1. Introduction
Forests are among the most important terrestrial ecosystems and are essential for human development [
1]. Well-managed forests provide renewable resources, protect biodiversity, maintain a stable energy cycle, and prevent soil degradation and erosion [
2]. Precise tree species surveys are crucial to forest inventory and management because they provide managers with a better understanding of forest species composition, changes in forest species, quantity of forest resources, and references for the formulation and adjustment of forestry policies [
3]. However, traditional survey methods are inefficient and their associated labor costs are high. Remote sensing-based methods are efficient when mapping forest types in areas with rough terrain or that are difficult to reach, and can significantly improve survey efficiency and reduce labor costs [
4].
Many remote sensing-based forest classification studies have considered multi-scale remote sensing data sources. Early developments used medium-spatial resolution satellite remote sensing data, such as Landsat Thematic Mapper imagery, for regional-scale forest classification [
5,
6]. However, because the spatial resolution of Landsat data is relatively low, individual trees cannot be precisely mapped. Spaceborne hyperspectral data have rarely been used for individual tree species classification. Most applications of hyperspectral data to date have been airborne [
7,
8,
9]. The high spatial resolution of airborne data meets the requirements for determining the locations of trees [
10,
11,
12,
13]; however, data acquisition costs are normally high and data processing is complex [
14,
15]. With the launch of IKONOS, QuickBird, GeoEye, and WorldView satellites, high-resolution optical images can be readily obtained and meet the requirements for locating individual trees. Using such high-resolution images to precisely classify tree species saves time and costs in tree distribution mapping.
