4D S.A.: Prilly, Switzerland, 2017; 305p. [Google Scholar]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from Motion Photogrammetry in Forestry: A Review. Curr. For. Rep. 2019, 5, 155–168. [Google Scholar] [CrossRef]
- Richards, J. Computer processing of remotely-sensed images: An introduction. Earth Sci. Rev. 1990, 27, 392–394. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Cruz, A.C.; Luvisi, A.; De Bellis, L.; Ampatzidis, Y. X-FIDO: An effective application for detecting olive quick decline syndrome with deep learning and data fusion. Front. Plant Sci. 2017, 8, 1741. [Google Scholar] [CrossRef] [PubMed]
- Hake, K.; Burch, T.; Mauney, J. Making Sense out of Stalks What Controls Plant Height and How it Affects Yield. Physiol. Today 1989, 4, 1–4. [Google Scholar]
- Sui, R.; Fisher, D.K.; NReddy, K. Cotton Yield Assessment Using Plant Height Mapping System. J. Agric. Sci. 2012, 5, 23. [Google Scholar] [CrossRef]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.N.; Heun, J.T.; Hunsaker, D.J.; et al. Field-based phenomics for plant genetics research. Field Crop. Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Patrick, A.; Li, C. High Throughput Phenotyping of Blueberry Bush Morphological Traits Using Unmanned Aerial Systems. Remote Sens. 2017, 9, 1250. [Google Scholar] [CrossRef]
- Su, W.; Zhang, M.; Bian, D.; Liu, Z.; Huang, J.; Wang, W.; Wu, J.; Guo, H. Phenotyping of Corn Plants Using Unmanned Aerial Vehicle (UAV) Images. Remote Sens. 2019, 11, 2021. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Dai, H.; Yang, H.; Xu, B.; Li, H.; Long, H.; Li, Z.; Yang, X.; Zhao, C. Combining self-organizing maps and biplot analysis to preselect maize phenotypic components based on UAV high-throughput phenotyping platform. Plant. Methods 2019, 15, 57. [Google Scholar] [CrossRef]
- Hu, P.; Chapman, S.C.; Wang, X.; Potgieter, A.; Duan, T.; Jordan, D.; Guo, Y.; Zheng, B. Estimation of plant height using a high throughput phenotyping platform based on unmanned aerial vehicle and self-calibration: Example for sorghum breeding. Eur. J. Agron. 2018, 95, 24–32. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Carter, A.H. Field-based crop phenotyping: Multispectral aerial imaging for evaluation of winter wheat emergence and spring stand. Comput. Electron. Agric. 2015, 118, 372–379. [Google Scholar] [CrossRef]
- Holman, F.; Riche, A.; Michalski, A.; Castle, M.; Wooster, M.; Hawkesford, M. High Throughput Field Phenotyping of Wheat Plant Height and Growth Rate in Field Plot Trials Using UAV Based Remote Sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Xie, T.; Li, J.; Yang, C.; Jiang, Z.; Chen, Y.; Guo, L.; Zhang, J. Crop height estimation based on UAV images: Methods, errors, and strategies. Comput. Electron. Agric. 2021, 185, 106155. [Google Scholar] [CrossRef]
- de Jesus Colwell, F.; Souter, J.; Bryan, G.J.; Compton, L.J.; Boonham, N.; Prashar, A. Development and Validation of Methodology for Estimating Potato Canopy Structure for Field Crop Phenotyping and Improved Breeding. Front. Plant Sci. 2021, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Varela, S.; Pederson, T.; Bernacchi, C.J.; Leakey, A.D.B. Understanding Growth Dynamics and Yield Prediction of Sorghum Using High Temporal Resolution UAV Imagery Time Series and Machine Learning. Remote Sens. 2021, 13, 1763. [Google Scholar] [CrossRef]
