2.1. Dynamic Time Warping
A similarity measure is a fundamental tool for k-nearest neighbor classification of SITS [
51,
52]. Currently, Euclidean distance and Dynamic Time Warping (DTW) [
53,
54] are the two most widely used similarity measure prototypes.
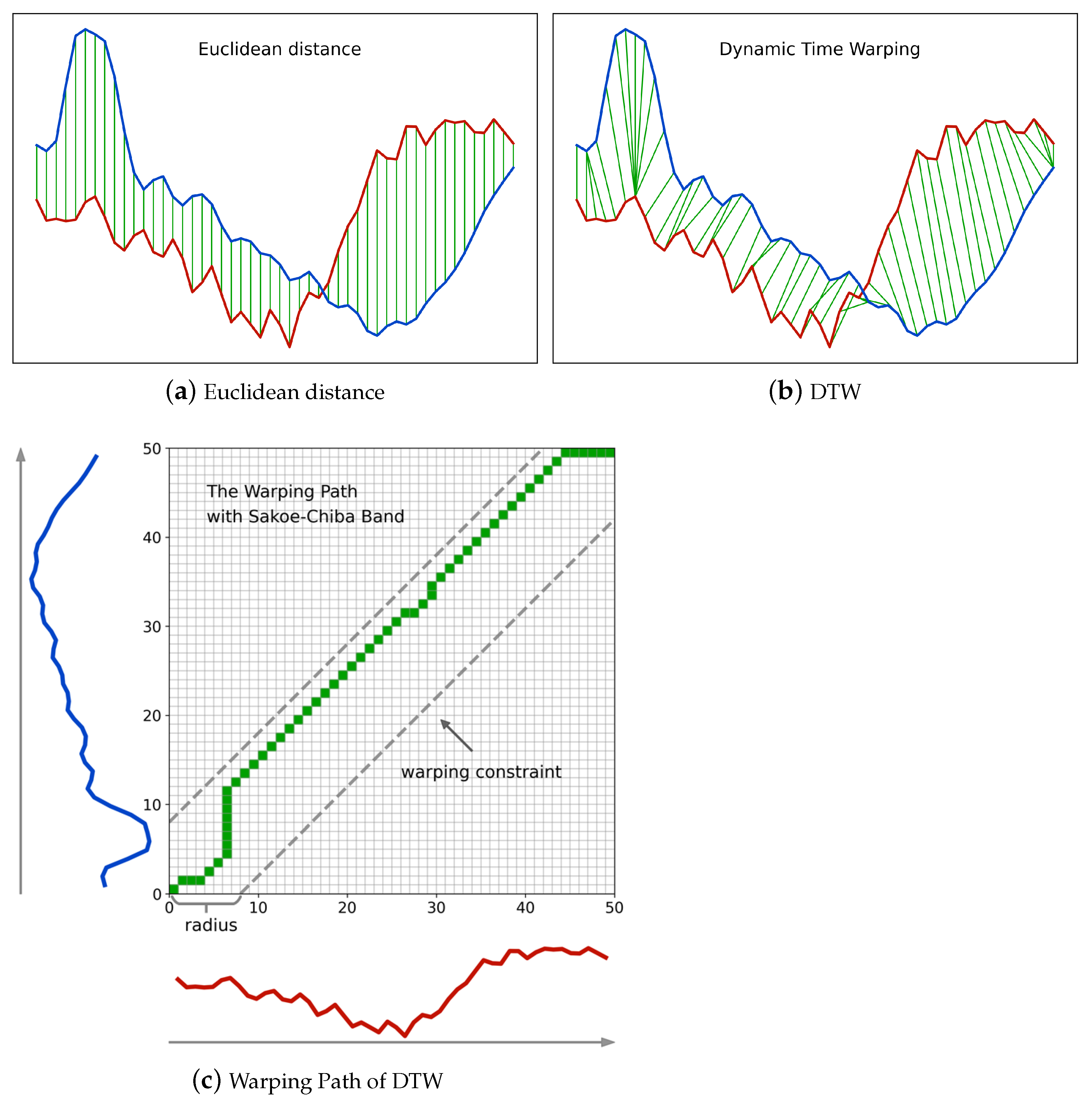
Figure 2a,b illustrates Euclidean distance and DTW, respectively. We observe that Euclidean distance imposes a temporally linear alignment between time series, while DTW is more flexible, and non-linear alignment is allowed in a temporally local range. The flexibility of DTW enables it to cope with time distortions effectively and makes DTW a more suitable similarity measure for complex SITS data.
To give a quantitative definition of Euclidean distance and DTW, let
and
be two time series of length
I and
J. The lowercase
and
with subscript
i and
j denote the
i-
and
j-
elements of time series
A and
B, respectively. The cost between the
i-
element of
A and the
j-
element of
B is denoted by
. In this setting, the Euclidean distance between
A and
B can be formulated as:
where
skips the usual square-root operation to save more calculation time, and the squared version does not influence the result of the k-nearest neighbor classifier.
As for DTW, its flexibility comes from its attempt to find an optimal alignment that achieves a minimum accumulated cost between two time series. Unlike the Euclidean distance, in DTW the k- element of A is not always aligned with the k- element of B, and thus a warping path is used to record each pair of aligned elements. The warping path is usually denoted by , where each time warp denotes a pair composed of and . The length of the warping path is the uppercase K.
Figure 2c shows the warping path in correspondence with
Figure 2b, where the x-coordinate and y-coordinate of each matrix cell in the warping path are also the indices of elements in the two time series, respectively. The final accumulated cost between the two time series, namely the DTW distance, is the sum of all pairwise costs. In this setting, DTW can be formulated as:
where
,
and
.
Equation (
2) is more of a conceptional definition of DTW than a solution. DTW, as defined by Equation (
2), is a typical dynamic programming problem that can be solved by a more straightforward recursive formula:
where
is the partial DTW distance between sub-sequences composed of the first
i elements of
A and the first
j elements of
B.
is the final DTW distance between the two entire time series.
Based on the fact that if two elements from different time series are temporally too far, their correlation tends to be weak and they should not be paired together, many constraints on DTW have been proposed to limit the warping path inside a warping window [
55,
56]. Among these constraints the Sakoe–Chiba band [
53] as shown in
Figure 2c is the most intuitive yet effective one. Given a radius
r of the Sakoe–Chiba band, the temporal difference of two time series elements
cannot exceed
r for any time warp
in a warping path. In addition, the Sakoe–Chiba constraint is a prerequisite for
LB_Keogh, a lower bound of DTW that will be used in our method and will be introduced in the next subsection. Thus, we adopt the Sakoe–Chiba constraint throughout this work.