Traditional classification methods, such as random forest (RF) and support vector machine (SVM), have been widely used to classify tree species [
16,
17,
18,
19]. These approaches generally require the artificial design and extraction of classification features, such as spectra, texture, and vegetation indices, in addition to linear transformations [
20,
21]. The classification accuracy depends largely on the rationality of the artificial feature design and selection, which is highly subjective; therefore, extensive professional knowledge is necessary.
The designed artificial features limit the information that can be used by the classifier [
22]. New classification technologies are necessary to improve classification efficiency and accuracy. As a promising classification technology, convolutional neural networks (CNNs) perform well in image classification tasks [
23,
24]. A CNN method does not require feature engineering, and its multi-layer structure can fully use the information in the data to automatically extract abstract and higher-level features for classification. As a result, CNN methods tend to result in accurate classifications.
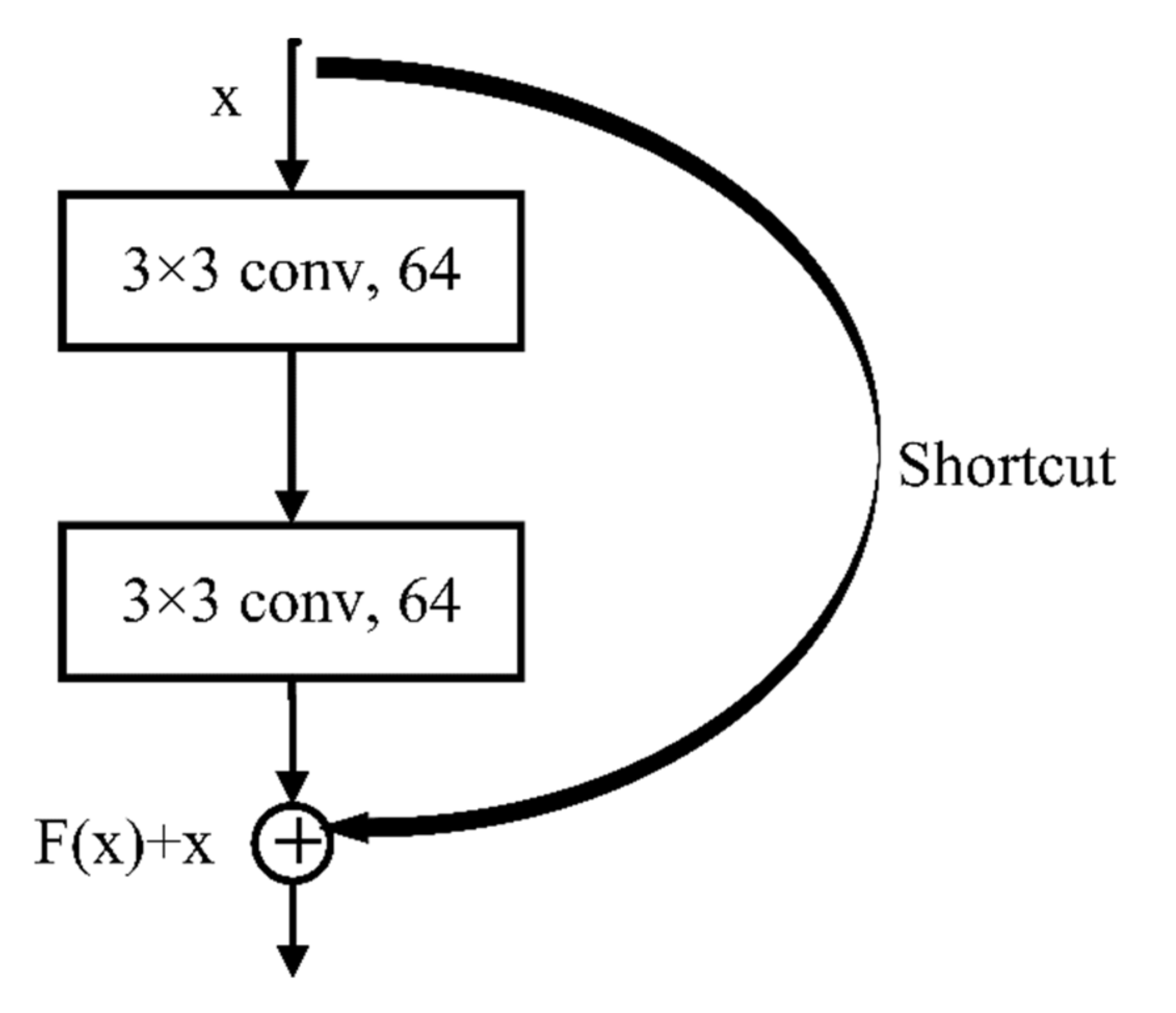
In recent years, CNNs have shown satisfactory results when applied to tree species classification [
25]. CNNs have been applied to classify three-dimensional point clouds of trees [
26,
27], airborne hyperspectral data [
28,
29], and high-resolution data combined with LiDAR data [
30]. These studies were almost all based on airborne imaging systems and multiple data sources, which are characterized by high data acquisition costs and complex data processing, preventing their wide application. To date, CNNs have rarely been applied to recognize individual tree species (ITS) from a single satellite data source. A method is needed for using satellite data to classify ITS for mapping forest tree species with a low data acquisition cost.