- Volpato, L.; Pinto, F.; González-Pérez, L.; Thompson, I.G.; Borém, A.; Reynolds, M.; Gérard, B.; Molero, G.; Rodrigues, F.A. High Throughput Field Phenotyping for Plant Height Using UAV-Based RGB Imagery in Wheat Breeding Lines: Feasibility and Validation. Front. Plant Sci. 2021, 12, 185. [Google Scholar] [CrossRef] [PubMed]
- Gano, B.; Dembele, J.S.B.; Ndour, A.; Luquet, D.; Beurier, G.; Diouf, D.; Audebert, A. Using UAV Borne, Multi-Spectral Imaging for the Field Phenotyping of Shoot Biomass, Leaf Area Index and Height of West African Sorghum Varieties under Two Contrasted Water Conditions. Agronomy 2021, 11, 850. [Google Scholar] [CrossRef]
- Sagoo, A.G.; Hannan, A.; Aslam, M.; Khan, E.A.; Hussain, A.; Bakhsh, I.; Arif, M.; Waqas, M. Development of Water Saving Techniques for Sugarcane (Saccharum officinarum L.) in the Arid Environment of Punjab, Pakistan. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; Volume 1, pp. 507–535. [Google Scholar] [CrossRef]
- Bréda, N.J.J. Leaf Area Index. In Encyclopedia of Ecology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 2148–2154. [Google Scholar] [CrossRef]
- Sankaran, S.; Zhou, J.; Khot, L.R.; Trapp, J.J.; Mndolwa, E.; Miklas, P.N. High-throughput field phenotyping in dry bean using small unmanned aerial vehicle based multispectral imagery. Comput. Electron. Agric. 2018, 151, 84–92. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Song, W.; Han, L.; Liu, X.; Sun, X.; Luo, M.; Chen, K.; Zhang, H.; Zhao, Y.; et al. Dynamic plant height QTL revealed in maize through remote sensing phenotyping using a high-throughput unmanned aerial vehicle (UAV). Sci. Rep. 2019, 9, 3458. [Google Scholar] [CrossRef]
- Stewart, E.L.; Wiesner-Hanks, T.; Kaczmar, N.; DeChant, C.; Wu, H.; Lipson, H.; Nelson, R.J.; Gore, M.A. Quantitative Phenotyping of Northern Leaf Blight in UAV Images Using Deep Learning. Remote Sens. 2019, 11, 2209. [Google Scholar] [CrossRef]
- Gracia-Romero, A.; Kefauver, S.; Fernandez-Gallego, J.; Vergara-Díaz, O.; Araus, J.; Nieto-Taladriz, M. UAV and ground image-based phenotyping: A proof of concept with durum wheat. Remote Sens. 2019, 11, 1244. [Google Scholar] [CrossRef]
- Camino, C.; Gonzalez-Dugo, V.; Hernandez, P.; Zarco-Tejada, P.J. Radiative transfer Vcmax estimation from hyperspectral imagery and SIF retrievals to assess photosynthetic performance in rainfed and irrigated plant phenotyping trials. Remote Sens. Environ. 2019, 231, 111186. [Google Scholar] [CrossRef]
- Gonzalez-Dugo, V.; Hernandez, P.; Solis, I.; Zarco-Tejada, P. Using High-Resolution Hyperspectral and Thermal Airborne Imagery to Assess Physiological Condition in the Context of Wheat Phenotyping. Remote Sens. 2015, 7, 13586–13605. [Google Scholar] [CrossRef]
- Sugiura, R.; Tsuda, S.; Tamiya, S.; Itoh, A.; Nishiwaki, K.; Murakami, N.; Shibuya, Y.; Hirafuji, M.; Nuske, S. Field phenotyping system for the assessment of potato late blight resistance using RGB imagery from an unmanned aerial vehicle. Biosyst. Eng. 2016, 148, 13586–13605. [Google Scholar] [CrossRef]
- Patrick, A.; Pelham, S.; Culbreath, A.; Corely Holbrook, C.; De Godoy, I.J.; Li, C. High throughput phenotyping of tomato spot wilt disease in peanuts using unmanned aerial systems and multispectral imaging. IEEE Instrum. Meas. Mag. 2017, 20, 4–12. [Google Scholar] [CrossRef]
