Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands
Abstract
:1. Introduction
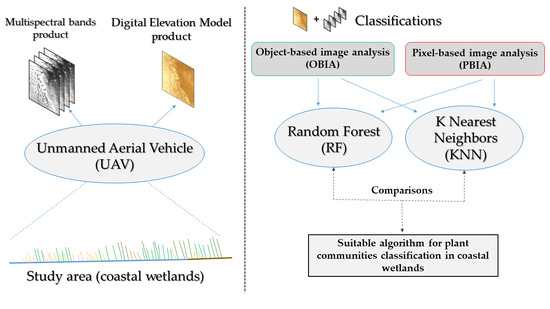
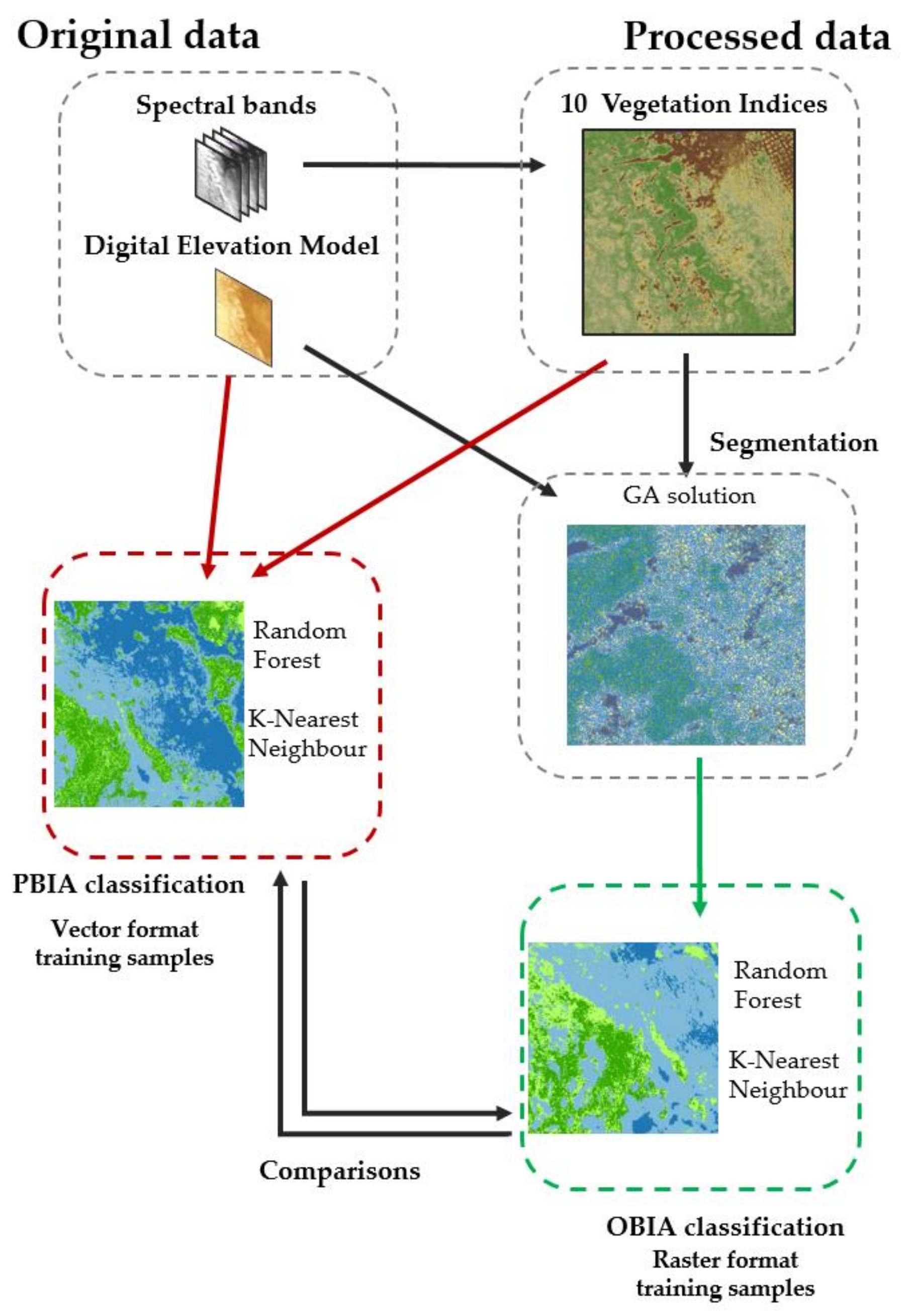
2. Materials and Methods
2.1. Study Areas
2.2. Data Collection