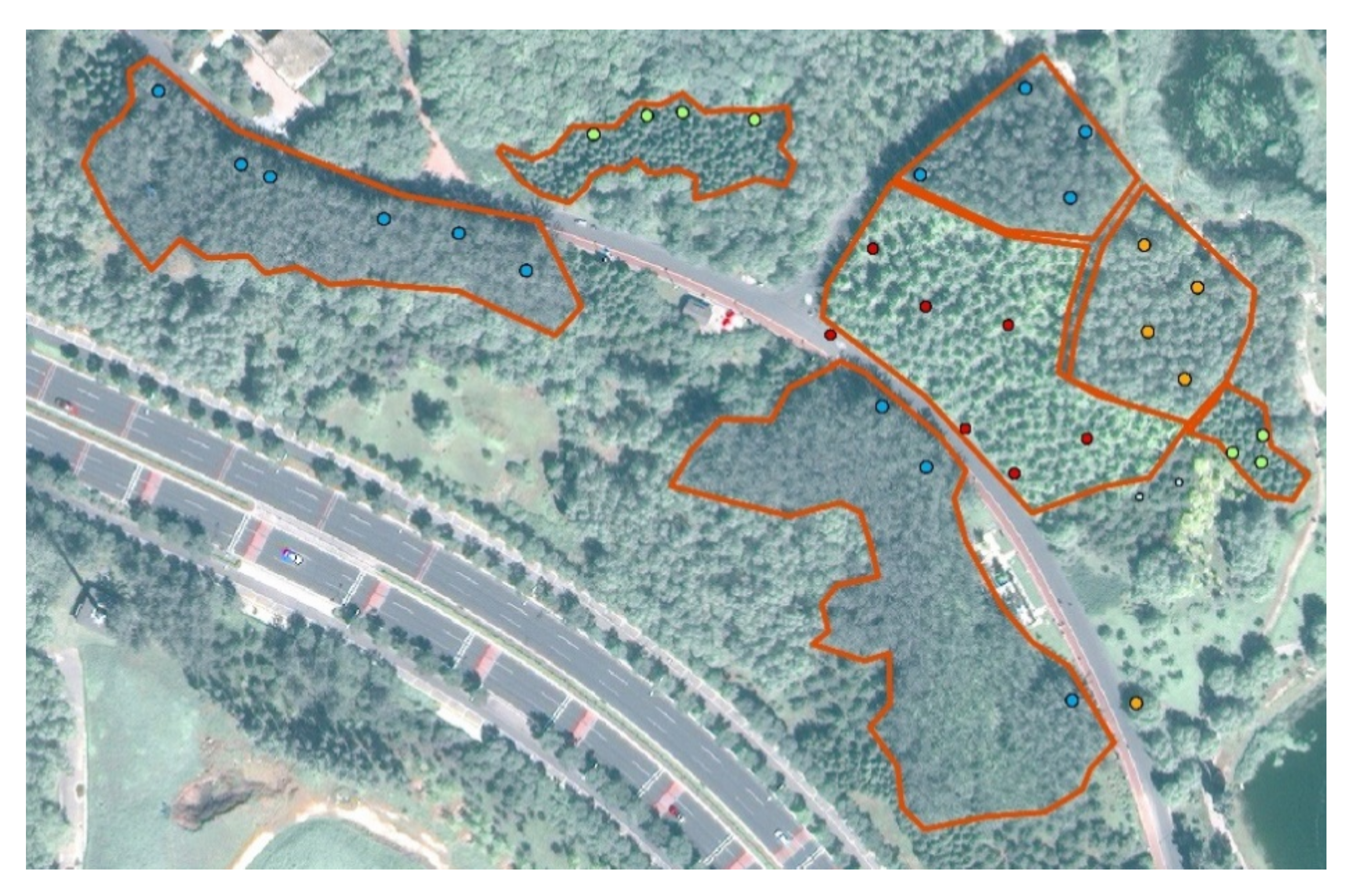
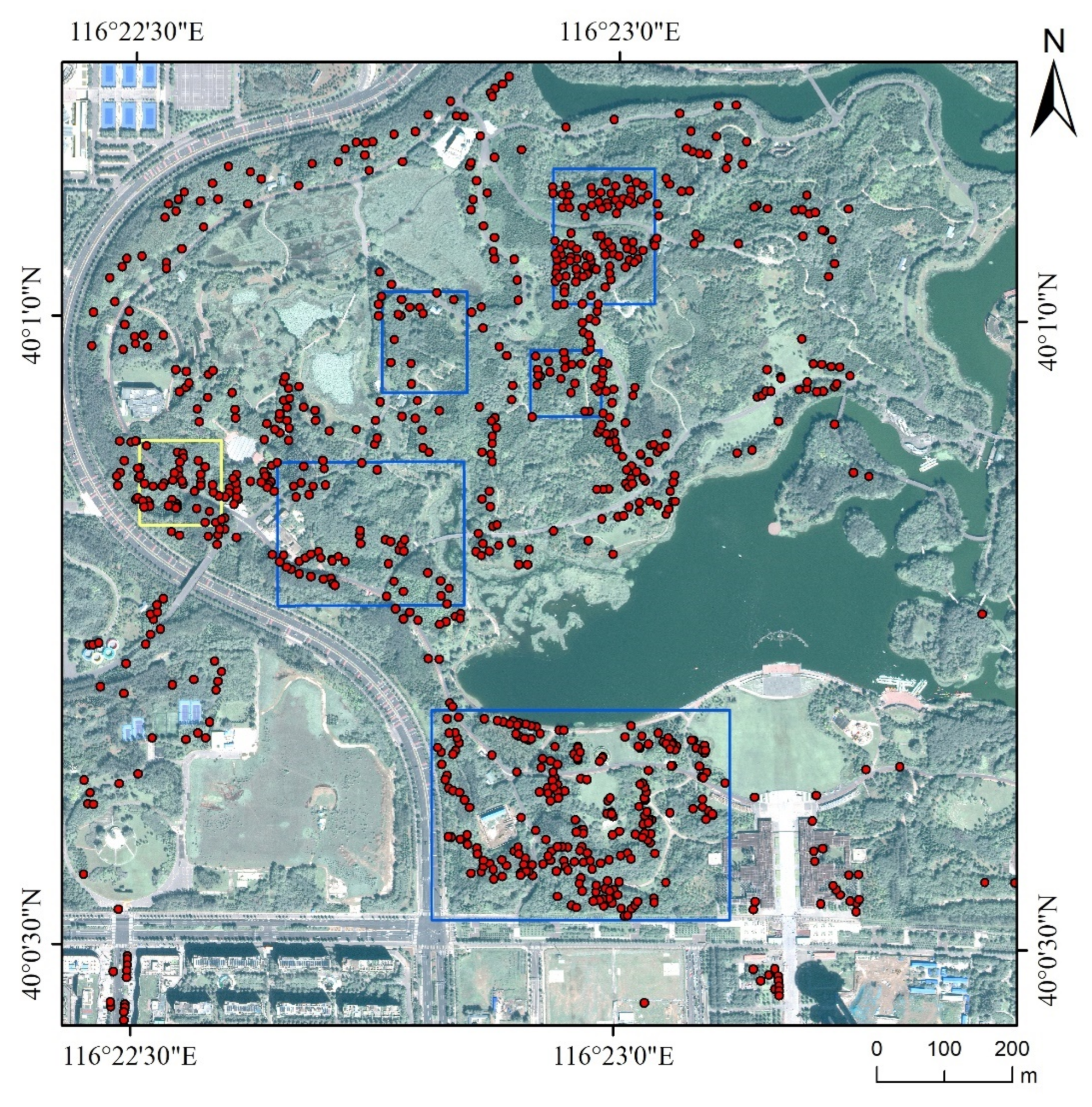
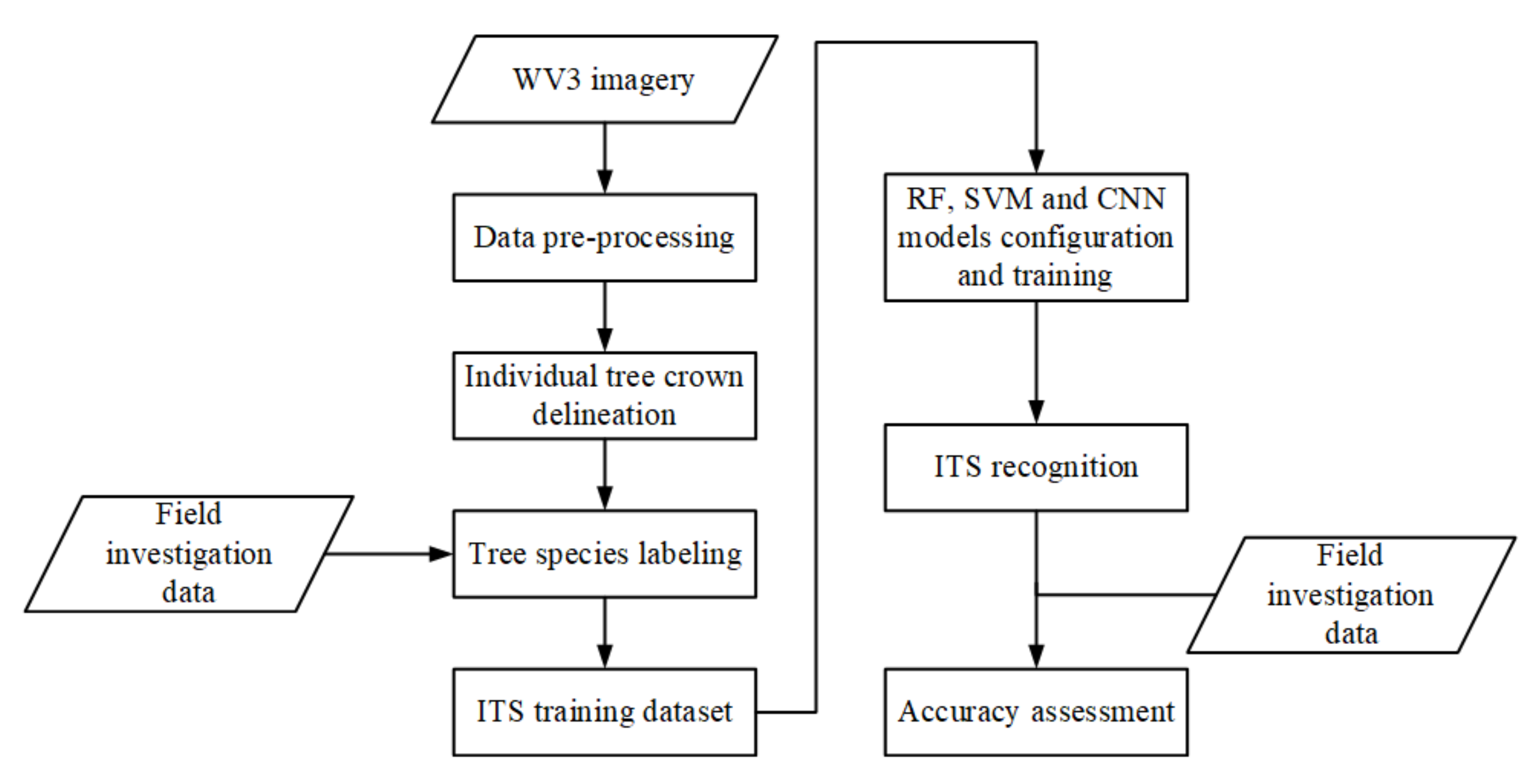
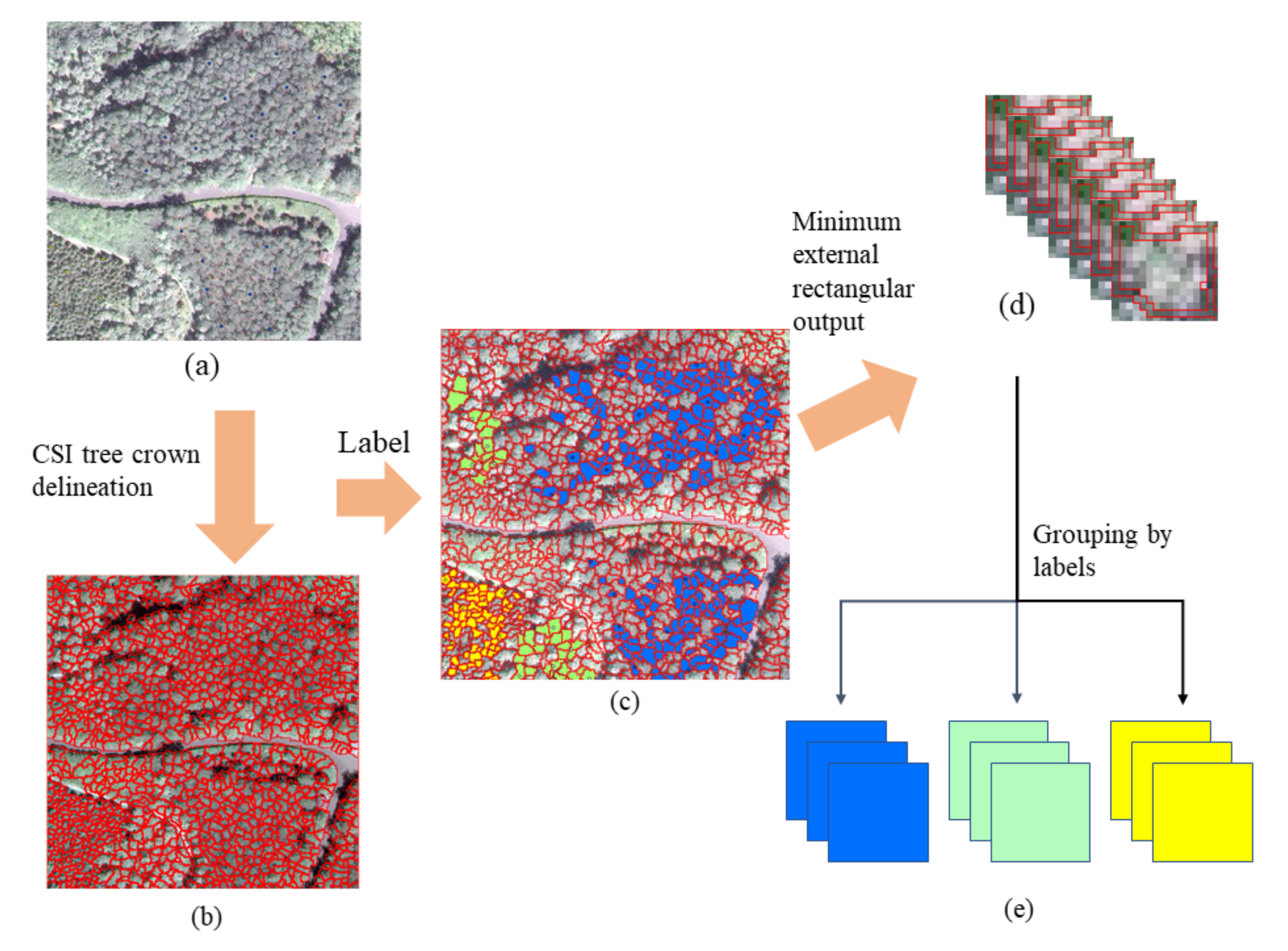
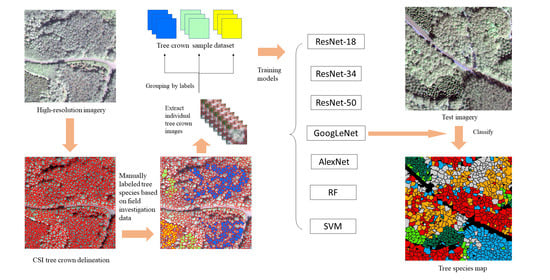
In this study, we explored the combination of high-resolution satellite remote sensing imagery and CNNs to recognize ITS. A CNN-based multi-scale ITS recognition (CMSIR) method was developed to improve the automation and accuracy of ITS mapping. In the CMSIR method, a tree crown delineation approach is used to quickly build an individual tree crown training dataset, and several popular CNN models are employed to automatically extract classification features and recognize ITS from a high-resolution satellite image. The multi-scale characteristic of different tree species was considered in tree crown delineation and ITS recognition.
