Near-Infrared Spectroscopy and Mode Cloning (NIR-MC) for In-Situ Analysis of Crude Protein in Bamboo
Abstract
:1. Introduction
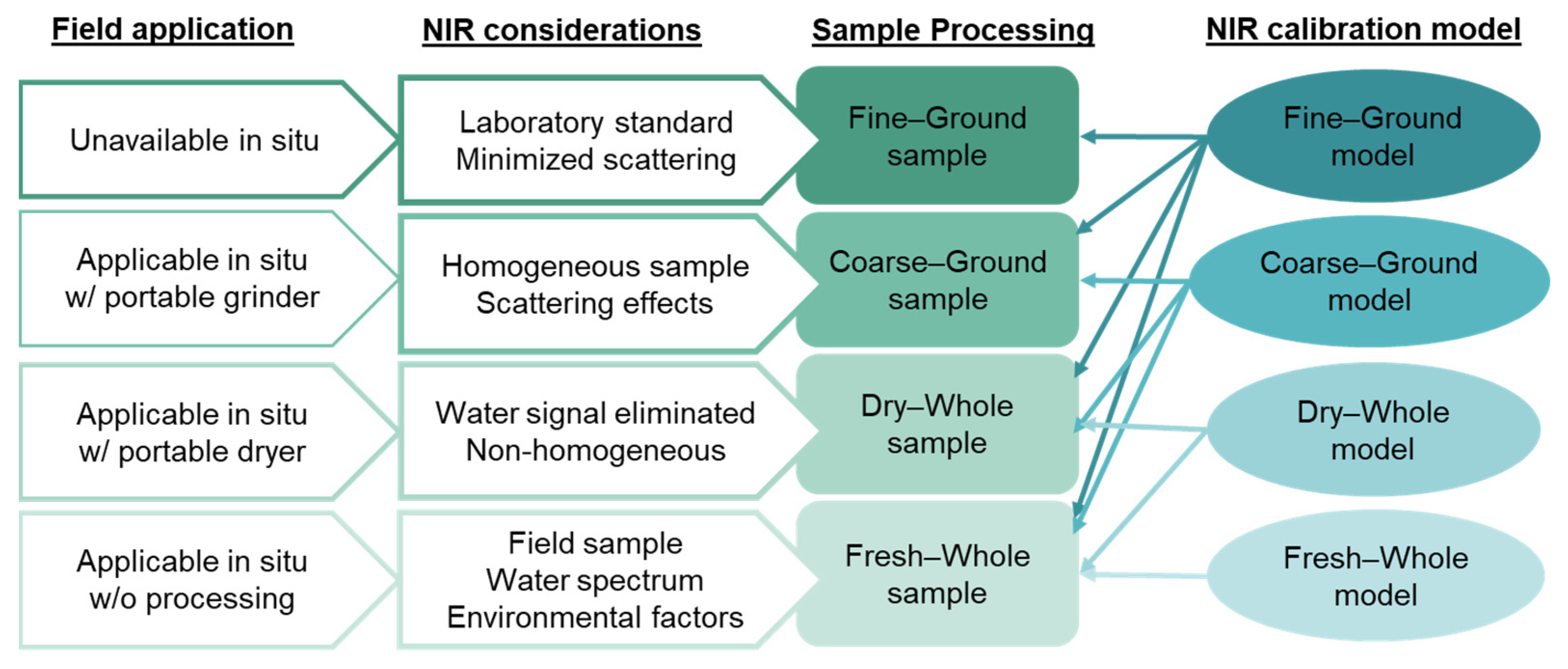
2. Materials and Methods
2.1. Bamboo Sampling and NIR Spectra Collection
2.2. Chemical Analysis
2.3. Chemometric Analysis
2.3.1. Calibration Models
2.3.2. NIR Mode Cloning (NIR-MC) Using Slope and Bias Correction (SBC)
3. Results
3.1. Sample Statistics
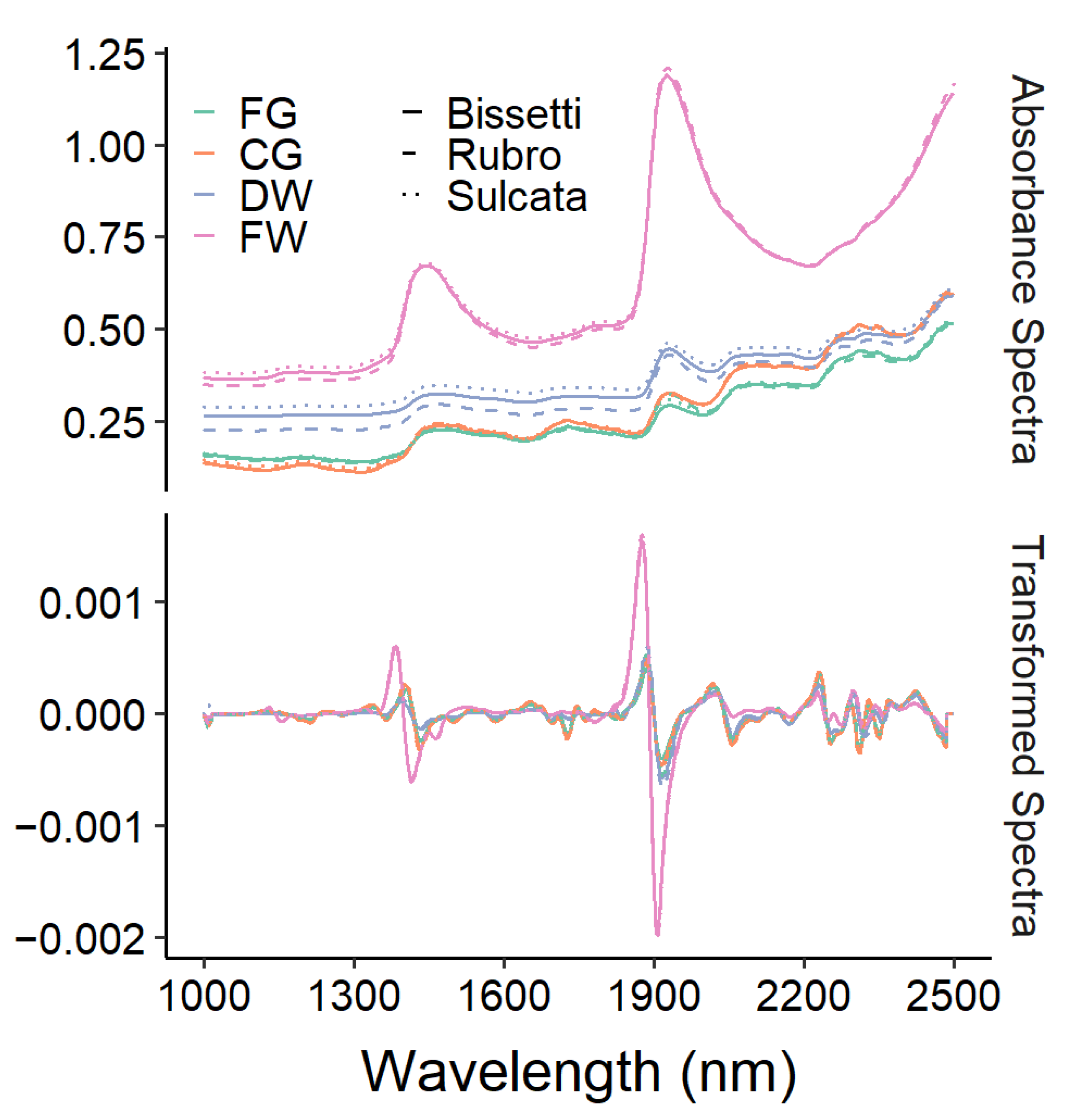
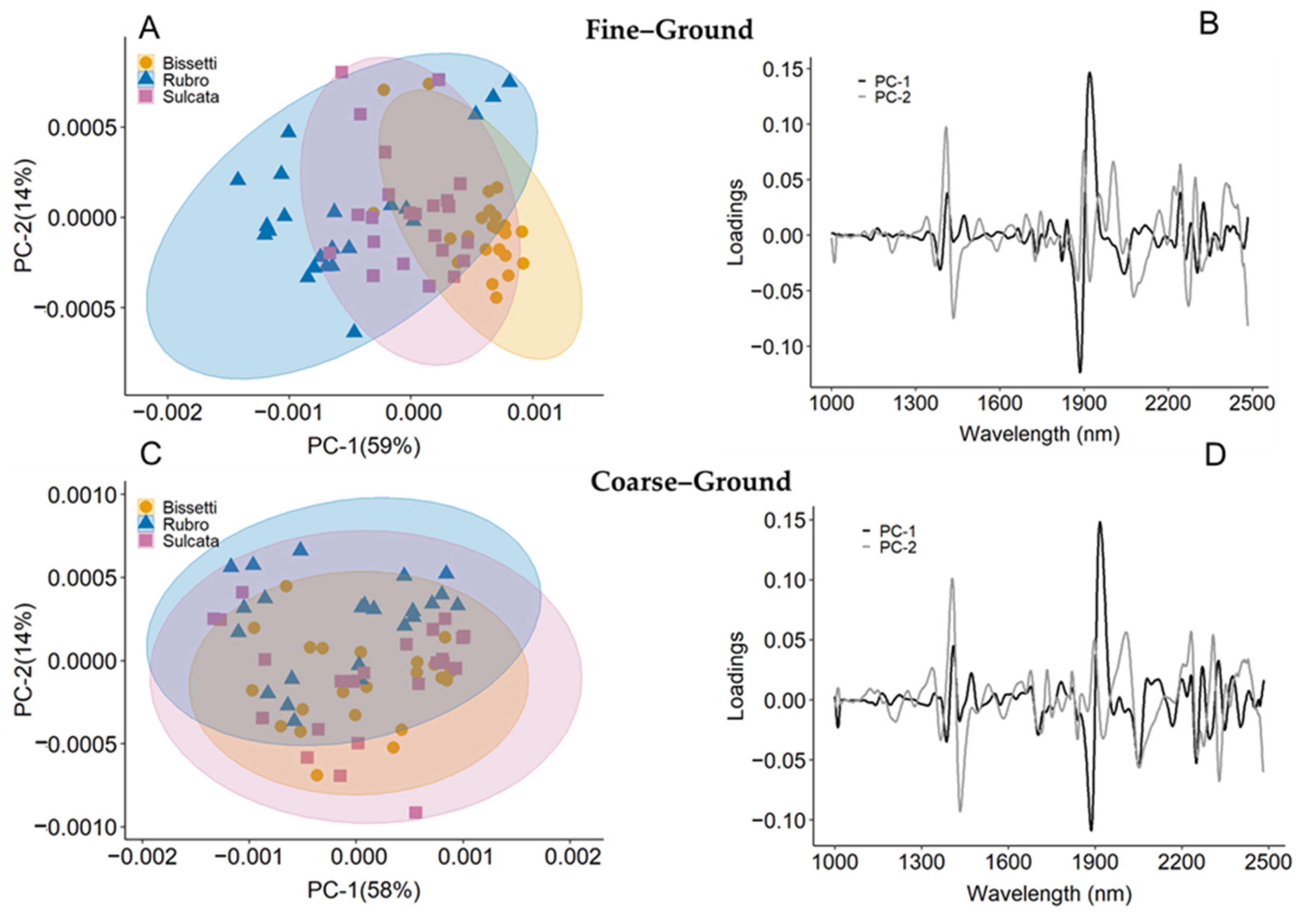
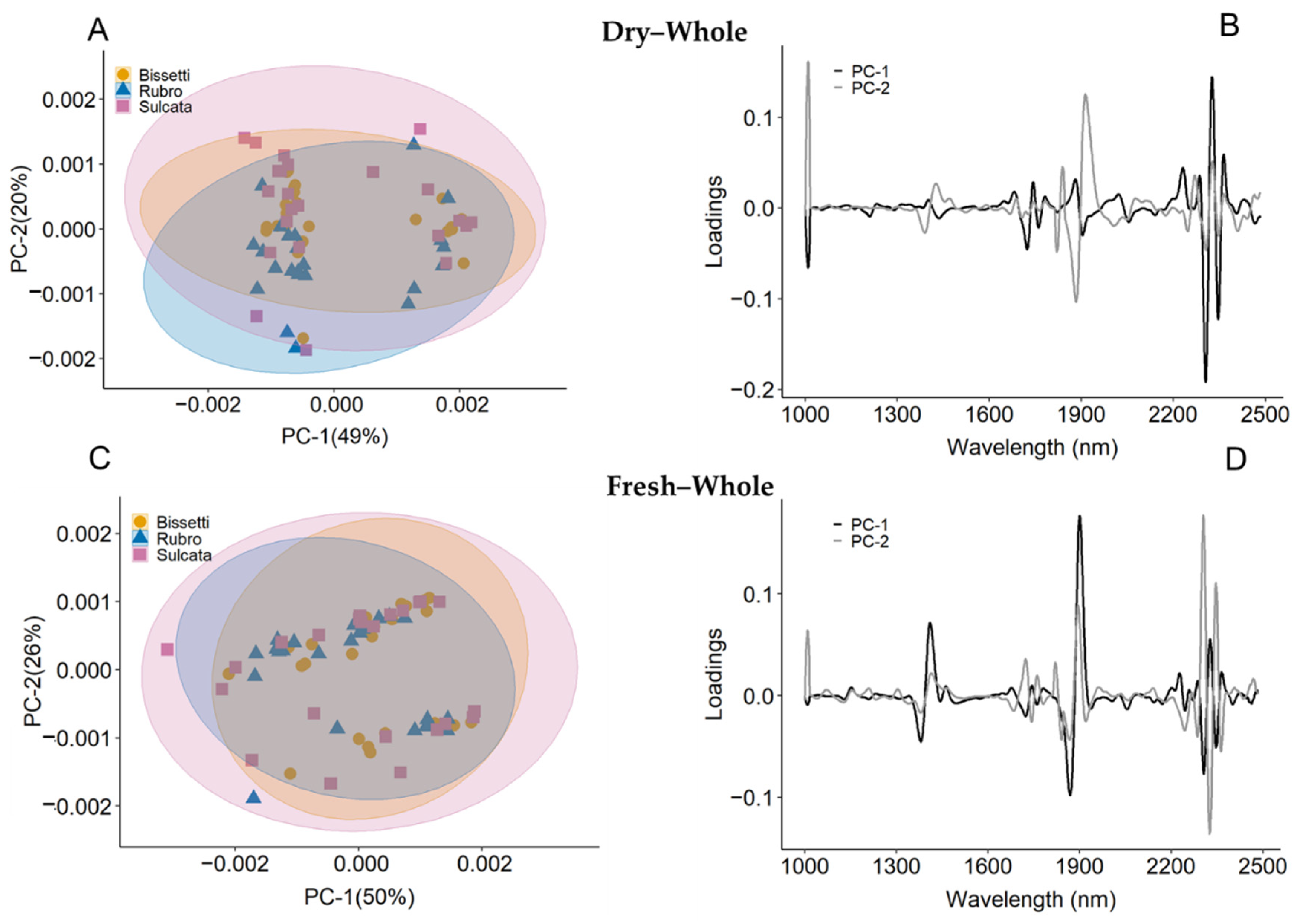
3.2. Spectral Characteristics
3.3. NIR Modeling across Species
3.4. NIR-MC and SBC across Processing Modes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Soest, P.J. Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1994; Volume 32, p. 7. [Google Scholar]
- Showers, A.; Scott, E.; Douglas, R.; Jerry, W.; James, C.; Ben, H.; Showers, S.E.; Tolleson, D.R.; Stuth, J.W.; Kroll, J.C.; et al. Predicting Diet Quality of White-Tailed Deer via NIRS Fecal Profiling Predicting Diet Quality of White-Tailed Deer via NIRS Fecal Profiling. Rangel. Ecol. Manag. 2021, 59, 300–307. [Google Scholar] [CrossRef]
- Dixon, R.; Coates, D. Near infrared spectroscopy of faeces to evaluate the nutrition and physiology of herbivores Review. J. Near Infrared Spectrosc. 2009, 17, 1–31. [Google Scholar] [CrossRef]
- Rothman, J.M.; Chapman, C.A.; Hansen, J.L.; Cherney, D.J.R.; Pell, A.N. Rapid assessment of the nutritional value of foods eaten by mountain gorillas: Applying near-infrared reflectance spectroscopy to primatology. Int. J. Primatol. 2009, 30, 729–742. [Google Scholar] [CrossRef]
- Givens, D.I.; De Boever, J.L.; Deaville, E.R. The principles, practices and some future applications of near infrared spectroscopy for predicting the nutritive value of foods for animals and humans. Nutr. Res. Rev. 1997, 10, 83–114. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Principles and Applications of Miniaturized Near-Infrared (NIR) Spectrometers. Chem.—A Eur. J. 2021, 27, 1514–1532. [Google Scholar] [CrossRef]
- Goi, A.; Simoni, M.; Righi, F.; Visentin, G.; De Marchi, M. Application of a handheld near-infrared spectrometer to predict gelatinized starch, fiber fractions, and mineral content of ground and intact extruded dry dog food. Animals 2020, 10, 1660. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Plewka, N.; Huck, C.W. Insect protein content analysis in handcrafted fitness bars by NIR spectroscopy. Gaussian process regression and data fusion for performance enhancement of miniaturized cost-effective consumer-grade sensors. Molecules 2021, 26, 6390. [Google Scholar] [CrossRef]