2.2. Lower Bounds of DTW
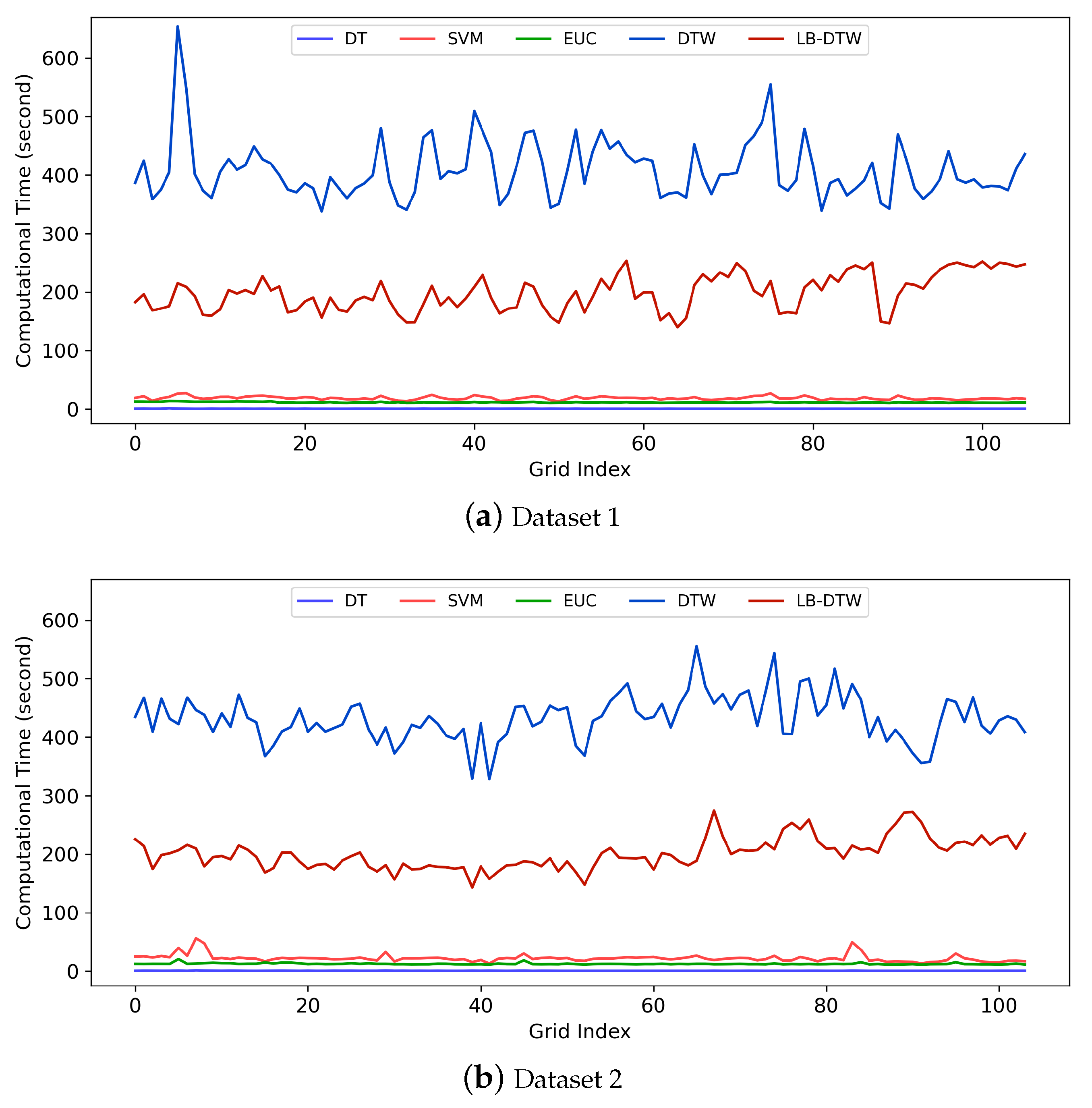
As the name implies, a lower bound of DTW is a value that is guaranteed to be smaller than DTW. If the computation speed of a lower bound is significantly faster than DTW, then the lower bound can be used to prune off unpromising candidates and thus speed up the k-nearest neighbor search of time series. For example, suppose the threshold distance for a candidate B to be the k-nearest neighbor of a time series A is . If a lower bound distance between A and B is and , then we can be sure that the DTW distance since is guaranteed, and because of , which means the distance is too far, the candidate can be pruned off, and the calculation of DTW can be safely skipped.
In this manner, a portion of time-consuming calculations of DTW can be replaced by the faster calculations of lower bounds, and thus the entire k-nearest neighbor classification process is accelerated. The use of lower bounds still maintains exactly the same result as the raw DTW rather than generating an approximate result.
Besides the computation speed, tightness is another important property for a lower bound. Tightness indicates how close the lower bound is to the original measure. If the tightness is high, more candidates will be pruned off and vice versa. Usually there is a tradeoff between the tightness and the computation speed of a lower bound, and thus it is difficult to find the best lower bound to use. Since each single lower bound has its weakness, a classic strategy is to use different kinds of lower bounds in a cascade. For example, we can first employ a fast lower bound to reject some obvious outliers and then employ a tight lower bound to maintain a high prune rate. In our method, we use
LB_Kim [
48] as the fast lower bound and
LB_Keogh [
44,
47] as the tight one for DTW.
Equation (
4) shows the definition of
LB_Kim and it has a computational complexity of
, which is the fastest possible situation.
