The Classification Method Study of Crops Remote Sensing with Deep Learning, Machine Learning, and Google Earth Engine
Abstract
:1. Introduction
2. Study Area and Datasets
2.1. Study Area
2.2. Satellite Data Processing
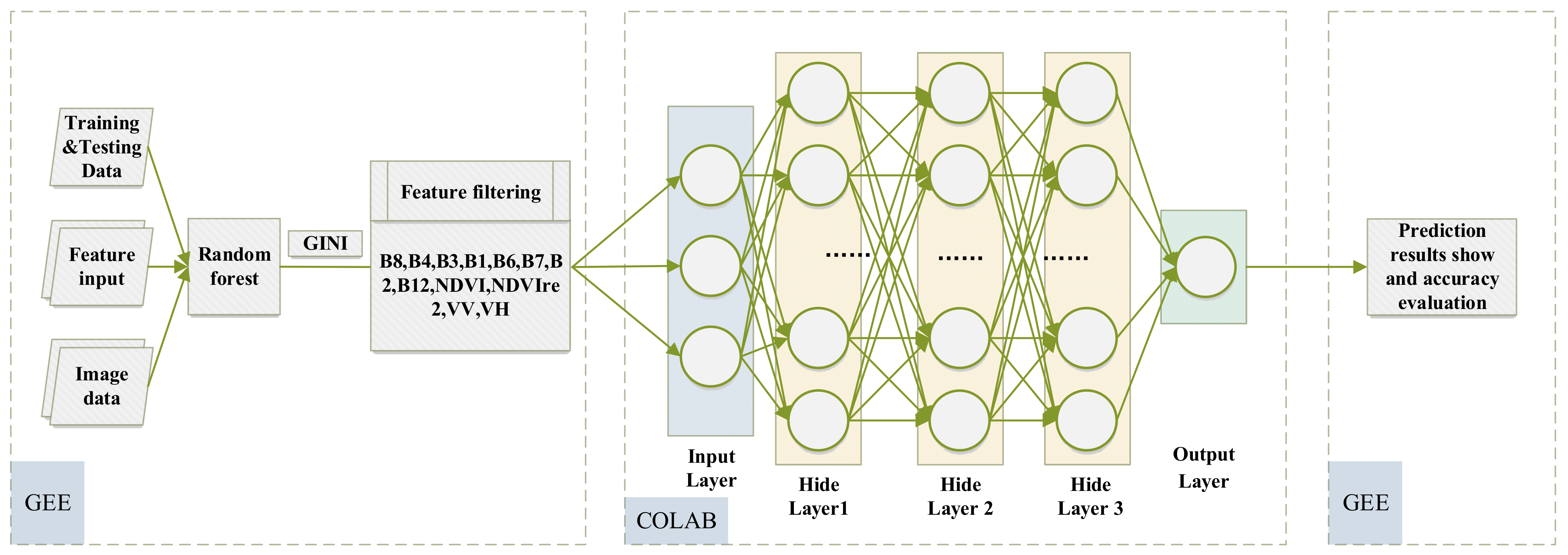
2.2.1. Data Processing Training Platform
2.2.2. Data Processing
- (1)
- Sentinel-2 Data
- (2)
- Sentinel-1 Data
2.3. Samples Selection and Set
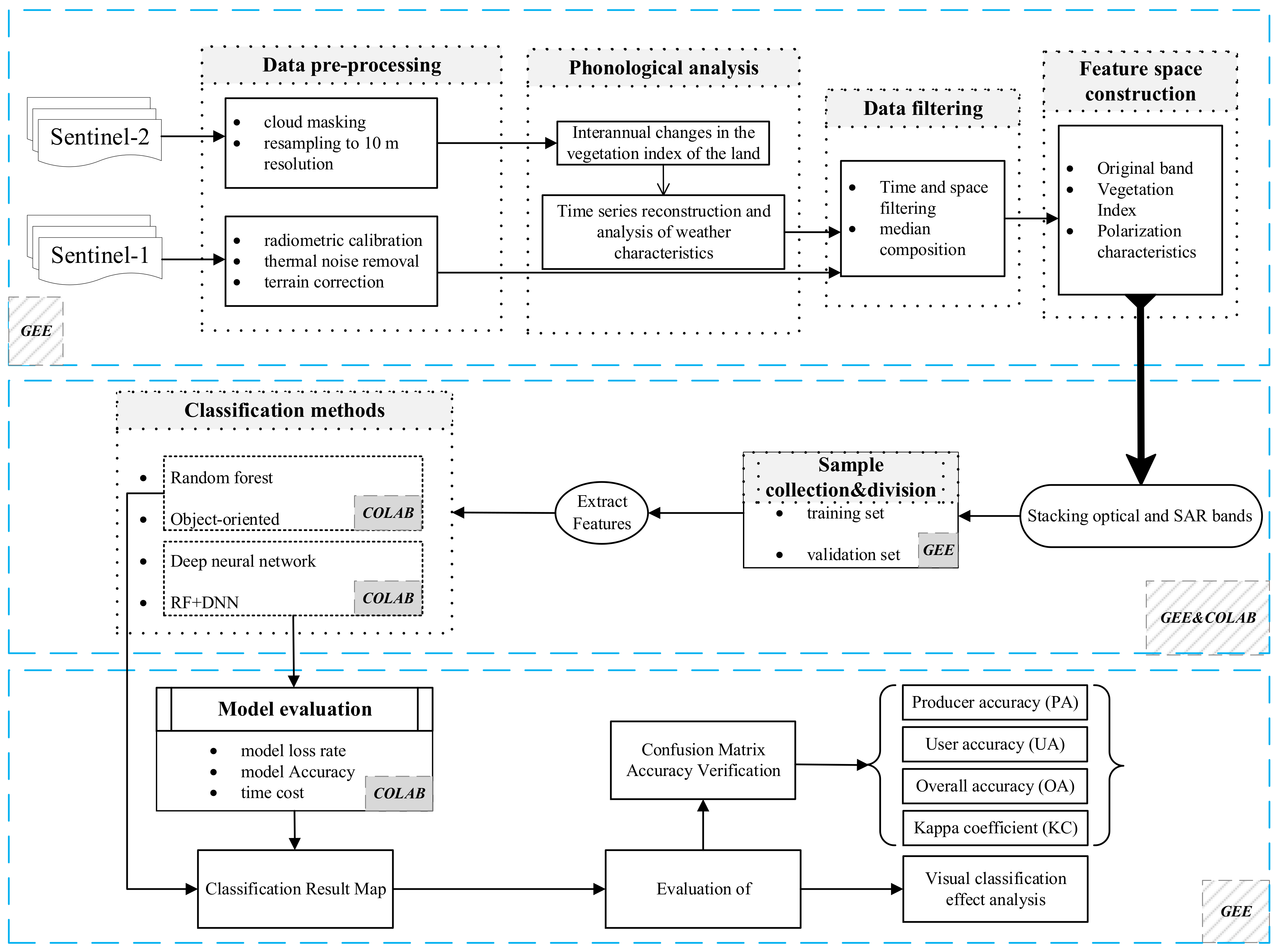
3. Method
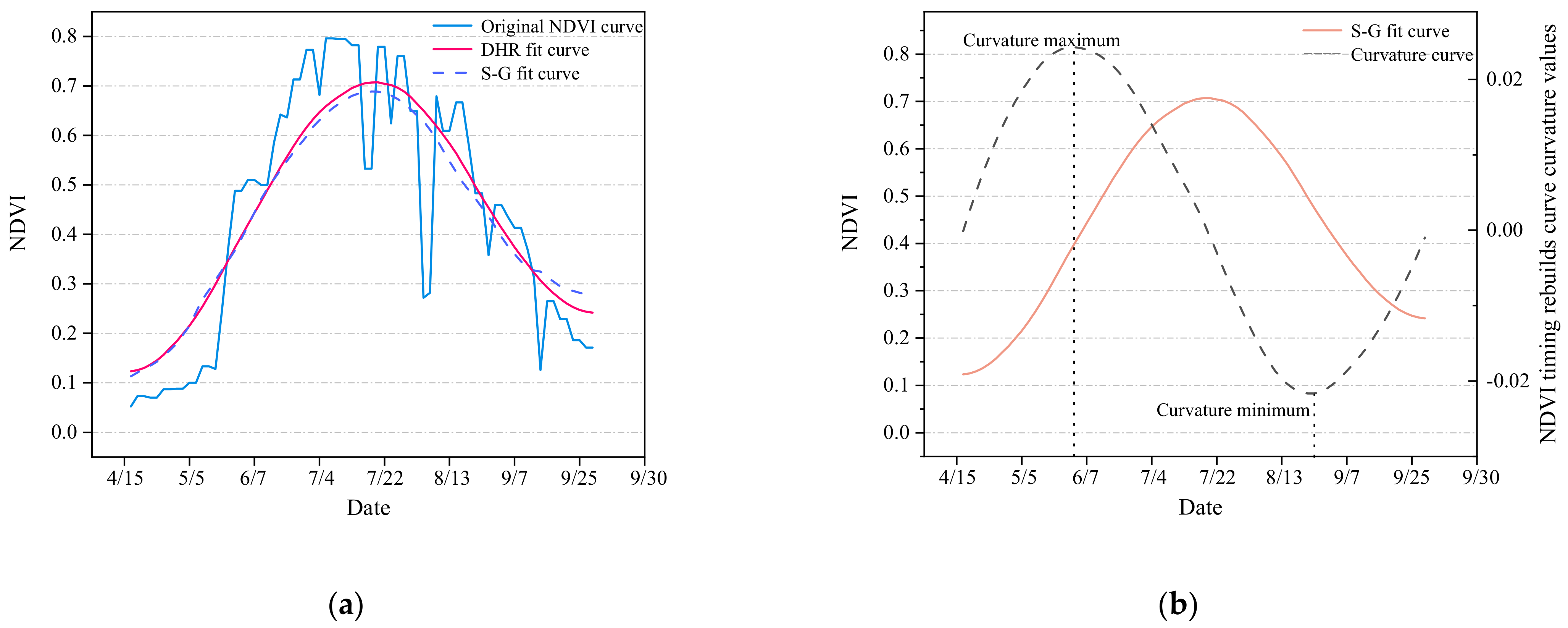
3.1. Crops Timing Characteristics
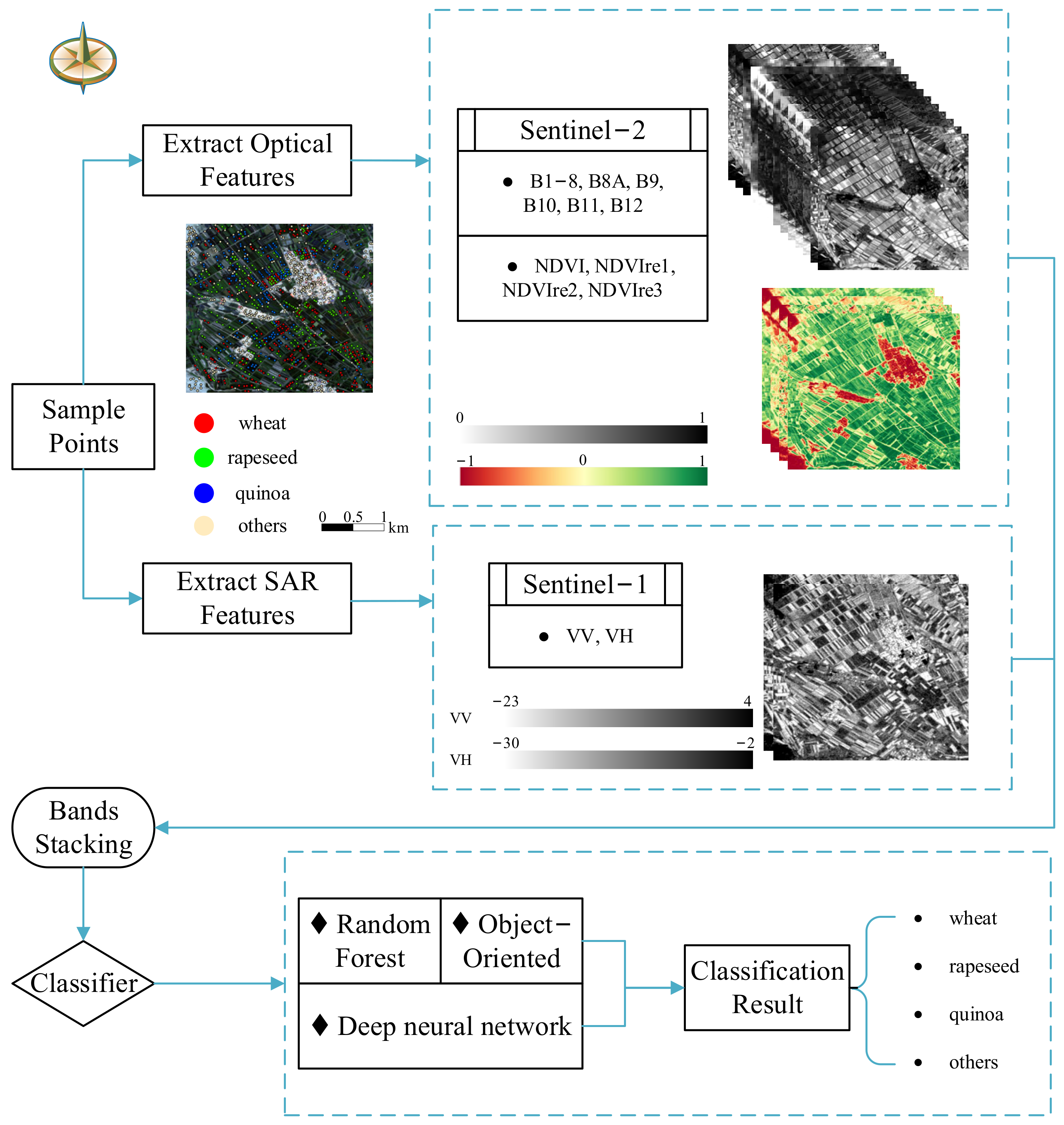
3.2. Feature Space Construction
3.2.1. Original Band and Vegetation Index Feature
3.2.2. Polarization Feature
3.3. Classification Method
3.3.1. Traditional Machine Learning
- (1)
- Random Forest Classification
- (2)
- Object-Oriented Classification
3.3.2. Deep Learning
- (1)
- Deep Neural Network Classification
- (2)
- RF+DNN Classification
3.4. Classification Features and Accuracy Assessment
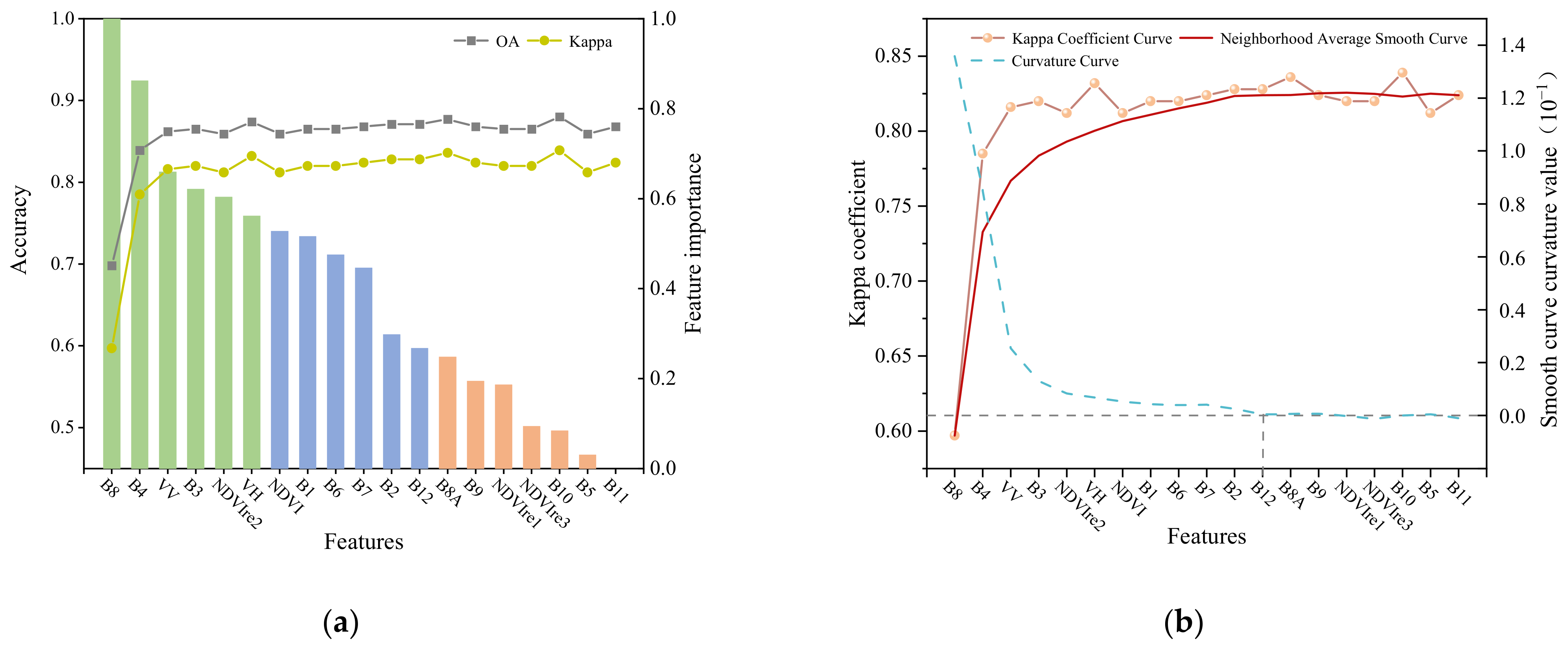
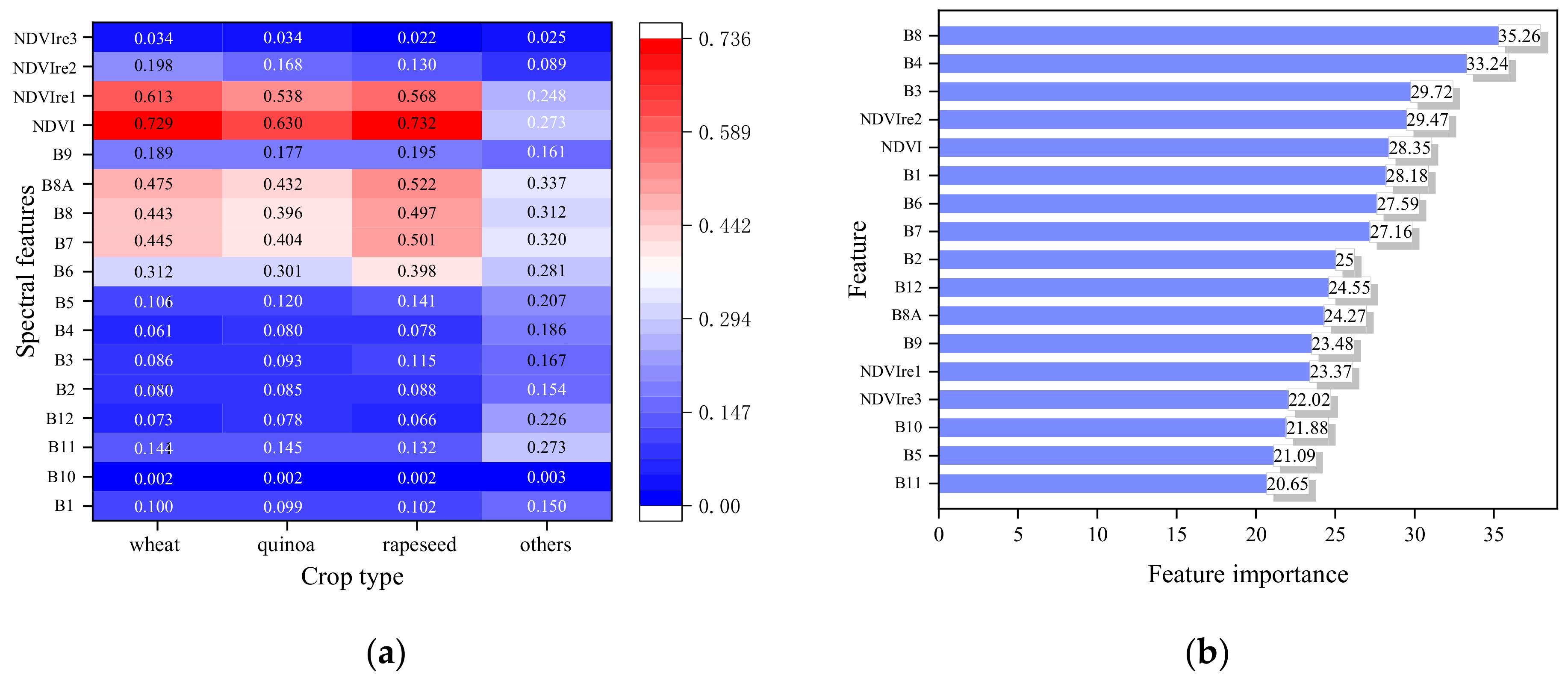
3.4.1. Assessment of Features Importance
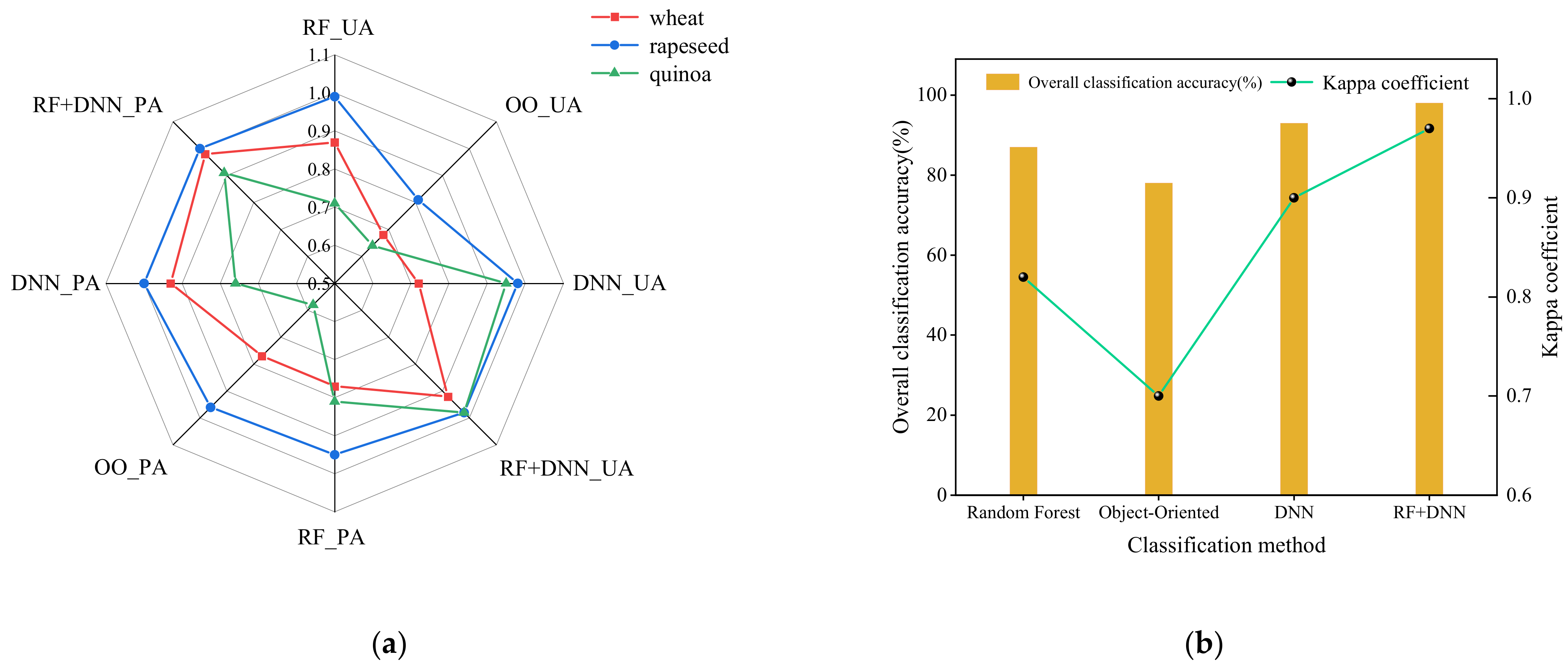
3.4.2. Accuracy Assessment
4. Results
4.1. Feature Space Analysis
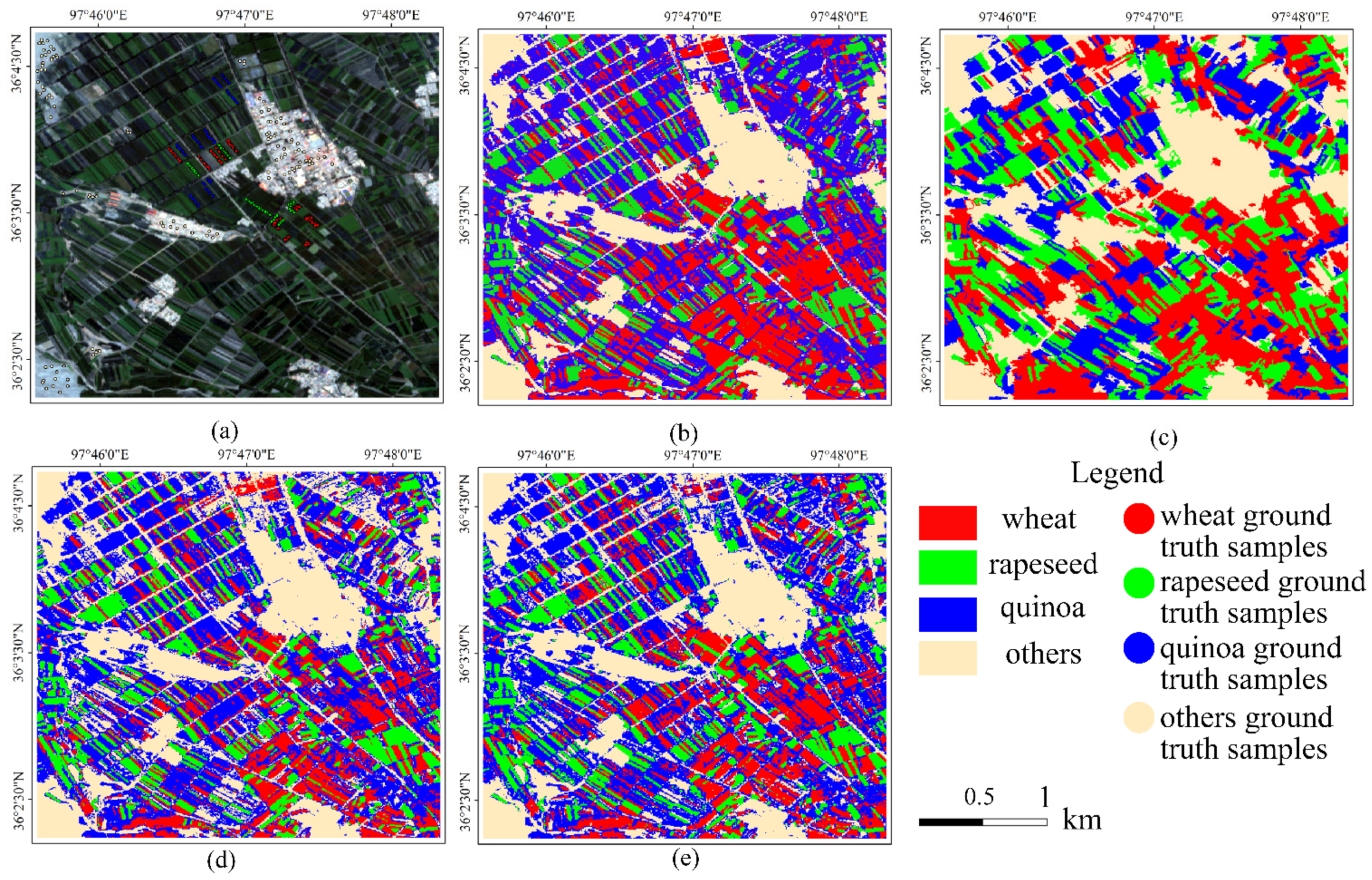
4.2. Analysis of Classification Results
5. Discussion
5.1. Model Transfer and Validation
5.2. Feature Contributions to Crops Classification
5.3. Advantages of Remote Sensing Cloud Platform in Crop Classification
5.4. Significance of RF+DNN for Crops Classification
5.5. Uncertainty of RF+DNN Classification Method
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, P.; Hu, S.; Li, W.; Zhang, C.; Cheng, P. Improving Parcel-Level Mapping of Smallholder Crops from VHSR Imagery: An Ensemble Machine-Learning-Based Framework. Remote Sens. 2021, 13, 2146. [Google Scholar] [CrossRef]