2.2.1. Field Sampling
2.2.2. Image Acquisition
2.3. Image Processing
2.3.1. Positional Accuracy
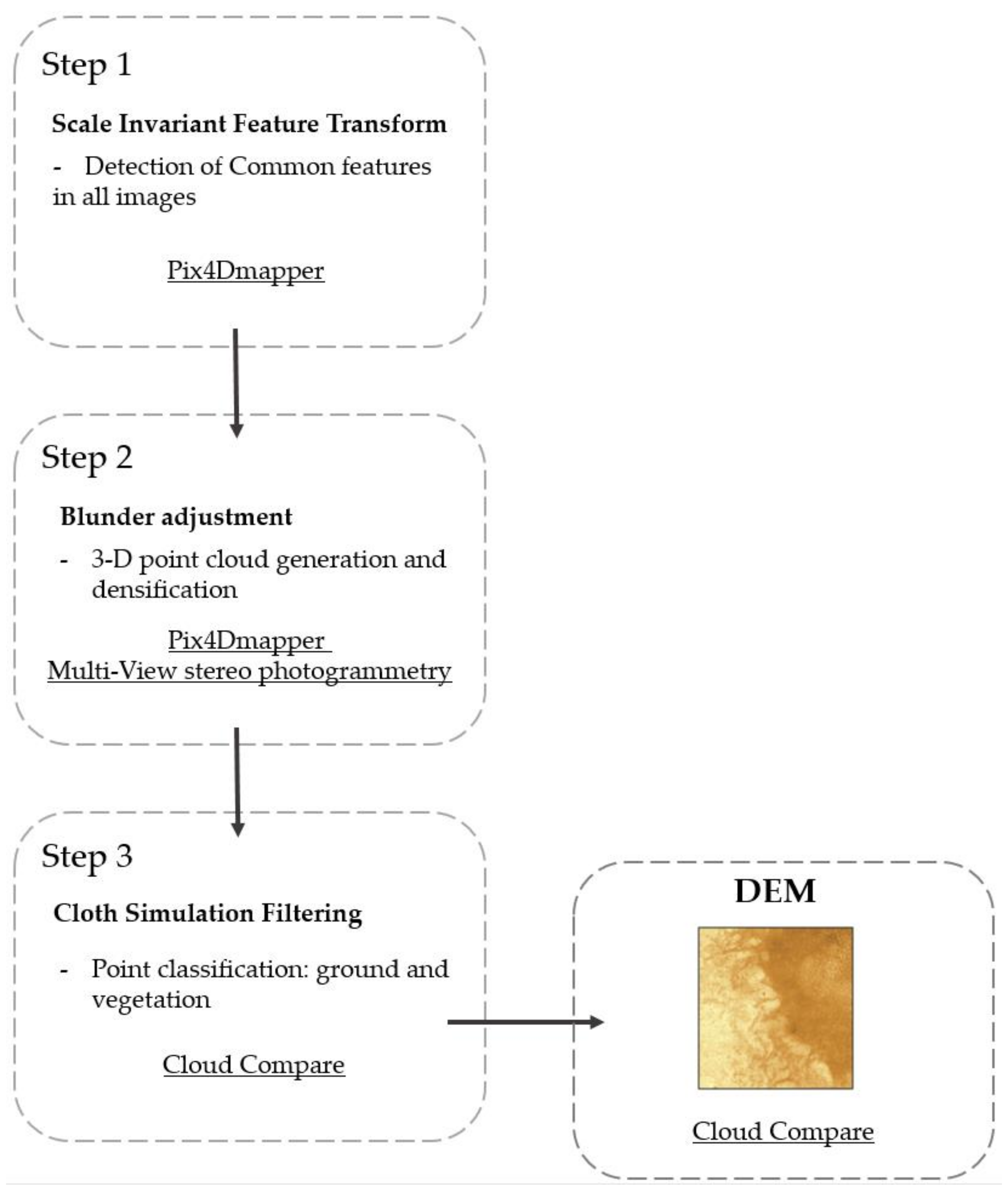