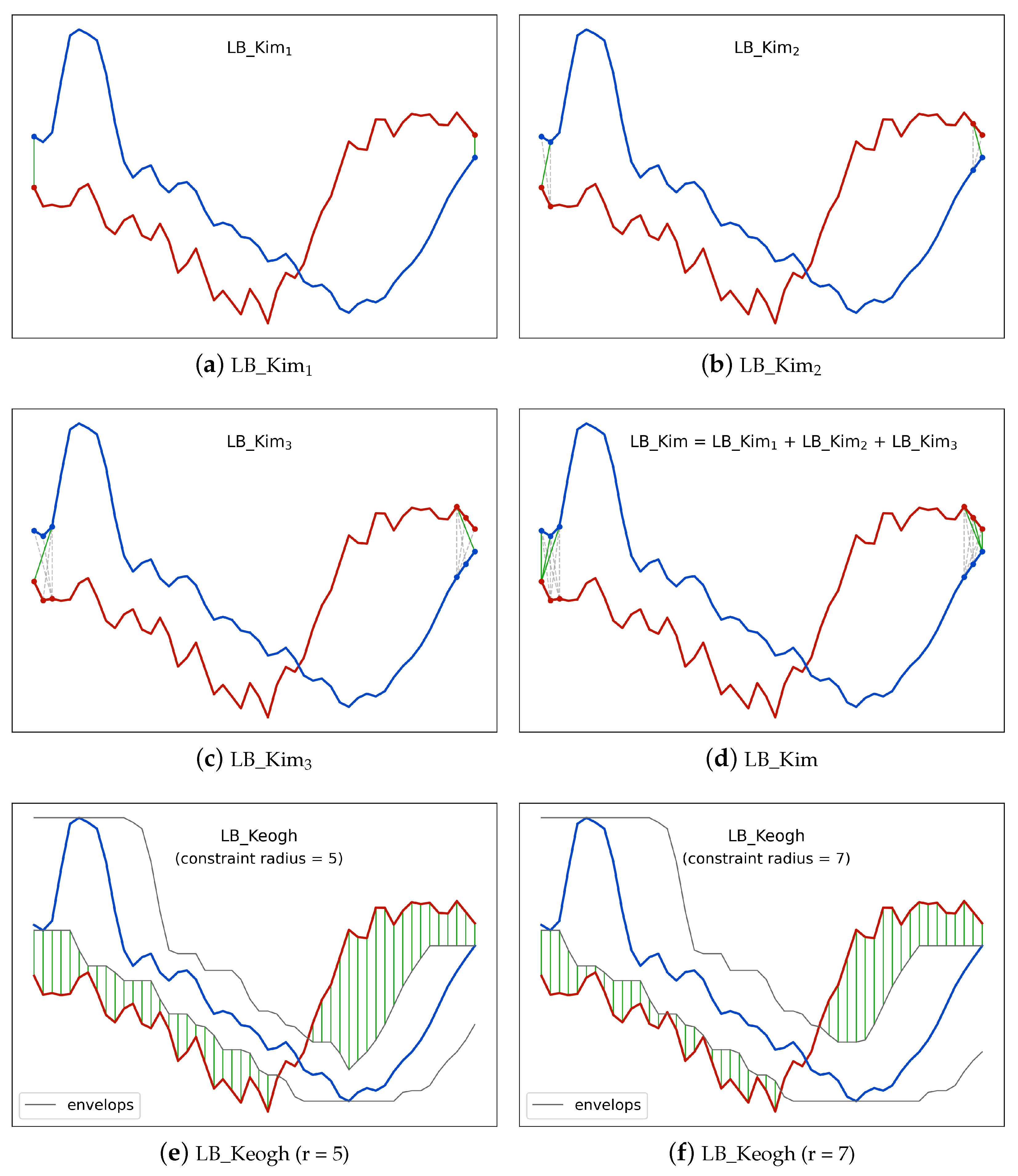
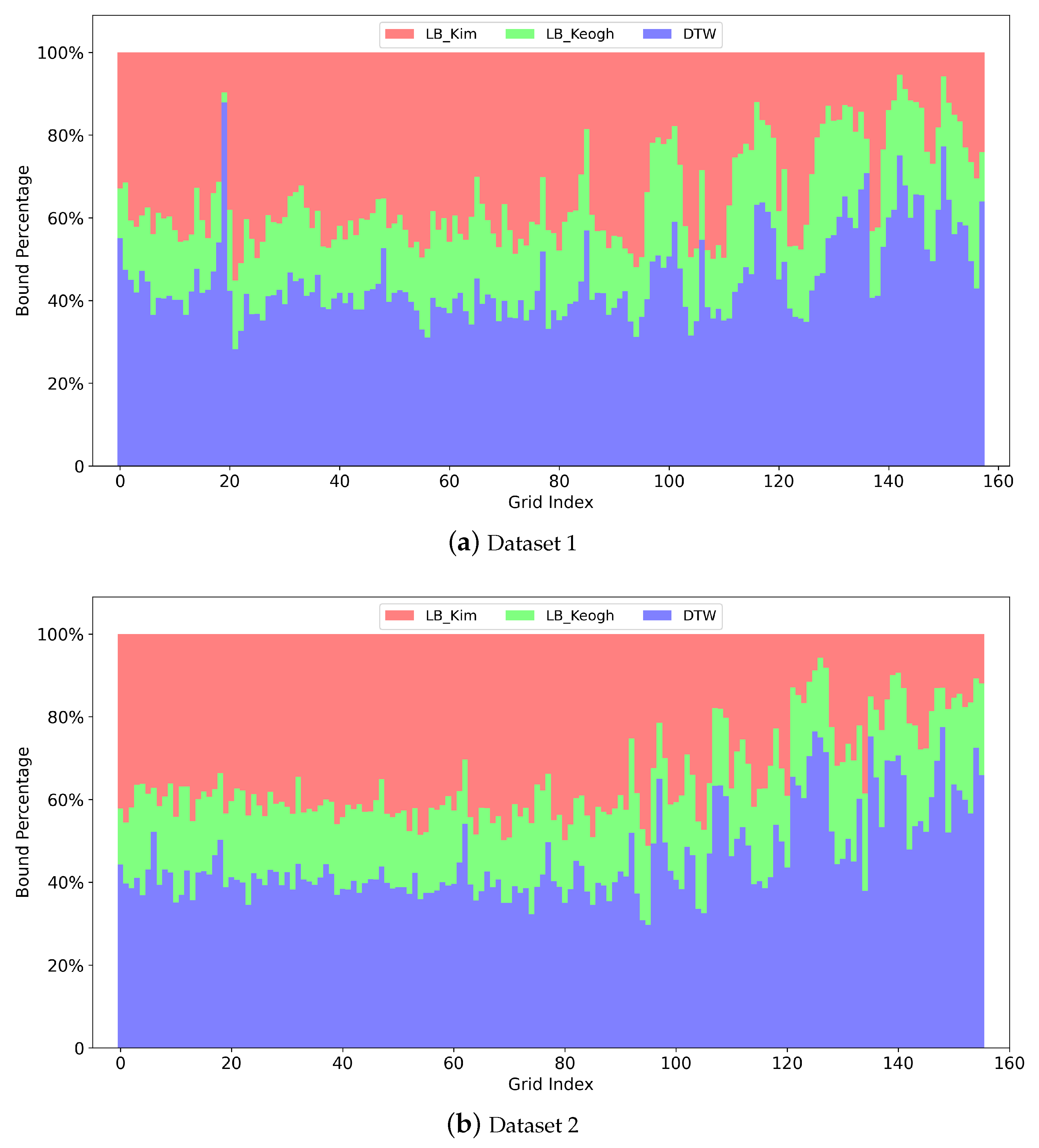
Figure 3 illustrates an examples of
LB_Kim. The full
LB_Kim is the sum of three parts. The first part
LB_Kim as shown in
Figure 3a considers the minimum possible cost among the starting elements and the ending elements of the two time series given the rule of DTW. Similarly, the second part
LB_Kim as shown in
Figure 3b further considers the minimum possible cost among the first two and the last two elements, and the third part
LB_Kim as shown in
Figure 3c considers the situation for the first three and the last three elements.