- Liu, Y.; Song, W.; Deng, X. Changes in crop type distribution in Zhangye City of the Heihe River Basin, China. Appl. Geogr. 2016, 76, 22–36. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Xiong, X.; Li, H.; Huang, J.; Ting, K.C.; Ying, Y.; Lin, T. Towards interpreting multi-temporal deep learning models in crop mapping. Remote Sens. Environ. 2021, 264, 112599. [Google Scholar] [CrossRef]
- Denize, J.; Hubert-Moy, L.; Betbeder, J.; Corgne, S.; Baudry, J.; Pottier, E. Evaluation of Using Sentinel-1 and -2 Time-Series to Identify Winter Land Use in Agricultural Landscapes. Remote Sens. 2018, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Lin, H.; Wang, G.; Sun, H.; Fu, J. Mapping Paddy Rice Using a Convolutional Neural Network (CNN) with Landsat 8 Datasets in the Dongting Lake Area, China. Remote Sens. 2018, 10, 1840. [Google Scholar] [CrossRef] [Green Version]
- Atzberger, C. Advances in Remote Sensing of Agriculture: Context Description, Existing Operational Monitoring Systems and Major Information Needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Boles, S.; Frolking, S.; Li, C.; Babu, J.Y.; Salas, W.; Moore, B. Mapping paddy rice agriculture in South and Southeast Asia using multi-temporal MODIS images. Remote Sens. Environ. 2006, 100, 95–113. [Google Scholar] [CrossRef]
- Xiao, X.; Boles, S.; Liu, J.; Zhuang, D.; Frolking, S.; Li, C.; Salas, W.; Moore, B. Mapping paddy rice agriculture in southern China using multi-temporal MODIS images. Remote Sens. Environ. 2005, 95, 480–492. [Google Scholar] [CrossRef]
- Zhang, H.; Du, H.; Zhang, C.; Zhang, L. An automated early-season method to map winter wheat using time-series Sentinel-2 data: A case study of Shandong, China. Comput. Electron. Agric. 2021, 182, 105962. [Google Scholar] [CrossRef]
- Jin, Z.; Azzari, G.; You, C.; Di Tommaso, S.; Aston, S.; Burke, M.; Lobell, D.B. Smallholder maize area and yield mapping at national scales with Google Earth Engine. Remote Sens. Environ. 2019, 228, 115–128. [Google Scholar] [CrossRef]
- Rudiyanto; Soh, N.C.; Shah, R.M.; Giap, S.G.E.; Setiawan, B.I.; Minasny, B. High-Resolution Mapping of Paddy Rice Extent and Growth Stages across Peninsular Malaysia Using a Fusion of Sentinel-1 and 2 Time Series Data in Google Earth Engine. Remote Sens. 2022, 14, 1875. [Google Scholar]