- Johansen, K.; Morton, M.J.L.; Malbeteau, Y.M.; Aragon, B.; Al-Mashharawi, S.K.; Ziliani, M.G.; Angel, Y.; Fiene, G.M.; Negrão, S.S.C.; Mousa, M.A.A.; et al. Unmanned Aerial Vehicle-Based Phenotyping Using Morphometric and Spectral Analysis Can Quantify Responses of Wild Tomato Plants to Salinity Stress. Front. Plant Sci. 2019, 10, 370. [Google Scholar] [CrossRef]
- Passioura, J.B. Phenotyping for drought tolerance in grain crops: When is it useful to breeders? Funct. Plant Biol. 2012, 39, 851. [Google Scholar] [CrossRef]
- Hu, Y.; Knapp, S.; Schmidhalter, U. Advancing High-Throughput Phenotyping of Wheat in Early Selection Cycles. Remote Sens. 2020, 12, 574. [Google Scholar] [CrossRef]
- Teke, M.; Deveci, H.S.; Haliloglu, O.; Gurbuz, S.Z.; Sakarya, U. A short survey of hyperspectral remote sensing applications in agriculture. In Proceedings of the 2013 6th International Conference on Recent Advances in Space Technologies (RAST), Istanbul, Turkey, 12–14 June 2013; pp. 171–176. [Google Scholar] [CrossRef]
- Thomas, S.; Behmann, J.; Steier, A.; Kraska, T.; Muller, O.; Rascher, U.; Mahlein, A.K. Quantitative assessment of disease severity and rating of barley cultivars based on hyperspectral imaging in a non-invasive, automated phenotyping platform. Plant Methods 2018, 14, 45. [Google Scholar] [CrossRef]
- tf.keras.metrics.MeanIoU—TensorFlow Core v2.6.0 n.d. Available online: https://www.tensorflow.org/api_docs/python/tf/keras/metrics/MeanIoU (accessed on 20 October 2021).
- López-Granados, F.; Torres-Sánchez, J.; Jiménez-Brenes, F.M.; Arquero, O.; Lovera, M.; De Castro, A.I. An efficient RGB-UAV-based platform for field almond tree phenotyping: 3-D architecture and flowering traits. Plant. Methods 2019, 15, 160. [Google Scholar] [CrossRef]
- Watanabe, K.; Guo, W.; Arai, K.; Takanashi, H.; Kajiya-Kanegae, H.; Kobayashi, M.; Yano, K.; Tokunaga, T.; Fujiwara, T.; Tsutsumi, N.; et al. High-throughput phenotyping of sorghum plant height using an unmanned aerial vehicle and its application to genomic prediction modeling. Front. Plant Sci. 2017, 8, 421. [Google Scholar] [CrossRef]
- Sarkar, S.; Cazenave, A.; Oakes, J.; McCall, D.; Thomason, W.; Abbot, L.; Balota, M. High-throughput measurement of peanut canopy height using digital surface models. Plant. Phenome J. 2020, 3, e20003. [Google Scholar] [CrossRef]
- Tilly, N.; Hoffmeister, D.; Schiedung, H.; Hütt, C.; Brands, J.; Bareth, G. Terrestrial laser scanning for plant height measurement and biomass estimation of maize. In The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences; International Society for Photogrammetry and Remote Sensing: Hannover, Germany, 2014; Volume 40, pp. 181–187. [Google Scholar] [CrossRef]
- Bronson, K.F.; French, A.N.; Conley, M.M.; Barnes, E.M. Use of an ultrasonic sensor for plant height estimation in irrigated cotton. Agron. J. 2021, 113, 2175–2183. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Robertson, J.S.; Sun, S.; Xu, R.; Paterson, A.H. GPhenoVision: A ground mobile system with multi-modal imaging for field-based high throughput phenotyping of cotton. Sci. Rep. 2018, 8, 1213. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-field High Throughput Phenotyping and Cotton Plant Growth Analysis Using LiDAR. Front. Plant Sci. 2018, 9, 16. [Google Scholar] [CrossRef]
- Comar, A.; Burger, P.; De Solan, B.; Baret, F.; Daumard, F.; Hanocq, J.F. A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions: Description and first results. Funct. Plant Biol. 2012, 39, 914–924. [Google Scholar] [CrossRef]