Figure 3d shows the combination of the three parts.
where
is the cost between the
i-
element of
A and the
j-
element of
B.
In contrast with
LB_Kim,
LB_Keogh achieves a higher tightness by comparing one time series with the upper and lower envelopes of the other time series. Equation (
5) defines the envelopes of time series and Equation (
6) defines
LB_Keogh. The upper or lower envelope consists of a sequence of local maximum or local minimum values for a sequence of sliding windows centered at each element of a time series. The length of the sliding window is
where the
r is the radius of the Sakoe–Chiba band of DTW. In this setting,
LB_Keogh is guaranteed to be smaller than DTW, and the proof can be found in [
47]. Given a pair of envelopes,
LB_Keogh sums the cost caused by elements larger than the upper envelope and elements smaller than the lower envelop.
Figure 3e,f illustrates the
LB_Keogh with different Sakoe–Chiba band radii.
where
r is the radius of the Sakoe–Chiba band of DTW, and
is the
i-
element of time series
A.
U and
L have the same length
I as
A and
.
where
,
and
are the
i-
element of time series
B, upper envelope
U and lower envelope
L, respectively.
For cascading lower bounds, we calculate LB_Kim first, and if LB_Kim is smaller than the current k-nearest neighbor threshold, we calculate LB_Keogh. If LB_Keogh is still smaller than the threshold, we calculate DTW. In this manner, we create two additional chances to skip the time-consuming calculation of DTW.
2.3. Early Abandoning of DTW
For a k-nearest neighbor classification problem, the similarity measure is used to decide whether a time series is close enough to be the neighbor of another time series. As soon as we know the distance between two time series is already too large to be neighbors, the calculation of distance can be abandoned to save more time. For many similarity measures we cannot know the interim results until the full calculation is completed; however, fortunately, DTW is calculated in an incremental manner, and we can obtain the interim result at each step.
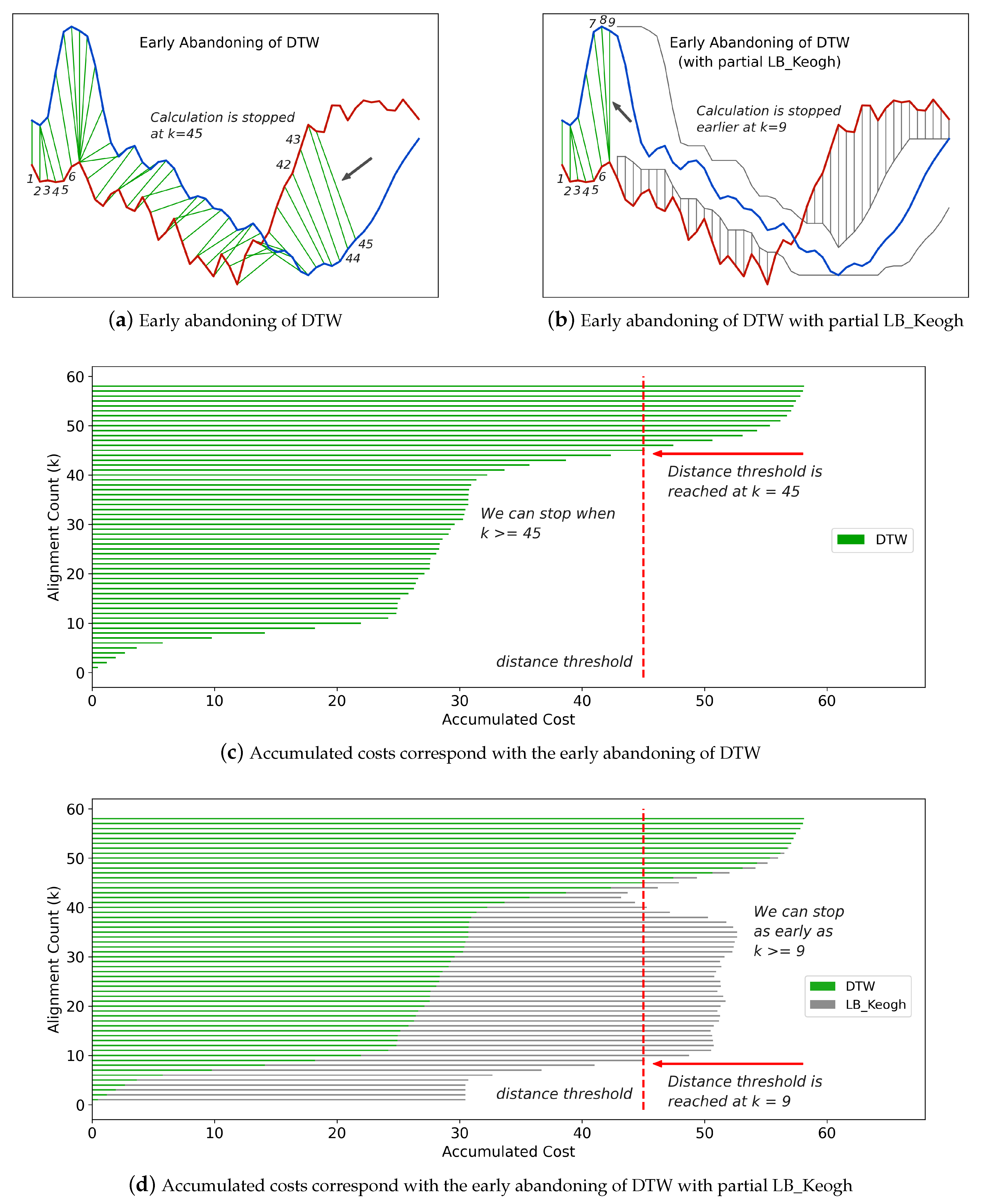
Figure 4a illustrates an example of early abandoning of DTW, and
Figure 4c shows the corresponding accumulated costs at each step. From
Figure 4c, we observe that the distance threshold is reached at the 45th step, and correspondingly in
Figure 4a, the calculation of DTW is abandoned at the 45th step.