- Chakhar, A.; Hernández-López, D.; Ballesteros, R.; Moreno, M.A. Improving the Accuracy of Multiple Algorithms for Crop Classification by Integrating Sentinel-1 Observations with Sentinel-2 Data. Remote Sens. 2021, 13, 243. [Google Scholar] [CrossRef]
- Felegari, S.; Sharifi, A.; Moravej, K.; Amin, M.; Golchin, A.; Muzirafuti, A.; Tariq, A.; Zhao, N. Integration of Sentinel 1 and Sentinel 2 Satellite Images for Crop Mapping. Appl. Sci. 2021, 11, 10104. [Google Scholar] [CrossRef]
- Friedl, M.A.; Brodley, C.E. Decision tree classification of land cover from remotely sensed data. Remote Sens. Environ. 1997, 61, 399–409. [Google Scholar] [CrossRef]
- Luo, C.; Qi, B.; Liu, H.; Guo, D.; Lu, L.; Fu, Q.; Shao, Y. Using Time Series Sentinel-1 Images for Object-Oriented Crop Classification in Google Earth Engine. Remote Sens. 2021, 13, 561. [Google Scholar] [CrossRef]
- Awad, M. Google Earth Engine (GEE) cloud computing based crop classification using radar, optical images and Support Vector Machine Algorithm (SVM). In Proceedings of the 2021 IEEE 3rd International Multidisciplinary Conference on Engineering Technology (IMCET), Beirut, Lebanon, 8–10 December 2021; pp. 71–76. [Google Scholar]
- Virnodkar, S.; Pachghare, V.K.; Patil, V.C.; Sunil, K.J. Performance Evaluation of RF and SVM for Sugarcane Classification Using Sentinel-2 NDVI Time-Series. In Progress in Advanced Computing and Intelligent Engineering; Springer: Singapore, 2021; pp. 163–174. [Google Scholar]
- Ponganan, N.; Horanont, T.; Artlert, K.; Pimpilai, N. Land Cover Classification using Google Earth Engine’s Object-oriented and Machine Learning Classifier. In Proceedings of the 2021 2nd International Conference on Big Data Analytics and Practices (IBDAP), Bangkok, Thailand, 26–27 August 2021; pp. 33–37. [Google Scholar]