3. Results
Table 8 shows confusion matrices and statistical measures of RF, SVM, AlexNet, GoogLeNet, and ResNet for ITS classification.
Table 9 shows the classification accuracies and kappa coefficients for six species based on different models. For CNN models, GoogLeNet achieved the best OA (82.7%) with the highest kappa coefficient (0.79), and was the only model that achieved an OA over 80%. It was followed by ResNet-18 (74.8%), ResNet-50 (71.7%), ResNet-34 (70.9%), and AlexNet, which achieved the lowest OA (52.0%) and kappa coefficient (0.41). GoogLeNet achieved the highest OA for each tree species, with almost all exceeding 80%. AlexNet misclassified almost all pines as poplars, and its classification accuracy for cypress (58.3%), pagoda (45.1%), and willow (34.9%) was about 30% lower than those of GoogLeNet and ResNet. All ResNet misclassified many ashes into willows; its classification accuracy for ash was significantly lower (20%) than that of GoogLeNet. ResNet-18 outperformed ResNet-34 and ResNet-50 of OA by 3.9% and 3.1%, respectively (74.8% > 70.9% > 71.7%).
Compared with RF and SVM, the OA of GoogLeNet (82.7%) was significantly higher than that of RF (44.1%) and SVM (48.8%); even AlexNet had higher OA (52.0%) than RF (44.1%) and SVM (48.8%). Likewise, CNN models all achieved higher kappa coefficients than RF (0.32) and SVM (0.40). RF classified almost all pines as cypresses, making pines almost invisible in the classification map. The classification accuracy of RF for pine, cypress, poplar, and ash ranged from 20% to 30%, which is much lower than that of GoogLeNet. The classification accuracies of RF for pagoda and willow were only about 20%. SVM classified almost all willows as pagodas, making willows almost invisible in the classification map. The classification accuracy of SVM for pine, poplar, ash, and pagoda ranged from 20% to 30%, much lower than that achieved by GoogLeNet.