- Wiedower, E.; Hansen, R.; Bissell, H.; Ouellette, R.; Kouba, A.; Stuth, J.; Rude, B.; Tolleson, D. Use of near infrared spectroscopy to discriminate between and predict the nutrient composition of different species and parts of bamboo: Application for studying giant panda foraging ecology. J. Near Infrared Spectrosc. 2009, 17, 265–273. [Google Scholar] [CrossRef]
- García, J.; Cozzolino, D. Use of near infrared reflectance (NIR) spectroscopy to predict chemical composition of forages in broad-based calibration models Uso de la espectroscopía de reflectancia en el infrarrojo cercano (NIR) para predecir la composición química de forrajes e. Agric. Tec. 2006, 66, 41–47. [Google Scholar]
- Deaville, E.R.; Flinn, P.C. Near infrared (NIR) spectroscopy: An alternative approach for the estimation of forage quality and voluntary intake. In Forage Evaluation in Ruminant Nutrition; CABI Publishing: Wallingford, UK, 2000; pp. 301–320. ISBN 0851993443. [Google Scholar]
- Alomar, D.; Fuchslocher, R.; De Pablo, M. Effect of preparation method on composition and NIR spectra of forage samples. Anim. Feed Sci. Technol. 2003, 107, 191–200. [Google Scholar] [CrossRef]
- Reeves, J.B., III. Use of Near Infrared Reflectance Spectroscopy. In Farm Animal Metabolism and Nutrition; CABI Publishing: Wallingford, UK, 2009; pp. 184–209. ISBN 0851993788. [Google Scholar]
- Bricklemyer, R.S.; Brown, D.J. On-the-go VisNIR: Potential and limitations for mapping soil clay and organic carbon. Comput. Electron. Agric. 2010, 70, 209–216. [Google Scholar] [CrossRef]
- Ely, D.R.; Thommes, M.; Carvajal, M.T. Analysis of the effects of particle size and densification on NIR spectra. Colloids Surfaces A Physicochem. Eng. Asp. 2008, 331, 63–67. [Google Scholar] [CrossRef]
- Duyens, L.N.M. The flattering of the absorption spectrum of suspensions, as compared to that of solutions. Biochim. Biophys. Acta 1956, 19, 1–12. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B.; Zhigalova, T.V.; Naqvi, K.R. Light absorption by isolated chloroplasts and leaves: Effects of scattering and “packing”. Photosynth. Res. 2009, 102, 31–41. [Google Scholar] [CrossRef]