- Svensgaard, J.; Roitsch, T.; Christensen, S. Development of a Mobile Multispectral Imaging Platform for Precise Field Phenotyping. Agronomy 2014, 4, 322–336. [Google Scholar] [CrossRef]
- Madec, S.; Baret, F.; de Solan, B.; Thomas, S.; Dutartre, D.; Jezequel, S.; Hemmerlé, M.; Colombeau, G.; Comar, A. High-Throughput Phenotyping of Plant Height: Comparing Unmanned Aerial Vehicles and Ground LiDAR Estimates. Front. Plant Sci. 2017, 8, 2002. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.T.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High Throughput Determination of Plant Height, Ground Cover, and Above-Ground Biomass in Wheat with LiDAR. Front. Plant Sci. 2018, 9, 237. [Google Scholar] [CrossRef]
- Khan, Z.; Chopin, J.; Cai, J.; Eichi, V.-R.; Haefele, S.; Miklavcic, S. Quantitative Estimation of Wheat Phenotyping Traits Using Ground and Aerial Imagery. Remote Sens. 2018, 10, 950. [Google Scholar] [CrossRef]
- Wang, X.; Singh, D.; Marla, S.; Morris, G.; Poland, J. Field-based high-throughput phenotyping of plant height in sorghum using different sensing technologies. Plant Methods 2018, 14, 53. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, K.; Guan, H.; Feng, J.; Yu, S.; Liu, G. High-Throughput Phenotyping Analysis of Potted Soybean Plants Using Colorized Depth Images Based on A Proximal Platform. Remote Sens. 2019, 11, 1085. [Google Scholar] [CrossRef]
- Thapa, S.; Zhu, F.; Walia, H.; Yu, H.; Ge, Y. A Novel LiDAR-Based Instrument for High-Throughput, 3D Measurement of Morphological Traits in Maize and Sorghum. Sensors 2018, 18, 1187. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Chai, H.; Shao, K.; Shen, M.; Wang, Q.; Wang, R.; Sui, Y.; Ma, Y. Image-Based Dynamic Quantification of Aboveground Structure of Sugar Beet in Field. Remote Sens. 2020, 12, 269. [Google Scholar] [CrossRef]
- Adams, T.; Bruton, R.; Ruiz, H.; Barrios-Perez, I.; Selvaraj, M.G.; Hays, D.B. Prediction of Aboveground Biomass of Three Cassava (Manihot esculenta) Genotypes Using a Terrestrial Laser Scanner. Remote Sens. 2021, 13, 1272. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Song, S.; Xu, K.; Hu, T.; Yang, Q.; Wu, F.; Xu, G.; Ma, Q.; Guan, H.; et al. Non-destructive estimation of field maize biomass using terrestrial lidar: An evaluation from plot level to individual leaf level. Plant Methods 2020, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Noguchi, N. Monitoring of Wheat Growth Status and Mapping of Wheat Yield’s within-Field Spatial Variations Using Color Images Acquired from UAV-camera System. Remote Sens. 2017, 9, 289. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A Direct Comparison of Remote Sensing Approaches for High-Throughput Phenotyping in Plant Breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef]
- Hunt, E.R.; Mccarty, G.W.; Fujikawa, S.J.; Yoel, D.W. Remote Sensing of Crop Leaf Area Index Using Unmanned Airborne Vehicles. In Proceedings of the Pecora 17—The Future of Land Imaging... Going Operational, Denver, CO, USA, 18–20 November 2008; American Society for Photogrammetry and Remote Sensing (ASPRS): Baton Rouge, LA, USA, 2008. [Google Scholar]