- Pott, L.P.; Amado, T.J.C.; Schwalbert, R.A.; Corassa, G.M.; Ciampitti, I.A. Satellite-based data fusion crop type classification and mapping in Rio Grande do Sul, Brazil. ISPRS J. Photogramm. Remote Sens. 2021, 176, 196–210. [Google Scholar] [CrossRef]
- Palchowdhuri, Y.; Valcarce-Diñeiro, R.; King, P.; Sanabria-Soto, M. Classification of multi-temporal spectral indices for crop type mapping: A case study in Coalville, UK. J. Agric. Sci. 2018, 156, 24–36. [Google Scholar] [CrossRef]
- Alibabaei, K.; Gaspar, P.D.; Lima, T.M. Crop Yield Estimation Using Deep Learning Based on Climate Big Data and Irrigation Scheduling. Energies 2021, 14, 3004. [Google Scholar] [CrossRef]
- Cai, Y.; Guan, K.; Peng, J.; Wang, S.; Seifert, C.; Wardlow, B.; Li, Z. A high-performance and in-season classification system of field-level crop types using time-series Landsat data and a machine learning approach. Remote Sens. Environ. 2018, 210, 35–47. [Google Scholar] [CrossRef]
- Emile, N.; Dinh, H.; Nicolas, B.; Dominique, C.; Laure, H. Deep Recurrent Neural Network for Agricultural Classification using multitemporal SAR Sentinel-1 for Camargue, France. Remote Sens. 2018, 10, 1217. [Google Scholar]
- Zhong, L.; Hu, L.; Zhou, H. Deep learning based multi-temporal crop classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Kavita, B.; Vijaya, M. Evaluation of Deep Learning CNN Model for Land Use Land Cover Classification and Crop Identification Using Hyperspectral Remote Sensing Images. J. Indian Soc. Remote Sens. 2019, 47, 1949–1958. [Google Scholar]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Yu, L.; Yuan, S.; Tao, W.; Pang, K.; Kanniah, D. Growing status observation for oil palm trees using Unmanned Aerial Vehicle (UAV) images. ISPRS J. Photogramm. Remote Sens. 2021, 173, 95–121. [Google Scholar] [CrossRef]
- Hu, X.; Wang, X.; Zhong, Y.; Zhang, L. S3ANet: Spectral-spatial-scale attention network for end-to-end precise crop classification based on UAV-borne H2 imagery. ISPRS J. Photogramm. Remote Sens. 2022, 183, 147–163. [Google Scholar] [CrossRef]