Another trick to make the abandoning happen even earlier is to adopt partial
LB_Keogh during the calculation of DTW [
48]. At any step
k, compared with
, the sum of
is a closer lower bound to the full DTW distance
. With the
LB_Keogh contribution from
to
I, we can always predict the full DTW distance and thus conclude whether the distance will exceed the threshold in an earlier step.
Figure 4b,d illustrates the use of partial
LB_Keogh and its corresponding accumulated costs at each step. We observe that with partial
LB_Keogh the abandoning happens at as early as the ninth step.
2.4. Seeded Classification of SITS
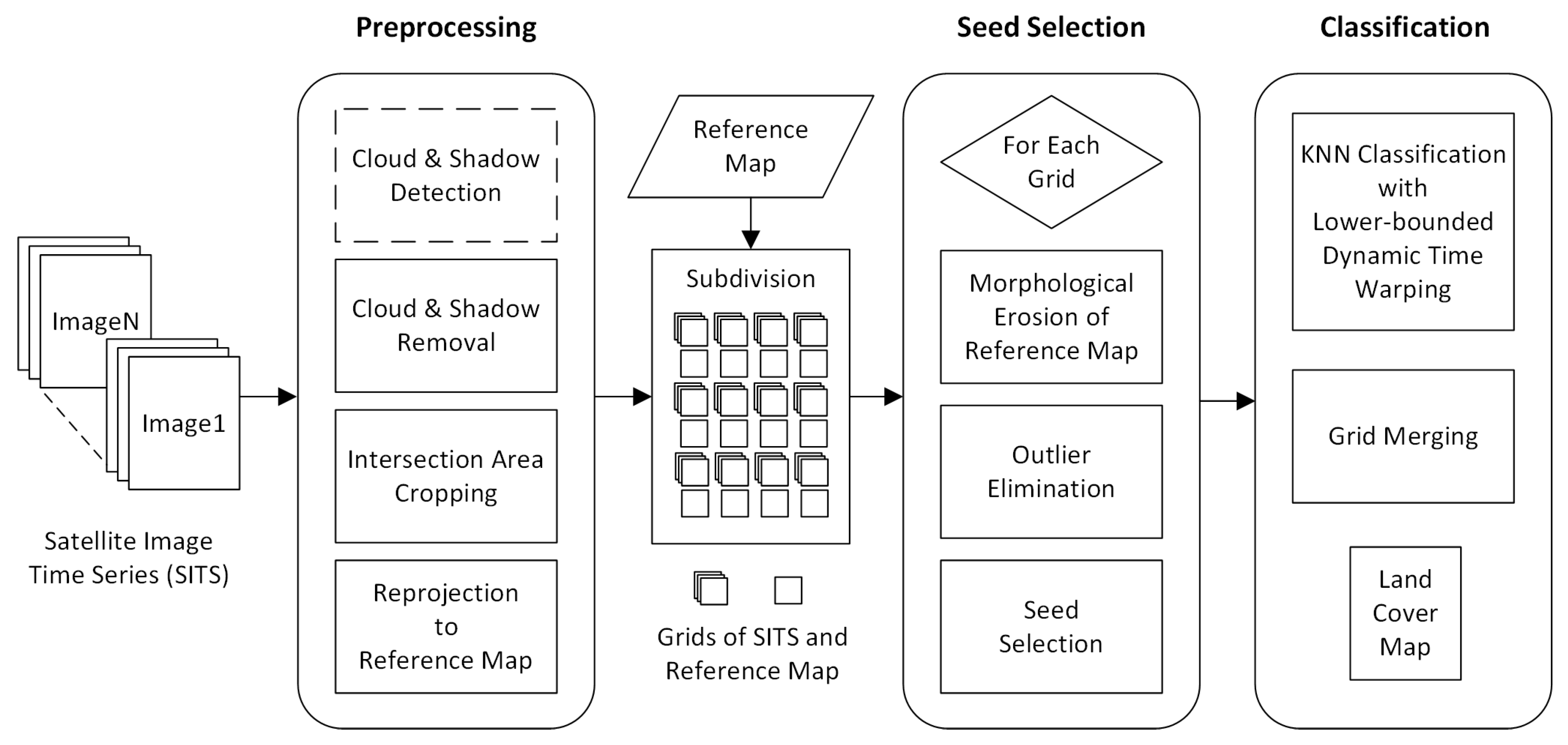
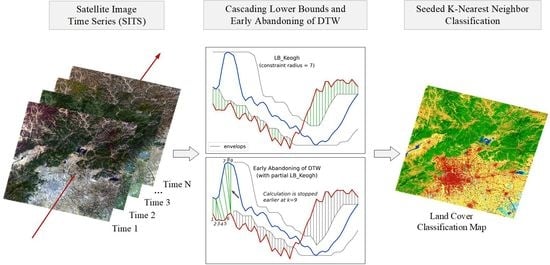
After the description of DTW based similarity measure combinations, in this subsection we present the whole process of the proposed seeded classification method for SITS.