2.3.2. Digital Elevation Models
2.3.3. Vegetation Indices
2.4. Classification of Images
2.4.1. Segmentation
2.4.2. ML Classifiers
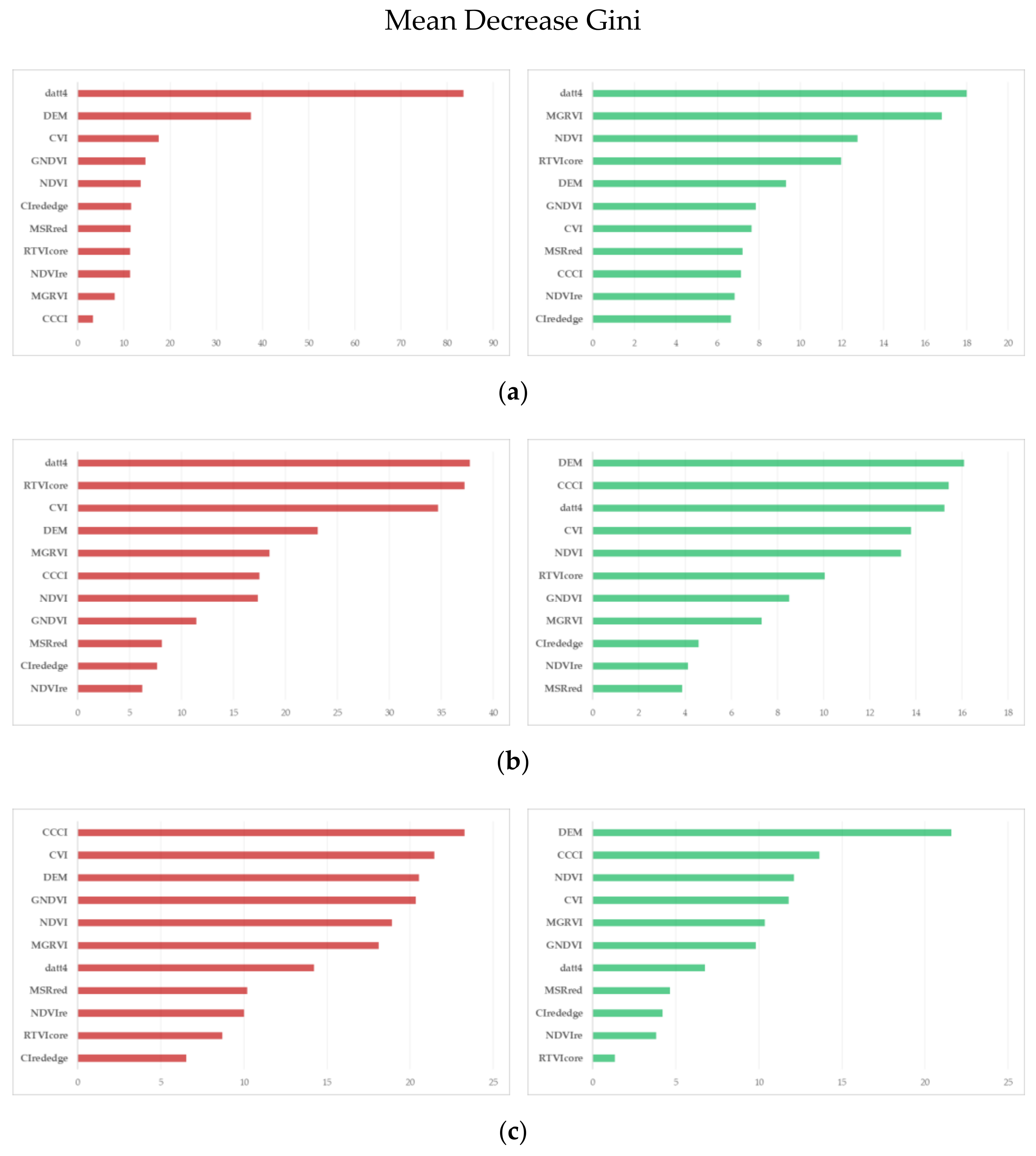
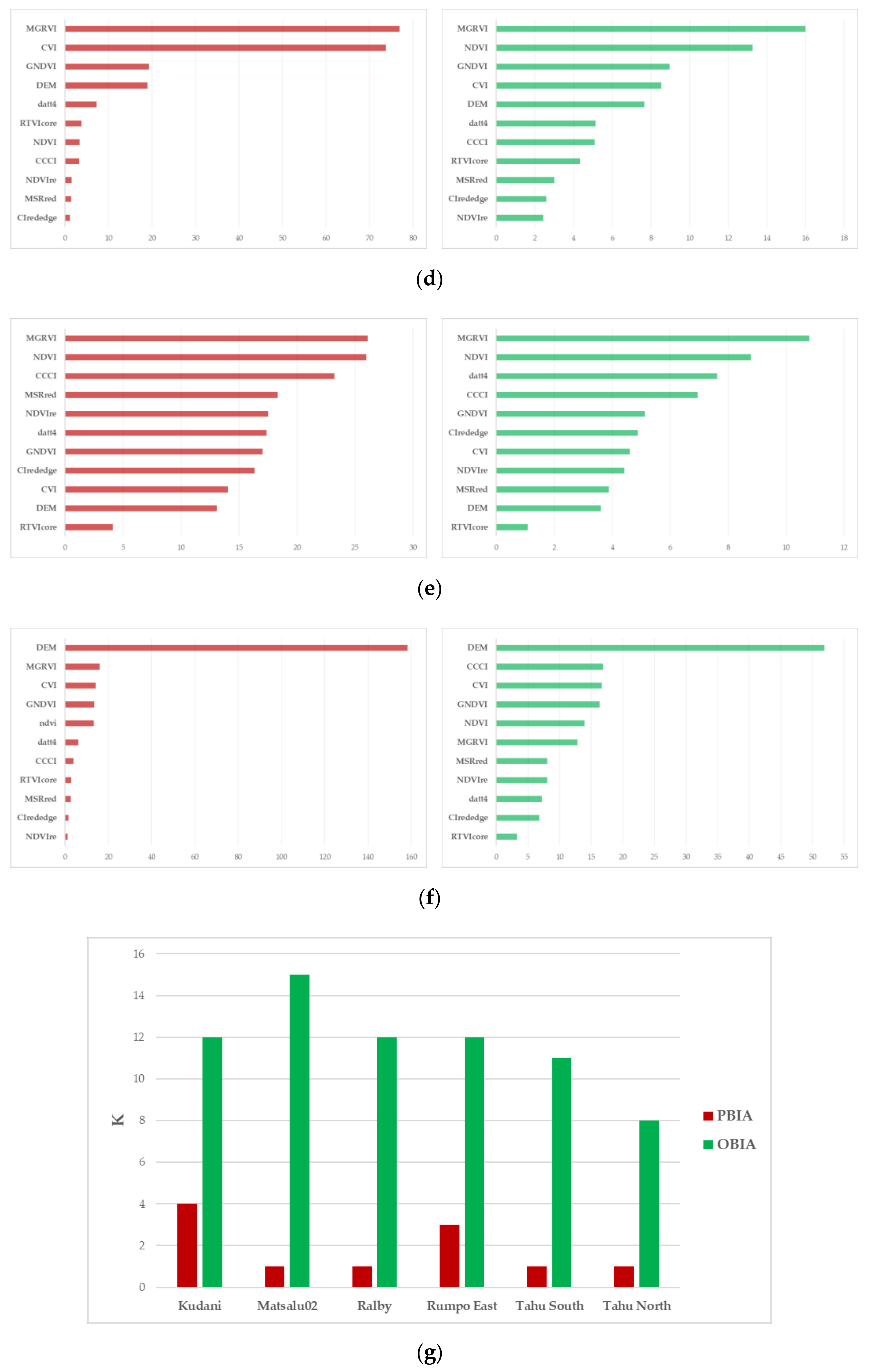