- Hassan, M.; Yang, M.; Fu, L.; Rasheed, A.; Xia, X.; Xiao, Y.; He, Z.; Zheng, B. Accuracy assessment of plant height using an unmanned aerial vehicle for quantitative genomic analysis in bread wheat. Plant Methods 2019, 15, 37. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Ewel, J.J.; Clark, D.B. Estimation of tropical rain forest aboveground biomass with small-footprint lidar and hyperspectral sensors. Remote Sens. Environ. 2011, 115, 2931–2942. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S.; Nelson, R. Lidar remote sensing of forest biomass: A scale-invariant estimation approach using airborne lasers. Remote Sens. Environ. 2009, 113, 182–196. [Google Scholar] [CrossRef]
- Swatantran, A.; Dubayah, R.; Roberts, D.; Hofton, M.; Blair, J.B. Mapping biomass and stress in the Sierra Nevada using lidar and hyperspectral data fusion. Remote Sens. Environ. 2011, 115, 2917–2930. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Xi, X.; Pan, F. Estimating FPAR of maize canopy using airborne discrete-return LiDAR data. Opt. Express 2014, 22, 5106. [Google Scholar] [CrossRef]
- Nie, S.; Wang, C.; Dong, P.; Xi, X. Estimating leaf area index of maize using airborne full-waveform lidar data. Remote Sens. Lett. 2016, 7, 111–120. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Pan, F.; Xi, X.; Li, G.; Nie, S.; Xia, S. Estimation of wetland vegetation height and leaf area index using airborne laser scanning data. Ecol. Indic. 2015, 48, 550–559. [Google Scholar] [CrossRef]
- Koester, R.P.; Skoneczka, J.A.; Cary, T.R.; Diers, B.W.; Ainsworth, E.A. Historical gains in soybean (Glycine max Merr.) seed yield are driven by linear increases in light interception, energy conversion, and partitioning efficiencies. J. Exp. Bot. 2014, 65, 3311–3321. [Google Scholar] [CrossRef]
- Purcell, L.C.; Specht, J.E. Physiological Traits for Ameliorating Drought Stress. In Soybeans: Improvement, Production, and Uses; Wiley: Hoboken, NJ, USA, 2016; pp. 569–620. [Google Scholar] [CrossRef]
- Fickett, N.D.; Boerboom, C.M.; Stoltenberg, D.E. Soybean Yield Loss Potential Associated with Early-Season Weed Competition across 64 Site-Years. Weed Sci. 2013, 61, 500–507. [Google Scholar] [CrossRef]
- Busemeyer, L.; Mentrup, D.; Möller, K.; Wunder, E.; Alheit, K.; Hahn, V.; Maurer, H.P.; Reif, J.C.; Würschum, T.; Müller, J.; et al. Breedvision—A multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors 2013, 13, 2830–2847. [Google Scholar] [CrossRef]
- Maresma, Á.; Ariza, M.; Martínez, E.; Lloveras, J.; Martínez-Casasnovas, J.A. Analysis of vegetation indices to determine nitrogen application and yield prediction in maize (Zea mays L.) from a standard UAV service. Remote. Sens. 2016; 8, 973. [Google Scholar] [CrossRef]
- Wang, F.; Wang, F.; Zhang, Y.; Hu, J.; Huang, J.; Xie, J. Rice Yield Estimation Using Parcel-Level Relative Spectral Variables From UAV-Based Hyperspectral Imagery. Front. Plant Sci. 2019, 10, 453. [Google Scholar] [CrossRef]
- Rischbeck, P.; Baresel, P.; Elsayed, S.; Mistele, B.; Schmidhalter, U. Development of a diurnal dehydration index for spring barley phenotyping. Funct. Plant Biol. 2014, 41, 1249. [Google Scholar] [CrossRef]
- Dyson, J.; Mancini, A.; Frontoni, E.; Zingaretti, P. Deep Learning for Soil and Crop Segmentation from Remotely Sensed Data. Remote Sens. 2019, 11, 1859. [Google Scholar] [CrossRef]
- Gnädinger, F.; Schmidhalter, U. Digital Counts of Maize Plants by Unmanned Aerial Vehicles (UAVs). Remote Sens. 2017, 9, 544. [Google Scholar] [CrossRef]
- Negin, B.; Moshelion, M. The advantages of functional phenotyping in pre-field screening for drought-tolerant crops. Funct. Plant Biol. 2017, 44, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef]