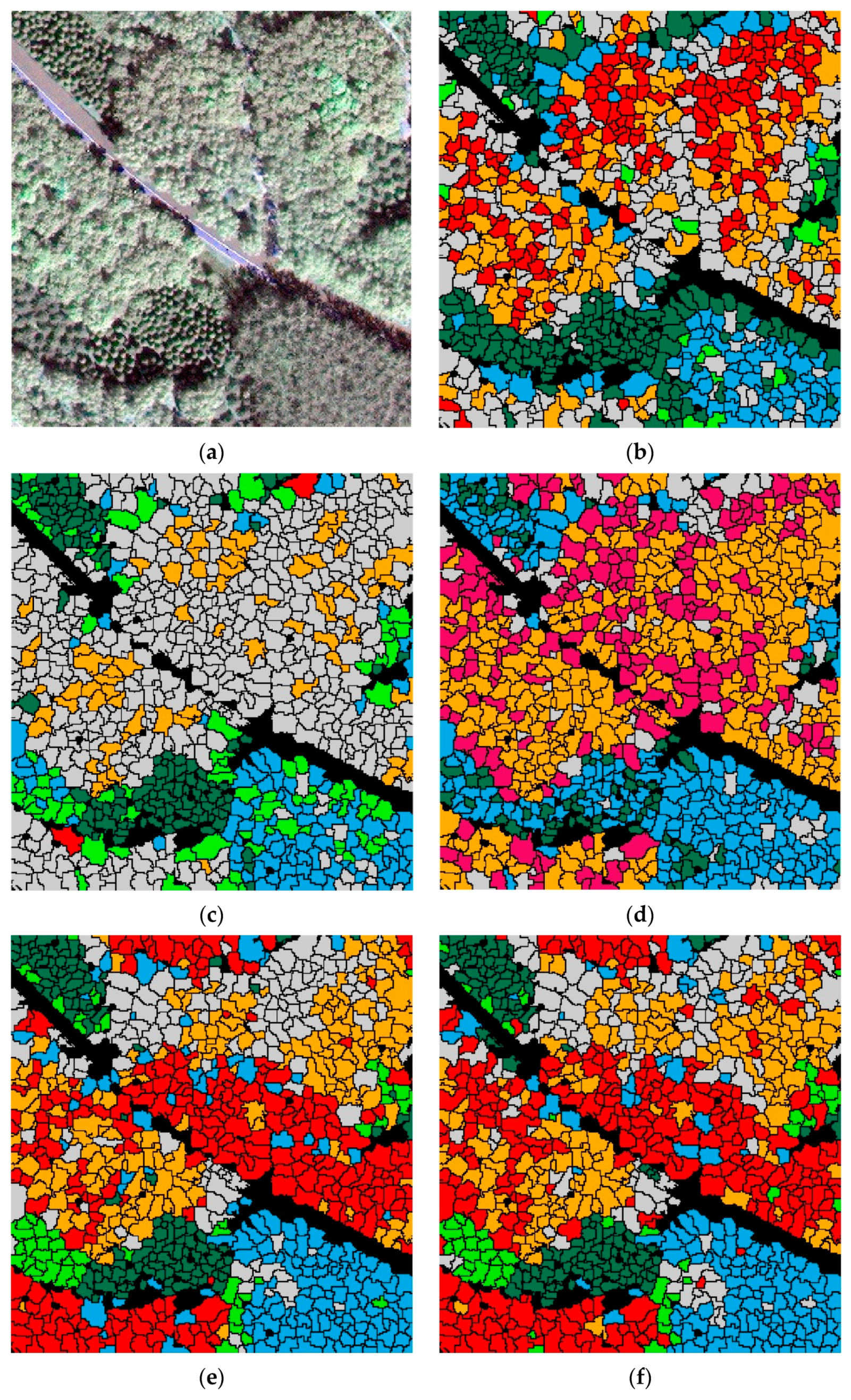
Figure 8 shows the ITS maps predicted by RF, SVM, and CNN. RF misclassified almost all pine as cypress. SVM did not separate willow from ash and pagoda. AlexNet classified many pines as poplars or cypresses.
The CSI tree crown delineation produced inevitable errors due to under- and over-segmentation, which are common problems in tree crown delineations [
35], especially when only two-dimensional information is used. Some non-crown segments produced by the under- and over-segmentation affected the classification map and reduced classification accuracies. Some tree crowns were misclassified, especially on both sides of the road and the edge of the image, because some non-crown pixels were included in test samples when taking the minimum outer cut rectangles, which changed the classification features of the test samples.
5. Conclusions
Based on practical application requirements, the CMSIR method focuses on two key issues in ITS recognition: fast and accurate construction of a training sample dataset and ITS classification methods. We proposed a method to construct ITC training samples suitable for CNN models. This method combines a multi-scale ITC delineation method, manual labeling of tree species, and sample enhancement techniques to build a training sample dataset. The CSI tree crown delineation algorithm was used to automatically describe tree crowns, which avoided manual sketching.
A method for ITS classification was explored using different CNN methods and high-resolution satellite remote sensing imagery. The data source was readily obtained and was low in cost and wide in scope. The CNN used in this study avoids manual feature extraction and its structures are relatively common, easy to build, and have a low training difficulty and fast training speed. GoogLeNet, which achieved the highest OA, can use multi-scale convolution filters to extract features in multi-scale tree crowns. The classification accuracies for tree crowns in different scales were very high (greater than 80%). Compared with other commonly used machine learning models, such as RF and SVM, the CNN models do not require manual feature extraction and achieve higher OA.