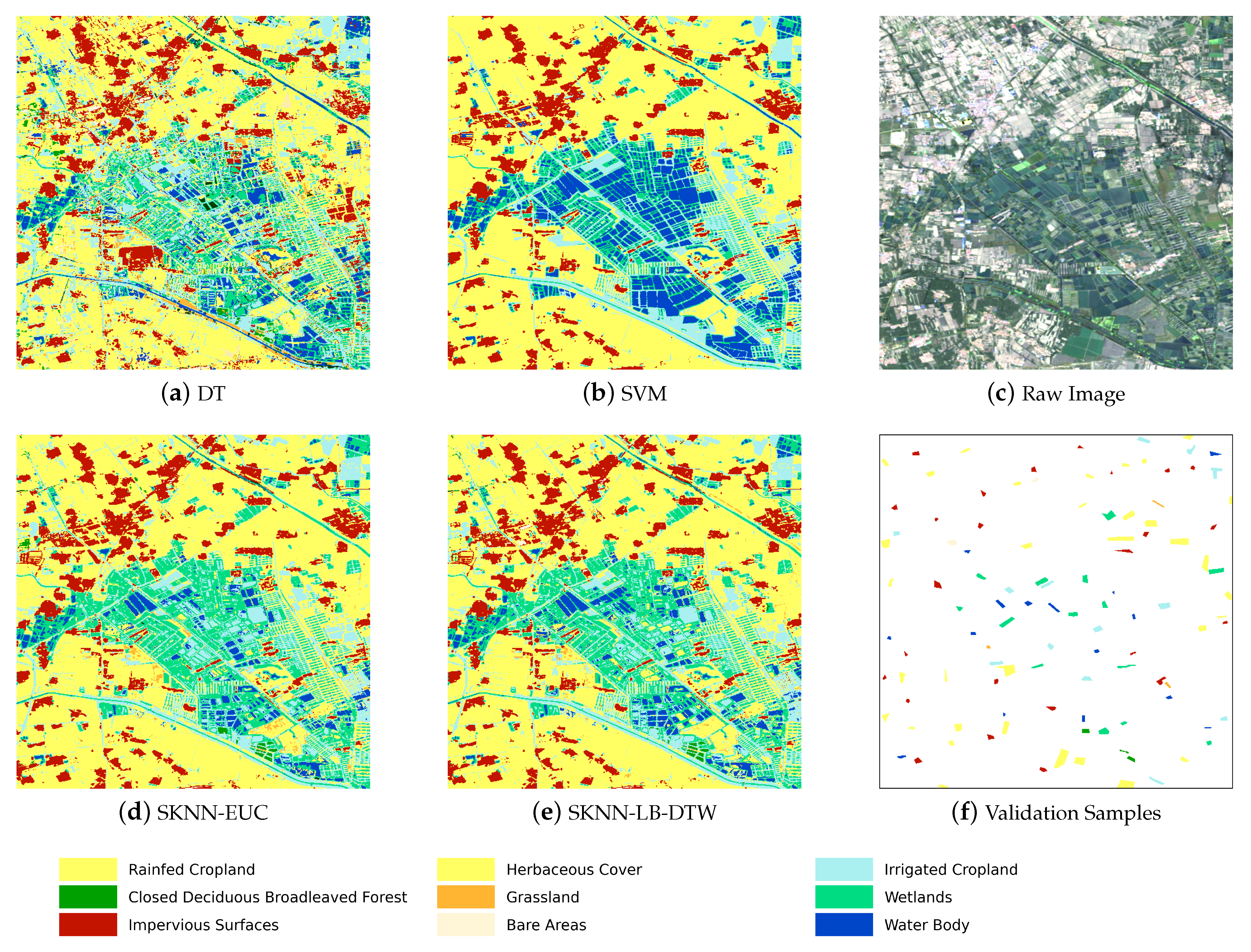
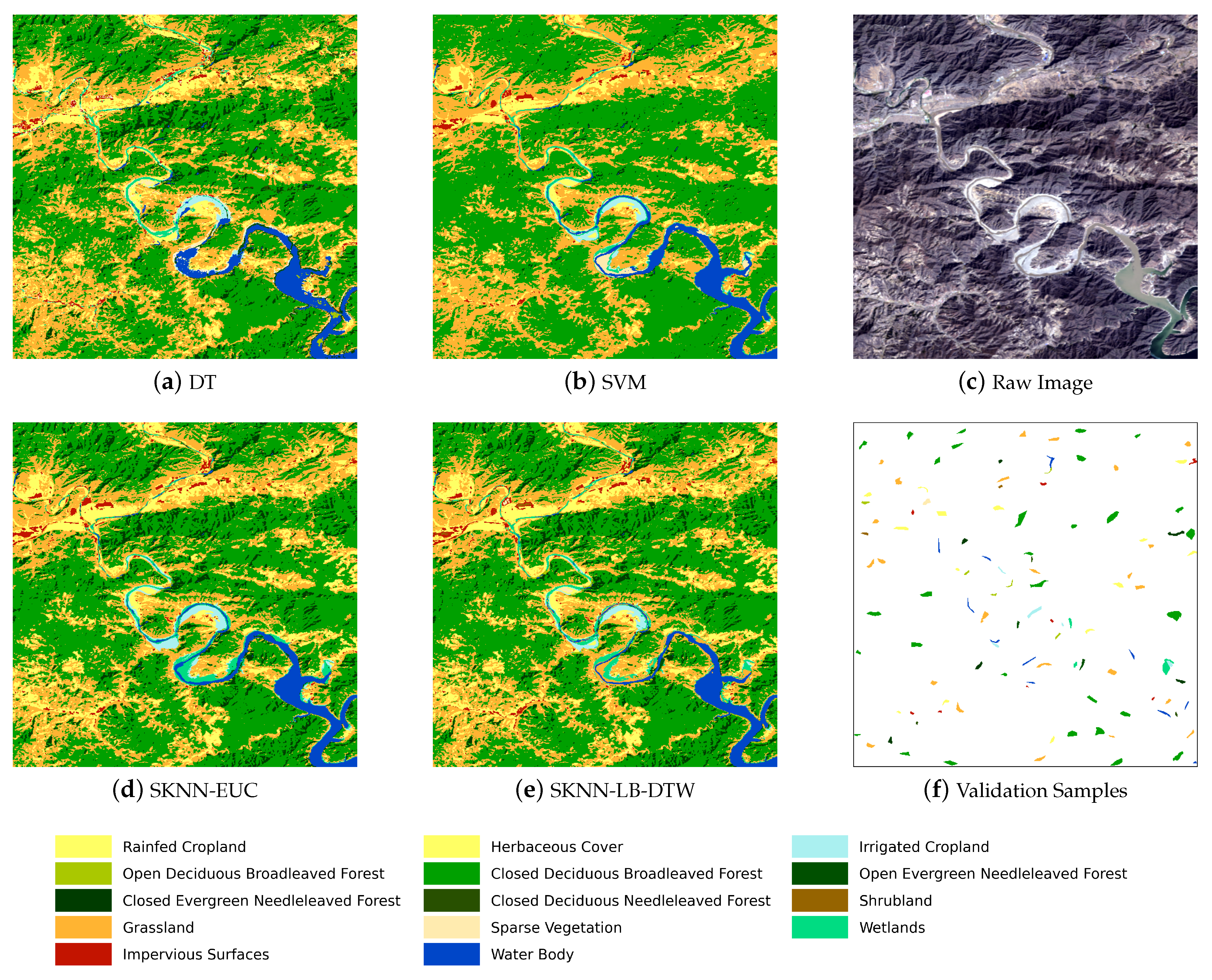
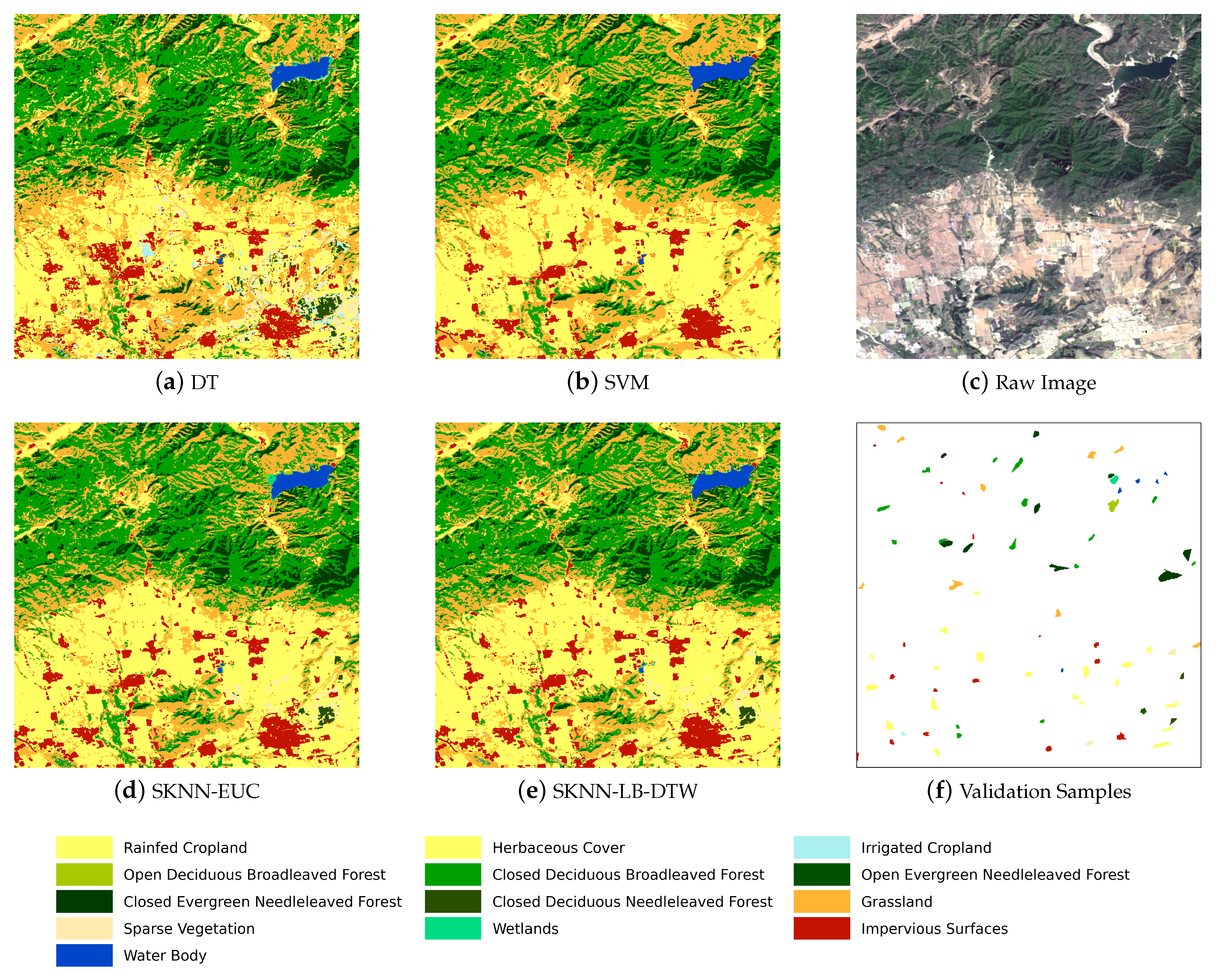
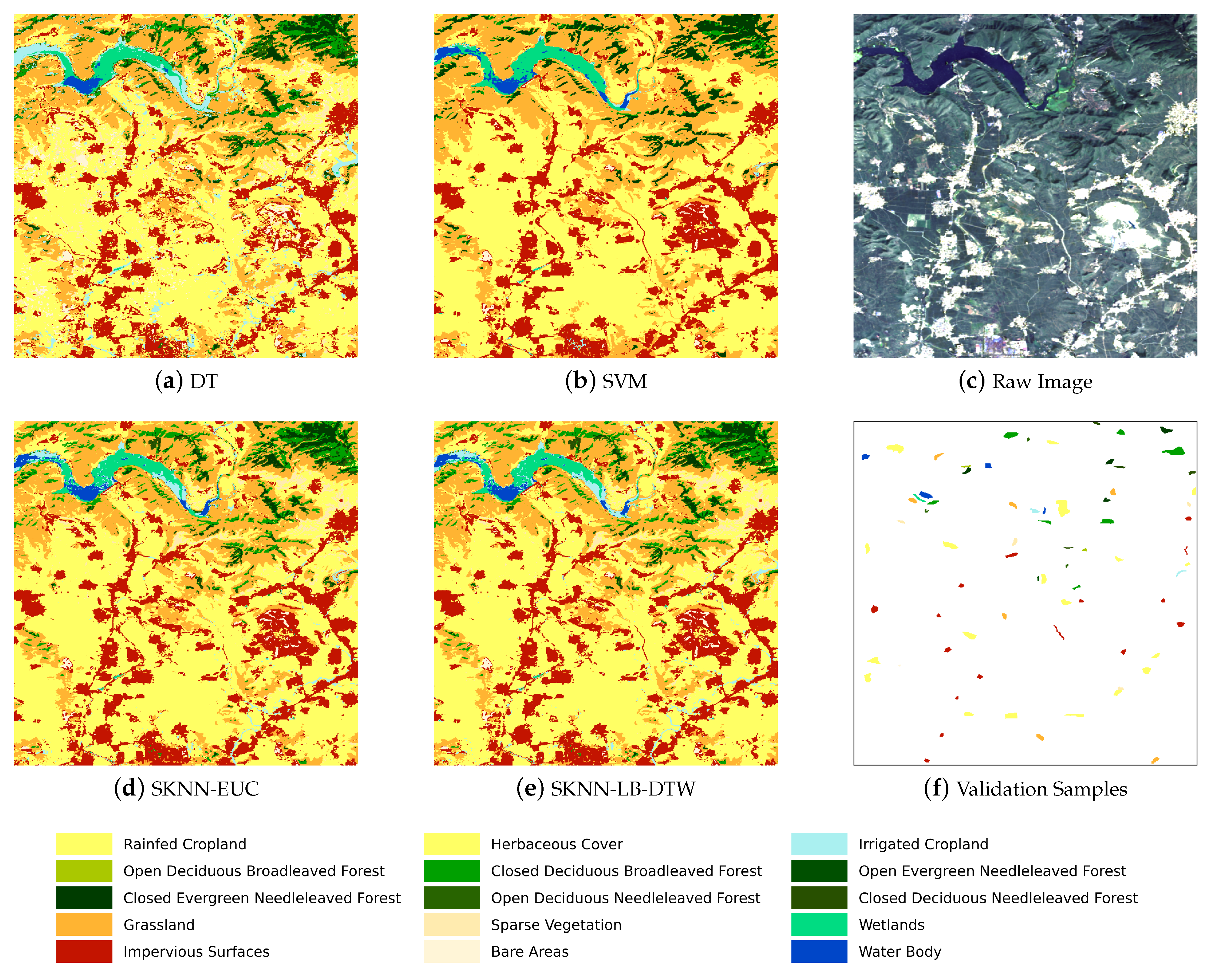
Figure 5 shows the complete flow chart with three main stages. The first stage is the preprocessing of SITS data. Optical satellite imagery usually contains massive cloud and shadow contaminated pixels. In order to ensure the continuity of temporal information, one major task is to recover cloud and shadow contaminated pixels for each image in a SITS.
If there is no cloud and shadow mask, cloud and shadow detection has to be conducted beforehand. Then, we use the information transfer method proposed in [
50] to recover contaminated pixels. For each cloud or shadow contaminated image patch, the method searches for a similar image patch from the same geographical location in other images of the SITS, and then Poisson blending [
57] is employed to make image patches transferred from other images fit seamlessly into the current image. For images that are too cloudy to recover, we must skip them.
For many satellites, such as Landsat 8 and GaoFen 1, images with the same tile number do not cover exactly the same spatial scope due to some linear offsets. Thus, we crop the intersection area of all images in a SITS to ensure pixels with the same coordinates in different images cover the same spatial scope and all time series have the same length. If a reference classification map with training labels is used for seed selection, we need to reproject all images to the same map projection system and spatial resolution as the reference map.
Due to the large size of satellite images, a full-size SITS with dozens of images will cost hundreds or thousands gigabytes of memory, which is difficult to satisfy by conventional computers. Therefore, images and the corresponding reference map have to be subdivided into multiple grids. In this work, the size of grids is .
The second stage of the process is the selection of labeled seeds for each land cover class. We select seeds from existing land cover classification products, such as FROM-GLC30 [