- Barthès, B.G.; Brunet, D.; Ferrer, H.; Chotte, J.L.; Feller, C. Determination of total carbon and nitrogen content in a range of tropical soils using near infrared spectroscopy: Influence of replication and sample grinding and drying. J. Near Infrared Spectrosc. 2006, 14, 341–348. [Google Scholar] [CrossRef]
- De La Haba, M.J.; Garrido-Varo, A.; Núñez, N.; Pérez-Marín, D.C.; Guerrero, J.E. Using calibrations developed for fine milled meat and bone meal on spectra measured on non-milled samples. J. Near Infrared Spectrosc. 2008, 16, 275–279. [Google Scholar] [CrossRef]
- Feudale, R.N.; Woody, N.A.; Tan, H.; Myles, A.J.; Brown, S.D.; Ferré, J. Transfer of multivariate calibration models: A review. Chemom. Intell. Lab. Syst. 2002, 64, 181–192. [Google Scholar] [CrossRef]
- Shenk, J.S.; Westerhaus, M.O. Optical Instrument Calibration System. U.S. Patent No. 4,866,644, 12 September 1989. [Google Scholar]
- Bouveresse, E.; Massart, D.L. Standardisation of near-infrared spectrometric instruments: A review. Vib. Spectrosc. 1996, 11, 3–15. [Google Scholar] [CrossRef]
- Salguero-Chaparro, L.; Palagos, B.; Peña-Rodríguez, F.; Roger, J.M. Calibration transfer of intact olive NIR spectra between a pre-dispersive instrument and a portable spectrometer. Comput. Electron. Agric. 2013, 96, 202–208. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Dong, J.; Li, Y.; Xu, H.; Tang, X. Maintaining the predictive abilities of egg freshness models on new variety based on VIS-NIR spectroscopy technique. Comput. Electron. Agric. 2019, 156, 669–676. [Google Scholar] [CrossRef]
- Abdelkader, M.F.; Cooper, J.B.; Larkin, C.M. Calibration transfer of partial least squares jet fuel property models using a segmented virtual standards slope-bias correction method. Chemom. Intell. Lab. Syst. 2012, 110, 64–73. [Google Scholar] [CrossRef]
- Shan, P.; Zhao, Y.; Wang, Q.; Wang, S.; Ying, Y.; Peng, S. A nonlinear calibration transfer method based on joint kernel subspace. Chemom. Intell. Lab. Syst. 2021, 210, 104247. [Google Scholar] [CrossRef]
- Schaller, G.B.; Hu, J.; Pan, W.; Zhu, J. The Giant Pandas of Wolong; The University of Chicago Press: Chicago, IL, USA, 1985. [Google Scholar]
- Johnson, K.G.; Schaller, G.B.; Jinchu, H. Comparative Behavior of Red and Giant Pandas in the Wolong Reserve, China. J. Mammal. 1988, 69, 552–564. [Google Scholar] [CrossRef]