- Lv, A.; Qi, S.; Wang, G. Multi-model driven by diverse precipitation datasets increases confidence in identifying dominant factors for runoff change in a subbasin of the Qaidam Basin of China. Sci. Total Environ. 2022, 802, 149831. [Google Scholar] [CrossRef]
- Zhu, W.; Lű, A.; Jia, S. Estimation of daily maximum and minimum air temperature using MODIS land surface temperature products. Remote Sens. Environ. 2013, 130, 62–73. [Google Scholar] [CrossRef]
- Kumar, L.; Mutanga, O. Google Earth Engine Applications Since Inception: Usage, Trends, and Potential. Remote Sens. 2018, 10, 1509. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Ghorbanian, A.; Kakooei, M.; Amani, M.; Mahdavi, S.; Mohammadzadeh, A.; Hasanlou, M. Improved land cover map of Iran using Sentinel imagery within Google Earth Engine and a novel automatic workflow for land cover classification using migrated training samples. ISPRS J. Photogramm. Remote Sens. 2020, 167, 276–288. [Google Scholar] [CrossRef]
- Amani, M.; Ghorbanian, A.; Ahmadi, S.A.; Kakooei, M.; Moghimi, A.; Mirmazloumi, S.M.; Moghaddam, S.H.A.; Mahdavi, S.; Ghahremanloo, M.; Parsian, S.; et al. Google Earth Engine Cloud Computing Platform for Remote Sensing Big Data Applications: A Comprehensive Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5326–5350. [Google Scholar] [CrossRef]
- Carneiro, T.; Medeiros Da Nobrega, R.V.; Nepomuceno, T.; Bian, G.-B.; De Albuquerque, V.H.C.; Filho, P.P.R. Performance Analysis of Google Colaboratory as a Tool for Accelerating Deep Learning Applications. IEEE Access 2018, 6, 61677–61685. [Google Scholar] [CrossRef]
- Tran, K.H.; Zhang, H.K.; McMaine, J.T.; Zhang, X.; Luo, D. 10 m crop type mapping using Sentinel-2 reflectance and 30 m cropland data layer product. Int. J. Appl. Earth Obs. Geoinf. 2022, 107, 102692. [Google Scholar] [CrossRef]