58] or GLC-FCS30 [
59]. With the aim of exploring the rich information contained in SITS data, rather than letting the training samples dominate the classification results, we strictly limit the number of seeds for each class.
Concretely, suppose the number of seeds for a class is denoted by S, the total number of samples of that class in the reference map is N, and the number of nearest neighbors is K for the classifier. Then, we make . The former part greatly condenses the number of samples, for example, , and it also makes the number of seeds proportional to the ground truth in general. The latter part allows tiny classes to exist by giving them the minimum number of samples required by the k-nearest neighbor classifier.
Since the numbers of seeds are limited, the quality of seeds becomes critical. We adopt two techniques, one morphological and one statistical, to select more correctly classified samples and enhance the reliability of seeds. We first morphologically erode [
60] all class labels in the reference map to keep only the central pixels of each land cover patch, because the central pixels usually have a higher possibility to be correctly classified. Then, we use the statistical isolation forest [
61,
62] algorithm to keep only the inliers, and finally seeds are randomly selected from the inliers according to the given quantity. If the number of selected seeds for a class is less than
, we randomly select non-repeated samples before the erosion for complements.
Given the seeds and SITS data, the third stage is the k-nearest neighbor classification. The seeds are used as the training samples and the combination of cascading lower bounds and early abandoning of DTW are used as the similarity measure. The classification is conducted by grids and results of all grids are finally merged into one land cover map.