- Wei, F.; Wang, W.; Zhou, A.; Hu, J.; Wei, Y. Preliminary study on food selection and feeding strategy of red pandas. Acta Theriol. Sin. 1995, 15, 259–266. [Google Scholar]
- Zhang, J.; Hull, V.; Huang, J.; Yang, W.; Zhou, S.; Xu, W.; Huang, Y.; Ouyang, Z.; Zhang, H.; Liu, J. Natural recovery and restoration in giant panda habitat after the Wenchuan earthquake. For. Ecol. Manag. 2014, 319, 1–9. [Google Scholar] [CrossRef]
- Roberts, B.M.S.; Gittleman, J.L. Ailurus fulgens. Mamm. Species 1984, Issue 222. 1–8. [Google Scholar] [CrossRef]
- Wei, F.; Feng, Z.; Wang, Z.; Zhou, A.; Hu, J. Use of the nutrients in bamboo by the red panda (Ailurus fulgens). J. Zool. 1999, 248, 535–541. [Google Scholar] [CrossRef]
- Dierenfeld, E.S.; Hintz, H.F.; Robertson, J.B.; Van Soest, P.J.; Oftedal, O.T. Utilization of bamboo by the giant panda. J. Nutr. 1982, 112, 636–641. [Google Scholar] [CrossRef]
- Finley, T.G.; Sikes, R.S.; Parsons, J.L.; Rude, B.J.; Bissell, H.A.; Ouellette, J.R. Energy digestibility of giant pandas on bamboo-only and on supplemented diets. Zoo Biol. 2011, 30, 121–133. [Google Scholar] [CrossRef]
- Wei, F.; Wang, Z.; Feng, Z.; Li, M.; Zhou, A. Seasonal energy utilization in bamboo by the red panda (Ailurus fulgens). Zoo Biol. Publ. Affil. Am. Zoo Aquar. Assoc. 2000, 19, 27–33. [Google Scholar] [CrossRef]
- Knott, K.K.; Christian, A.L.; Falcone, J.F.; Vance, C.K.; Bauer, L.L.; Fahey, G.C.; Kouba, A.J. Phenological changes in bamboo carbohydrates explain the preference for culm over leaves by giant pandas (Ailuropoda melanoleuca) during spring. PLoS ONE 2017, 12, e0177582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.; Zhang, Z.; Raubenheimer, D.; Elser, J.J.; Wei, W.; Wei, F. Obligate herbivory in an ancestrally carnivorous lineage: The giant panda and bamboo from the perspective of nutritional geometry. Funct. Ecol. 2015, 29, 26–34. [Google Scholar] [CrossRef]
- Dairy, N. Kjeldahl Nitrogen Analysis as a Reference Method for Protein Determination in Dairy Products. J. AOAC Int. 1999, 82, 1389–1398. [Google Scholar]
- Bernhard, T.; Truberg, B.; Friedt, W.; Snowdon, R.; Wittkop, B. Development of Near-Infrared Reflection Spectroscopy Calibrations for Crude Protein and Dry Matter Content in Fresh and Dried Potato Tuber Samples. Potato Res. 2016, 59, 149–165. [Google Scholar] [CrossRef]