- Mibei, E.K.; Ambuko, J.; Giovannoni, J.J.; Onyango, A.N.; Owino, W.O. Carotenoid profiling of the leaves of selected African eggplant accessions subjected to drought stress. Food Sci. Nutr. 2017, 5, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Strzałka, K.; Kostecka-Gugała, A.; Latowski, D. Carotenoids and Environmental Stress in Plants: Significance of Carotenoid-Mediated Modulation of Membrane Physical Properties. Russ. J. Plant Physiol. 2003, 50, 168–173. [Google Scholar] [CrossRef]
| Journals | Number of Publications | Percentage of Total Publication |
|---|---|---|
| Remote Sensing | 59 | 29 |
| Frontiers in Plant Science | 25 | 12 |
| Computers and Electronics in Agriculture | 20 | 10 |
| Field Crops Research | 12 | 6 |
| Plant Methods | 11 | 5 |
| Sensors | 9 | 4 |
| Journal of Experimental Botany | 8 | 4 |
| Agronomy-Basel | 5 | 2 |
| ISPRS Journal of Photogrammetry and Remote Sensing | 4 | 2 |
| Remote Sensing of Environment | 4 | 2 |
| IEEE Access | 4 | 2 |
| PLOS One | 3 | 1 |
| Scientific Reports | 3 | 1 |
| Precision Agriculture | 3 | 1 |
| Others | 35 | 17 |
| Total | 205 | 100 |
| Crop Phenotype or Trait | HTPP Platform | Crop | Sensor | Evaluation Method | Reference | ||
|---|---|---|---|---|---|---|---|
| r2 | RMSE | Accuracy | |||||
| Plant height | Aerial (sUAS) | Cotton | RGB | 0.98 | [53] | ||
| Multispectral | 0.90–0.96 | 5.50–10.10% | [52] | ||||
| 0.78 | [122] | ||||||
| Soybean | RGB | 0.70 | [69] | ||||
| >0.70 | [69] | ||||||
| Maize | RGB | 0.78 | 0.168 | [122] | |||
| 0.95 | [135] | ||||||
| Rice | 0.71 | [54] | |||||
| Sorghum | RGB | 0.57–0.62 | [149] | ||||
| NIR-GB | 0.58–0.62 | ||||||
| RGB | 0.69–0.73 | ||||||
| 0.34 | [124] | ||||||
| Bioenergy grass (Arundo donax) | LiDAR | 0.73 | [102] | ||||
| Peanut | RGB | 0.95 | [150] | ||||
| 0.86 | [150] | ||||||
| Blueberry | RGB | 0.92 | [121] | ||||
| Terrestrial or ground platform | Maize | Laser scanner | 0.93 | [151] | |||
| Cotton | Ultrasonic | 0.87 | 3.10 cm | [152] | |||
| RGB-D | 0.99 | 0.34 cm | [153] | ||||
| LiDAR | 0.97 | [154] | |||||
| 0.98 | 6.50 cm | [155] | |||||
| Triticale | RGB | 0.97 | [156] | ||||
| Wheat | LiDAR | 0.86 | 7.90 cm | [24] | |||
| 0.90 | [157] | ||||||
| 0.99 | 1.70 cm | [158] | |||||
| RGB | 0.95 | 3.95 cm | [159] | ||||
| Sorghum | Ultrasonic | 0.93 | [160] | ||||
| LiDAR | 0.88–0.9 | ||||||
| Canopy cover and leaf area index | sUAS | Blueberry | RGB | 0.70–0.83 | [121] | ||
| Soybean | RGB | >0.70 | [69] | ||||
| Cotton | Multispectral | 0.33–0.57 | [52] | ||||
| Citrus | Multispectral | 0.85% | [87] | ||||
| Rice | Hyperspectral | 0.82 | 0.10 | [70] | |||
| Soybean | RGB and Multispectral fusion | 0.059 | [50] | ||||
| Maize | RGB | 0.75 | 0.34 | [122] | |||
| Terrestrial or ground platform | Cotton | LiDAR | 0.97 | [154] | |||
| Wheat | LiDAR | 0.92 | [158] | ||||
| Soybean | RGB | 0.89 | [161] | ||||
| HSI | 0.80 | [161] | |||||
| Maize | LiDAR | 0.92 | [162] | ||||
| Sorghum | LiDAR | 0.94 | [162] | ||||
| Biomass or dry matter production | sUAS | Soybean | Multispectral and thermal fusion | 0.10 | [50] | ||
| Rice | Hyperspectral | 0.79 | 0.11 | [70] | |||
| Barley, triticale, and wheat | Multispectral | 0.44–0.59 | [57] | ||||
| Maize | Hyperspectral | 0.47 | [108] | ||||
| LiDAR | 0.83 | ||||||
| Hyperspectral and LiDAR fusion | 0.88 | ||||||