- Tuvdendorj, B.; Zeng, H.; Wu, B.; Elnashar, A.; Zhang, M.; Tian, F.; Nabil, M.; Nanzad, L.; Bulkhbai, A.; Natsagdorj, N. Performance and the Optimal Integration of Sentinel-1/2 Time-Series Features for Crop Classification in Northern Mongolia. Remote Sens. 2022, 14, 1830. [Google Scholar] [CrossRef]
- Griffiths, P.; Nendel, C.; Hostert, P. Intra-annual reflectance composites from Sentinel-2 and Landsat for national-scale crop and land cover mapping. Remote Sens. Environ. 2019, 220, 135–151. [Google Scholar] [CrossRef]
- Singha, M.; Dong, J.; Zhang, G.; Xiao, X. High resolution paddy rice maps in cloud-prone Bangladesh and Northeast India using Sentinel-1 data. Sci. Data 2019, 6, 26. [Google Scholar] [CrossRef]
- Zhou, B.; Okin, G.S.; Zhang, J. Leveraging Google Earth Engine (GEE) and machine learning algorithms to incorporate in situ measurement from different times for rangelands monitoring. Remote Sens. Environ. 2020, 236, 111521. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Bhavsar, D.; Pandey, K.; Gupta, S.; Roy, A. Red Edge Index as an Indicator of Vegetation Growth and Vigor Using Hyperspectral Remote Sensing Data. Proc. Natl. Acad. Sci. India Sect. A Phys. Sci. 2017, 87, 879–888. [Google Scholar] [CrossRef]
- Mariette, V.; Wolfgang, W.; Isabella, P.; Irene, T.; Bernhard, B.-M.; Christoph, R.; Peter, S. Sensitivity of Sentinel-1 Backscatter to Vegetation Dynamics: An Austrian Case Study. Remote Sens. 2018, 10, 1396. [Google Scholar]
- Yang, G.; Zhao, Y.; Xing, H.; Fu, Y.; Liu, G.; Kang, X.; Mai, X. Understanding the changes in spatial fairness of urban greenery using time-series remote sensing images: A case study of Guangdong-Hong Kong-Macao Greater Bay. Sci. Total Environ. 2020, 715, 136763. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Y.; Chen, X.; Chen, Y.; Lin, Y. Object-oriented classification of remote sensing data for change detection. In Geoinformatics 2006: Remotely Sensed Data and Information, Proceedings of the Geoinformatics 2006: GNSS and Integrated Geospatial Applications Wuhan, China, 28–29 October 2006; SPIE: Bellingham, WA, USA, 2006. [Google Scholar]