- Williams, P.C. Implementation of near-infrared technology. In Near-Infrared Technology in the Agricultural and Food Industries; Williams, P., Norris, K., Eds.; American Association of Cereal Chemists Inc.: Saint Paul, MN, USA, 1987; pp. 145–169. [Google Scholar]
- Kuang, B.; Mouazen, A.M. Non-biased prediction of soil organic carbon and total nitrogen with vis-NIR spectroscopy, as affected by soil moisture content and texture. Biosyst. Eng. 2013, 114, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Büning-Pfaue, H. Analysis of water in food by near infrared spectroscopy. Food Chem. 2003, 82, 107–115. [Google Scholar] [CrossRef]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Nayar, R.; Carpenter, J.F.; Manning, M.C. Noninvasive determination of protein conformation in the solid state using near infrared (NIR) spectroscopy. J. Pharm. Sci. 2005, 94, 2030–2038. [Google Scholar] [CrossRef]
- Shenk, J.S.; Workman, J.J., Jr.; Westerhaus, M.O. Application of NIR Spectroscopy to Agricultural Products. In Handbook of Near-Infrared Analysis; Burns, D.A., Ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 347–386. [Google Scholar]
- Chen, J.; Ren, X.; Zhang, Q.; Diao, X.; Shen, Q. Determination of protein, total carbohydrates and crude fat contents of foxtail millet using effective wavelengths in NIR spectroscopy. J. Cereal Sci. 2013, 58, 241–247. [Google Scholar] [CrossRef]
- Lu, Y.; Du, C.; Yu, C.; Zhou, J. Fast and nondestructive determination of protein content in rapeseeds (Brassica napus L.) using Fourier transform infrared photoacoustic spectroscopy (FTIR-PAS). J. Sci. Food Agric. 2014, 94, 2239–2245. [Google Scholar] [CrossRef]
- Halvorson, J.J.; Cassida, K.A.; Turner, K.E.; Belesky, D.P. Nutritive value of bamboo as browse for livestock. Renew. Agric. Food Syst. 2010, 26, 161–170. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Renvoize, S.A.; Chonghaile, G.N.; Stapleton, C.M.A.; Chase, M.W. A comparison of ITS nuclear rDNA sequence data and AFLP markers for phylogenetic studies in Phyllostachys (Bambusoideae, Poaceae). J. Plant Res. 2000, 113, 259–269. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, C.; Li, D. Scanning electron microscopy of the leaf epidermis in Arundinarieae (Poaceae: Bambusoideae): Evolutionary implications of selected micromorphological features. Bot. J. Linn. 2014, 176, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Minasny, B.; McBratney, A.B.; Bellon-Maurel, V.; Roger, J.M.; Gobrecht, A.; Ferrand, L.; Joalland, S. Removing the effect of soil moisture from NIR diffuse reflectance spectra for the prediction of soil organic carbon. Geoderma 2011, 167–168, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Yang, P.; Chen, J.; Wu, Z.; Jia, Y.; Ma, C.; Zhan, X. A new calibration model transferring strategy maintaining the predictive abilities of NIR multivariate calibration model applied in different batches process of extraction. Infrared Phys. Technol. 2019, 103, 103046. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Ma, S.; Ma, T.; Shan, L.; Wang, X.; Nie, Y.; Ning, Z.; Yan, L.; Xiu, Y.; et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. USA 2017, 114, 1081–1086. [Google Scholar] [CrossRef] [Green Version]
- Nie, Y.; Wei, F.; Zhou, W.; Hu, Y.; Senior, A.M.; Wu, Q.; Yan, L.; Raubenheimer, D. Giant Pandas are Macronutritional Carnivores. Curr. Biol. 2019, 29, 1677–1682.e2. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, J.; Yang, J.; Li, M.; Wei, F. Food habits and space-use of red pandas Ailurus fulgens in the Fengtongzhai Nature Reserve, China: Food effects and behavioural responses. Acta Theriol. 2009, 54, 225–234. [Google Scholar] [CrossRef]
- Morgan, L.R.; Marsh, K.J.; Tolleson, D.R.; Youngentob, K.N. The application of nirs to determine animal physiological traits for wildlife management and conservation. Remote Sens. 2021, 13, 3699. [Google Scholar] [CrossRef]



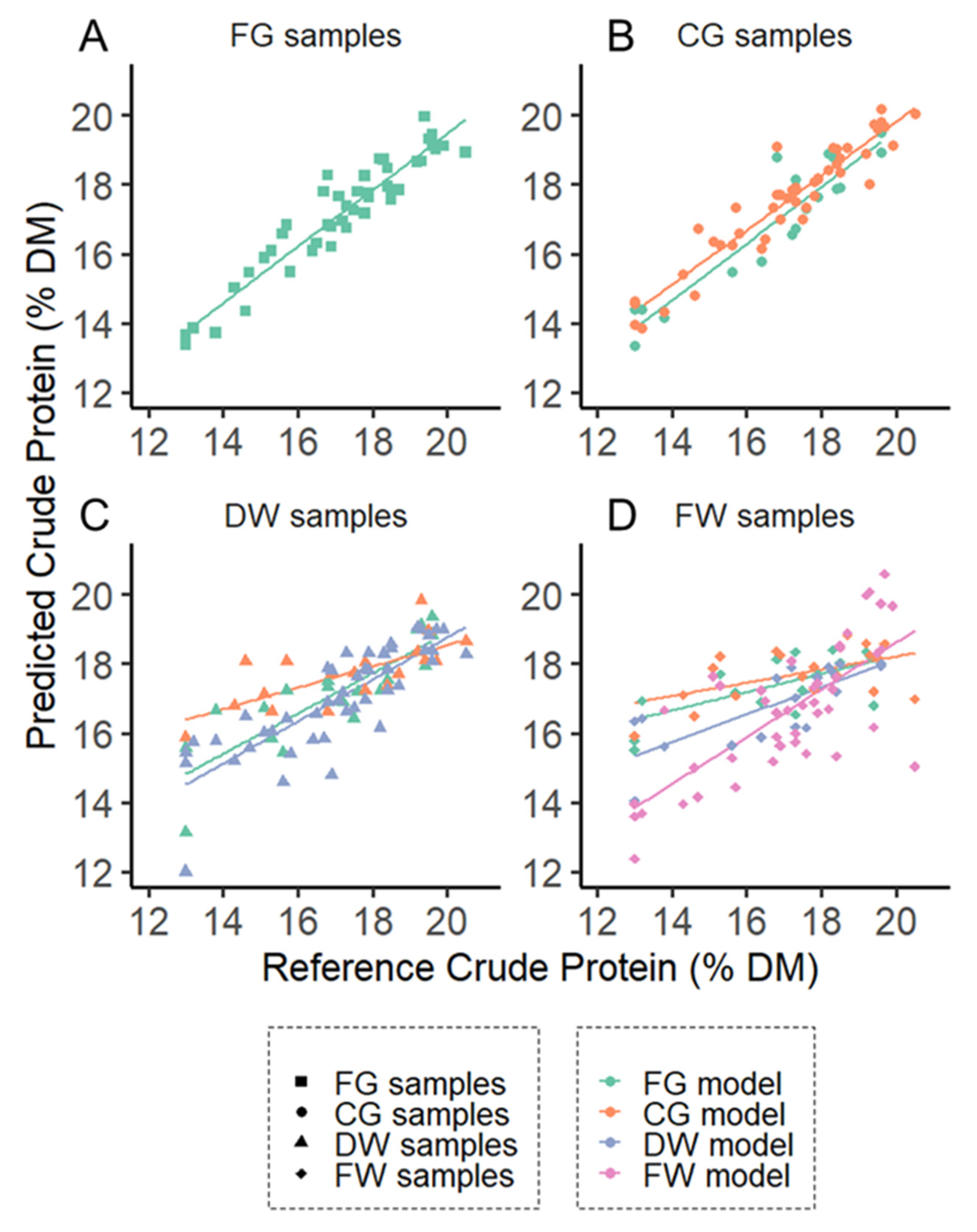
| Database | Species | Calibration for Each Processing Mode | Validation for Each Processing Mode | External Validation for Each Processing Mode |
|---|---|---|---|---|
| 1 | Rubro | 23 | 6 | 0 |
| Bissetii | 23 | 6 | 0 | |
| Sulcata | 23 | 6 | 0 | |
| 2 | Rubro | 23 | 6 | 0 |
| Sulcata | 23 | 6 | 0 | |
| Bissetii | 0 | 0 | 47 | |
| 3 | Rubro | 31 | 8 | 0 |
| Bissetii | 31 | 8 | 0 | |
| Sulcata | 0 | 0 | 29 | |
| 4 | Bissetii | 23 | 6 | 0 |
| Sulcata | 23 | 6 | 0 | |
| Rubro | 0 | 0 | 39 |
| Predicted Species | Calibration Model | PCs | Calibration | Cross-Validation | Validation | External-Validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSEC | SEC | RPD | R2 | RMSECV | SECV | RPD | R2 | RMSEP | SEP | RPD | R2 | RMSEP | SEP | RPD | |||