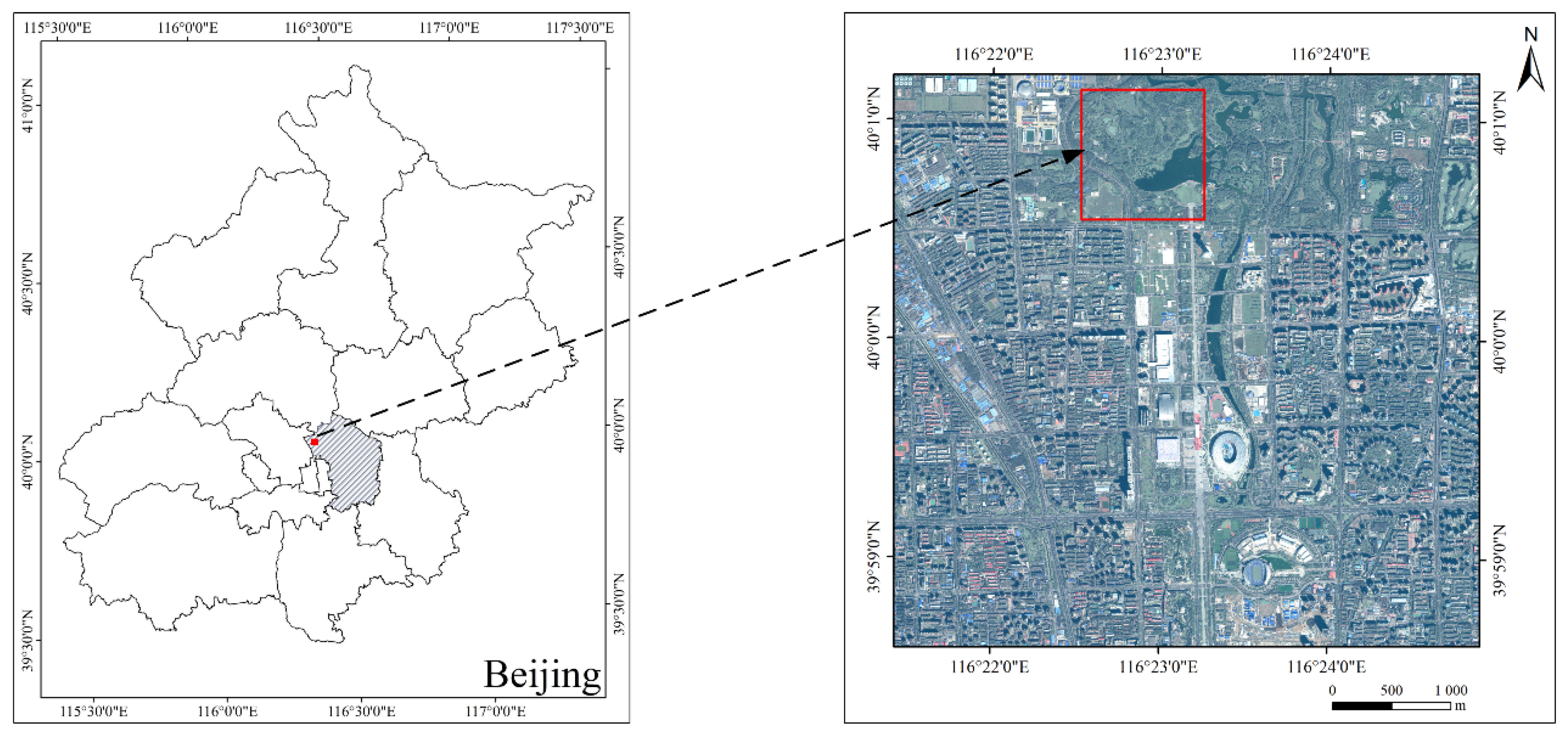
Our study area was located in a park, where trees were manually planted and pruned regularly by gardeners. The distribution of trees had a certain pattern. The crowns of the same trees were similar in size. There were intervals between the crowns, which made it easier to delineate the crowns. Thus, we expected higher accuracies in the manually planted forest. The CMSIR method could be improved in future research in the following ways: (1) Natural forests have a larger number of tree species and the distribution of tree species is more random. The tree crown delineation significantly influences the accuracy of the subsequent classification. Researchers need to effectively delineate tree crowns, especially in dense natural forests. (2) We used some classic CNN models, which were originally used for the ILSVRC dataset. Compared with ILSVRC images, tree crown images have smaller scales and multiple bands. Therefore, a network model suitable for small-scale and multiband images should be constructed for ITS classification.