- Yang, L.; Wang, L.; Abubakar, G.A.; Huang, J. High-Resolution Rice Mapping Based on SNIC Segmentation and Multi-Source Remote Sensing Images. Remote Sens. 2021, 13, 1148. [Google Scholar] [CrossRef]
- Atli, B.J.; Martino, P.; Kolbeinn, A. Classification and Feature Extraction for Remote Sensing Images from Urban Areas Based on Morphological Transformations. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1940–1949. [Google Scholar]
- Zhang, F.; Yang, X. Improving land cover classification in an urbanized coastal area by random forests: The role of variable selection. Remote Sens. Environ. 2020, 251, 112105. [Google Scholar] [CrossRef]
- Menze, B.H.; Kelm, B.M.; Masuch, R.; Himmelreich, U.; Bachert, P.; Petrich, W.; Hamprecht, F.A. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009, 10, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonetti, E.; Simonetti, D.; Preatoni, D. Phenology-Based Land Cover Classification Using Landsat 8 Time Series; Publications Office of the European Union: Luxembourg, 2014; ISBN 978-92-79-40844-1. [Google Scholar]
- Aptoula, E. Weed and Crop Classification with Domain Adaptation for Precision Agriculture. In Proceedings of the 2021 29th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 9–11 June 2021. [Google Scholar]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Zhao, Y.; Dong, R.; Yu, L. Cross-regional oil palm tree counting and detection via a multi-level attention domain adaptation network. ISPRS J. Photogramm. Remote Sens. 2020, 167, 154–177. [Google Scholar] [CrossRef]
- Hamrouni, Y.; Paillassa, E.; Chéret, V.; Monteil, C.; Sheeren, D. From local to global: A transfer learning-based approach for mapping poplar plantations at national scale using Sentinel-2. ISPRS J. Photogramm. Remote Sens. 2021, 171, 76–100. [Google Scholar] [CrossRef]




| Category | Type | Number | Total Number | T/V Ratio |
|---|---|---|---|---|
| G 1 | 60 | 1183 | 0.7:0.3 | |
| Wheat | F 1 | 242 | ||
| Sum | 302 | |||
| G 1 | 60 | |||