| Database 1 —3 species combined | FG model | 6 | 0.97 | 0.34 | 0.34 | 6.29 | 0.96 | 0.43 | 0.43 | 5.00 | 0.88 | 0.69 | 0.70 | 2.89 | ||||
| CG model | 9 | 0.97 | 0.38 | 0.38 | 5.70 | 0.94 | 0.57 | 0.57 | 3.79 | 0.82 | 0.88 | 0.89 | 2.26 | |||||
| DW model | 9 | 0.90 | 0.63 | 0.64 | 3.25 | 0.80 | 0.94 | 0.94 | 2.20 | 0.42 | 1.48 | 1.52 | 1.34 | |||||
| FW model | 9 | 0.96 | 0.36 | 0.36 | 5.28 | 0.83 | 0.75 | 0.76 | 2.50 | 0.22 | 1.78 | 1.84 | 1.11 | |||||
| Database 2 —Bissetti | FG model | 7 | 0.98 | 0.25 | 0.25 | 8.15 | 0.96 | 0.40 | 0.41 | 5.09 | 0.92 | 0.59 | 0.61 | 3.60 | 0.91 | 0.65 | 0.65 | 3.37 |
| CG model | 9 | 0.98 | 0.21 | 0.21 | 9.10 | 0.96 | 0.47 | 0.47 | 3.98 | 0.81 | 0.90 | 0.93 | 2.36 | 0.91 | 0.82 | 0.70 | 2.65 | |
| DW model | 6 | 0.85 | 0.77 | 0.78 | 2.58 | 0.71 | 1.12 | 1.13 | 1.78 | 0.75 | 1.06 | 1.11 | 2.00 | 0.68 | 1.26 | 1.16 | 1.73 | |
| FW model | 9 | 0.96 | 0.34 | 0.34 | 5.03 | 0.89 | 0.65 | 0.66 | 2.61 | 0.21 | 1.96 | 2.05 | 1.09 | 0.39 | 1.93 | 1.82 | 1.13 | |
| Database 3 —Sulcata | FG model | 7 | 0.96 | 0.29 | 0.29 | 4.92 | 0.93 | 0.39 | 0.39 | 3.64 | 0.93 | 0.47 | 0.46 | 3.58 | 0.96 | 0.70 | 0.70 | 3.59 |
| CG mode | 9 | 0.97 | 0.27 | 0.27 | 5.86 | 0.92 | 0.45 | 0.46 | 3.50 | 0.95 | 0.36 | 0.38 | 4.59 | 0.76 | 1.34 | 1.37 | 1.86 | |
| DW model | 7 | 0.92 | 0.42 | 0.42 | 3.53 | 0.72 | 0.79 | 0.79 | 1.87 | 0.25 | 1.61 | 1.65 | 1.04 | 0.61 | 1.53 | 1.56 | 1.63 | |
| FW model | 9 | 0.97 | 0.21 | 0.21 | 6.19 | 0.88 | 0.47 | 0.48 | 2.78 | 0.37 | 1.47 | 1.40 | 1.13 | 0.29 | 2.30 | 2.33 | 1.09 | |
| Database 4 —Rubro | FG model | 6 | 0.97 | 0.40 | 0.41 | 5.75 | 0.93 | 0.62 | 0.63 | 3.70 | 0.99 | 0.29 | 0.26 | 12.80 | 0.82 | 0.65 | 0.65 | 2.14 |
| CG model | 6 | 0.97 | 0.41 | 0.41 | 5.86 | 0.94 | 0.59 | 0.60 | 4.06 | 0.76 | 1.88 | 1.91 | 1.95 | 0.84 | 1.20 | 0.56 | 1.16 | |
| DW model | 6 | 0.91 | 0.57 | 0.58 | 3.33 | 0.61 | 1.26 | 1.29 | 1.51 | 0.44 | 2.75 | 2.83 | 1.33 | 0.30 | 1.43 | 1.45 | 0.97 | |
| FW model | 11 | 0.96 | 0.46 | 0.46 | 5.05 | 0.58 | 1.44 | 1.46 | 1.61 | 0.80 | 2.01 | 2.10 | 1.82 | 0.34 | 1.44 | 1.38 | 0.97 | |
| Calibration Model | Slope | Offset | Correlation | R2 | RMSEP | SEP | Bias | RPD |
|---|---|---|---|---|---|---|---|---|
| FG sample–FG model | 0.85 | 2.59 | 0.96 | 0.91 | 0.65 | 0.65 | 0.02 | 3.37 |