Concretely, for each input sample to be classified, we first find the K closest training examples of it, and then it is classified by a plurality vote of the K closest neighbors. To search for the K closest training examples, we iterate through all training samples and check whether a training sample is close enough to be the top K closest ones. However, we do not calculate the DTW distance directly because of its high computational complexity. Instead, we calculate the lower bounds of DTW first to prune off unnecessary calculations of DTW.
If a lower bound is already too large to make a training sample one of the K closest neighbors, then the calculation of DTW for this training sample is unnecessary because DTW is guaranteed to be larger than its lower bounds. The threshold to decide whether a training sample belongs to the top K closest ones is the DTW distance between the current k- closest training sample and the input sample. If lower bounds are smaller than the threshold, we calculate the DTW and compare the DTW distance with the threshold. To accelerate each single calculation of DTW, the early abandoning strategy can be adopted because DTW can be calculated incrementally.
During DTW calculation, we observe the interim result at each step and as soon as the threshold is reached, we know this training sample is already too far to be the top K closest neighbors and the calculation can be abandoned safely. If a training sample is closer than the threshold, then it will replace one of the old K closest neighbors, and the threshold should be updated by the new k- closest distance. In the entire classification process, we do not abandon the classification of any sample but we abandon the unnecessary calculations during the search for the K closest neighbors. All samples will be classified as the standard k-nearest neighbor classification.
It is worth noting that the process as a whole is actually unsupervised because the seed selection is unsupervised, and the succeeding classification turns unsupervised without manually training sample preparation. This enables the automatic classification of SITS in a large or even global scale.