| Dry bean | Multispectral and thermal | (−0.67)–(−0.91) | [51] | ||||
| Bioenergy grass (Arundo donax) | LiDAR | 0.71 | [102] | ||||
| Terrestrial or ground platform | Wheat | LiDAR | 0.92–0.93 | [158] | |||
| Sugar beet | RGB | 0.82–0.88 | [163] | ||||
| Cassava | LiDAR | 0.73 | [164] | ||||
| Maize | LiDAR | 0.68–0.80 | [165] | ||||
| Canopy temperature and yield | sUAS | Wheat | RGB | 0.94 | [166] | ||
| Multispectral | 0.60 | [144] | |||||
| 0.63 | [144] | ||||||
| 0.65 | |||||||
| 0.43 | |||||||
| 0.57 | |||||||
| Rice | RGB | 0.73–076 | [167] | ||||
| Multispectral | 0.82 | [144] | |||||
| Multispectral and thermal | |||||||
| Terrestrial or ground platform | Wheat | 0.52 | |||||
| 0.36 | [168] | ||||||
| 0.51 | |||||||
| Platform | Initial Cost | Maintenance | Training | Human Resources | Payload | Coverage Area |
| sUAS | Moderate (≤USD 20K) | Low to Moderate | Moderate to High | Low | Low (≤1 kg) | Moderate to High (>5 ha) |
| Terrestrial handheld | Low to Moderate (USD 100–20K) | Low | Low | High | Low to Moderate (1–20 kg) | Low (<2 ha) |
| Terrestrial cart | Moderate (≤USD 20K) | Low to Moderate | Moderate | Low to Moderate | Moderate (≤20 kg) | Low to Moderate (≤2 ha) |
| Terrestrial tractor | High (≥USD 40K) | High | Moderate to High | Moderate | High (>20 kg) | Moderate (≤5 ha) |
| Sensor | Initial Cost | Maintenance | Deployment | Platform | Data Processing | Computational Resources |
| RGB camera | Low (≤USD 1K) | Low | Easy | sUAS * and Terrestrial | Easy to Moderate | Moderate |
| Multispectral camera | Moderate (≤USD 20K) | Moderate | Moderate | sUAS * and Terrestrial | Moderate | Moderate |
| Thermal camera | Moderate (≤USD 20K) | Moderate | Moderate to Difficult | sUAS * and Terrestrial | Moderate | Moderate to High |
| Hyperspectral camera | High (≥USD 40K) | Moderate | Moderate to Difficult | sUAS | Moderate | Moderate to High |
| LiDAR | High (≥USD 40K) | Moderate to High | Difficult | sUAS and Terrestrial * | Moderate to Difficult | Moderate to High |
| Infrared thermometer | Low (≤USD 1K) | Low | Easy | Terrestrial | Easy | Low |
| Multispectral radiometer | Low (≤USD 1K) | Low | Easy | Terrestrial | Easy | Low |
| Ultrasonic transducer | Low (≤USD 1K) | Low | Easy | Terrestrial | Easy | Low |
| Laser scanner | Low to Moderate (USD 100–20K) | Low to Moderate | Moderate | Terrestrial | Easy to Moderate | Low to Moderate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayankojo, I.T.; Thorp, K.R.; Thompson, A.L. Advances in the Application of Small Unoccupied Aircraft Systems (sUAS) for High-Throughput Plant Phenotyping. Remote Sens. 2023, 15, 2623. https://doi.org/10.3390/rs15102623
Ayankojo IT, Thorp KR, Thompson AL. Advances in the Application of Small Unoccupied Aircraft Systems (sUAS) for High-Throughput Plant Phenotyping. Remote Sensing. 2023; 15(10):2623. https://doi.org/10.3390/rs15102623
Chicago/Turabian StyleAyankojo, Ibukun T., Kelly R. Thorp, and Alison L. Thompson. 2023. "Advances in the Application of Small Unoccupied Aircraft Systems (sUAS) for High-Throughput Plant Phenotyping" Remote Sensing 15, no. 10: 2623. https://doi.org/10.3390/rs15102623
APA StyleAyankojo, I. T., Thorp, K. R., & Thompson, A. L. (2023). Advances in the Application of Small Unoccupied Aircraft Systems (sUAS) for High-Throughput Plant Phenotyping. Remote Sensing, 15(10), 2623. https://doi.org/10.3390/rs15102623