| CG sample−FG model—before SBC | 0.96 | 0.51 | 0.93 | 0.87 | 0.83 | 0.82 | −0.23 | 2.62 |
| CG sample–FG model—after SBC | 0.82 | 3.19 | 0.93 | 0.87 | 0.79 | 0.80 | 0.16 | 2.73 |
| CG sample–CG model | 0.80 | 3.93 | 0.95 | 0.91 | 0.82 | 0.70 | 0.44 | 2.65 |
| DW sample–FG model—before SBC | 0.60 | 8.43 | 0.85 | 0.73 | 1.99 | 1.19 | 1.62 | 1.10 |
| DW sample–FG model—after SBC | 0.58 | 7.26 | 0.85 | 0.73 | 1.19 | 1.20 | 0.20 | 1.84 |
| DW sample–CG model—before SBC | 0.41 | 12.51 | 0.72 | 0.51 | 2.67 | 1.53 | 2.21 | 0.80 |
| DW sample–CG model—after SBC | 0.31 | 12.40 | 0.72 | 0.51 | 1.62 | 1.62 | 0.38 | 1.32 |
| DW sample–DW model | 0.57 | 7.27 | 0.83 | 0.68 | 1.26 | 1.26 | −0.16 | 1.73 |
| FW sample–FG model—before SBC | 0.70 | −9.30 | 0.66 | 0.44 | 14.42 | 1.77 | −14.32 | 0.15 |
| FW sample–FG model—after SBC | 0.26 | 12.96 | 0.66 | 0.44 | 1.72 | 1.67 | 0.56 | 1.22 |
| FW sample–CG model—before SBC | 0.68 | −13.33 | 0.53 | 0.28 | 19.05 | 2.45 | −18.91 | 0.11 |
| FW sample–CG model—after SBC | 0.19 | 14.41 | 0.53 | 0.28 | 1.85 | 1.86 | 0.36 | 1.16 |
| FW sample–DW model—before SBC | 0.50 | −6.15 | 0.77 | 0.59 | 14.59 | 1.42 | −14.52 | 0.15 |
| FW sample–DW model—after SBC | 0.40 | 10.22 | 0.77 | 0.59 | 1.46 | 1.50 | 0.09 | 1.49 |
| FW sample–FW model | 0.57 | 6.68 | 0.62 | 0.39 | 1.93 | 1.82 | −0.68 | 1.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, Q.; Santos-Rivera, M.; Ouyang, X.; Kouba, A.J.; Vance, C.K. Near-Infrared Spectroscopy and Mode Cloning (NIR-MC) for In-Situ Analysis of Crude Protein in Bamboo. Remote Sens. 2022, 14, 1302. https://doi.org/10.3390/rs14061302
Sheng Q, Santos-Rivera M, Ouyang X, Kouba AJ, Vance CK. Near-Infrared Spectroscopy and Mode Cloning (NIR-MC) for In-Situ Analysis of Crude Protein in Bamboo. Remote Sensing. 2022; 14(6):1302. https://doi.org/10.3390/rs14061302
Chicago/Turabian StyleSheng, Qingyu, Mariana Santos-Rivera, Xiaoguang Ouyang, Andrew J. Kouba, and Carrie K. Vance. 2022. "Near-Infrared Spectroscopy and Mode Cloning (NIR-MC) for In-Situ Analysis of Crude Protein in Bamboo" Remote Sensing 14, no. 6: 1302. https://doi.org/10.3390/rs14061302
APA StyleSheng, Q., Santos-Rivera, M., Ouyang, X., Kouba, A. J., & Vance, C. K. (2022). Near-Infrared Spectroscopy and Mode Cloning (NIR-MC) for In-Situ Analysis of Crude Protein in Bamboo. Remote Sensing, 14(6), 1302. https://doi.org/10.3390/rs14061302