| Rapeseed | F 1 | 244 | ||
| Sum | 304 | |||
| G 1 | 60 | |||
| Quinoa | F 1 | 196 | ||
| Sum | 256 | |||
| G 1 | 120 | |||
| Others | F 1 | 201 | ||
| Sum | 321 |
| Index | Raw Data | HDR Data | S-G Data |
|---|---|---|---|
| Mean | 0.4389 | 0.4458 | 0.4389 |
| Mean square root error | 0.1072 | 0.1135 | |
| Correlation coefficient | 0.9158 | 0.9109 |
| Vegetation Index | Formula (Sentinel-2) |
|---|---|
| NDVI | (B8 1 − B4 1)/(B8 1 + B4 1) |
| NDVIre1 | (B8A 1 − B5 1)/(B8A 1 + B5 1) |
| NDVIre2 | (B8A 1 − B6 1)/(B8A 1 + B6 1) |
| NDVIre3 | (B8A 1 − B7 1)/(B8A 1 + B7 1) |
| Type | Model Loss | Model Accuracy | Training Time | Prediction Time |
|---|---|---|---|---|
| DNN 1 | 0.2957 | 0.8674 | 36 min 11 s | 58 s |
| RF+DNN 1 | 0.2386 | 0.9042 | 17 min 23 s | 11 s |
| Validation Area | Model Loss | Model Accuracy | OA 1 | KC 1 |
|---|---|---|---|---|
| Validation Area i | 0.1947 | 0.9341 | 0.95 | 0.93 |
| Validation Area ii | 0.0976 | 0.9812 | 0.98 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, J.; Wu, J.; Xiao, C.; Zhang, Z.; Li, J. The Classification Method Study of Crops Remote Sensing with Deep Learning, Machine Learning, and Google Earth Engine. Remote Sens. 2022, 14, 2758. https://doi.org/10.3390/rs14122758
Yao J, Wu J, Xiao C, Zhang Z, Li J. The Classification Method Study of Crops Remote Sensing with Deep Learning, Machine Learning, and Google Earth Engine. Remote Sensing. 2022; 14(12):2758. https://doi.org/10.3390/rs14122758
Chicago/Turabian StyleYao, Jinxi, Ji Wu, Chengzhi Xiao, Zhi Zhang, and Jianzhong Li. 2022. "The Classification Method Study of Crops Remote Sensing with Deep Learning, Machine Learning, and Google Earth Engine" Remote Sensing 14, no. 12: 2758. https://doi.org/10.3390/rs14122758
APA StyleYao, J., Wu, J., Xiao, C., Zhang, Z., & Li, J. (2022). The Classification Method Study of Crops Remote Sensing with Deep Learning, Machine Learning, and Google Earth Engine. Remote Sensing, 14(12), 2758. https://doi.org/10.3390/rs14122758