2.4.3. Classification Accuracy and Variable Importance
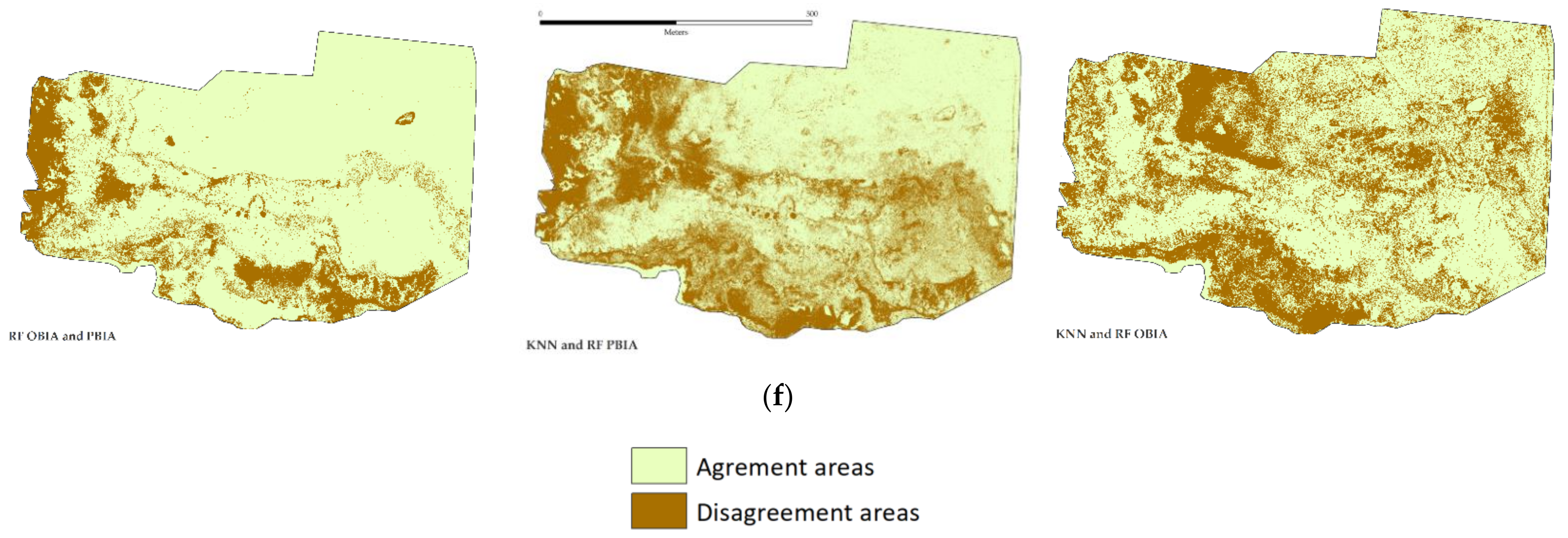
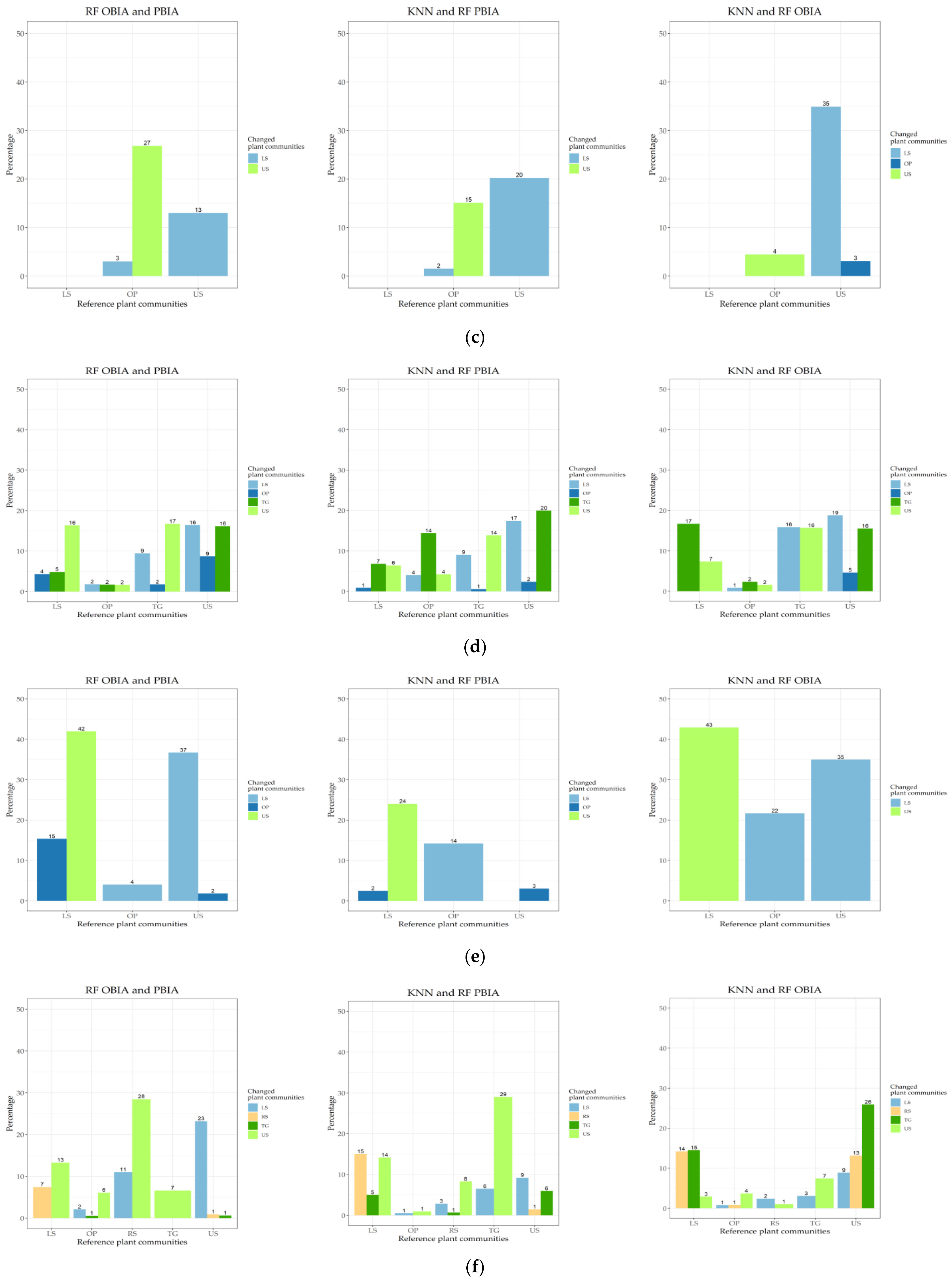
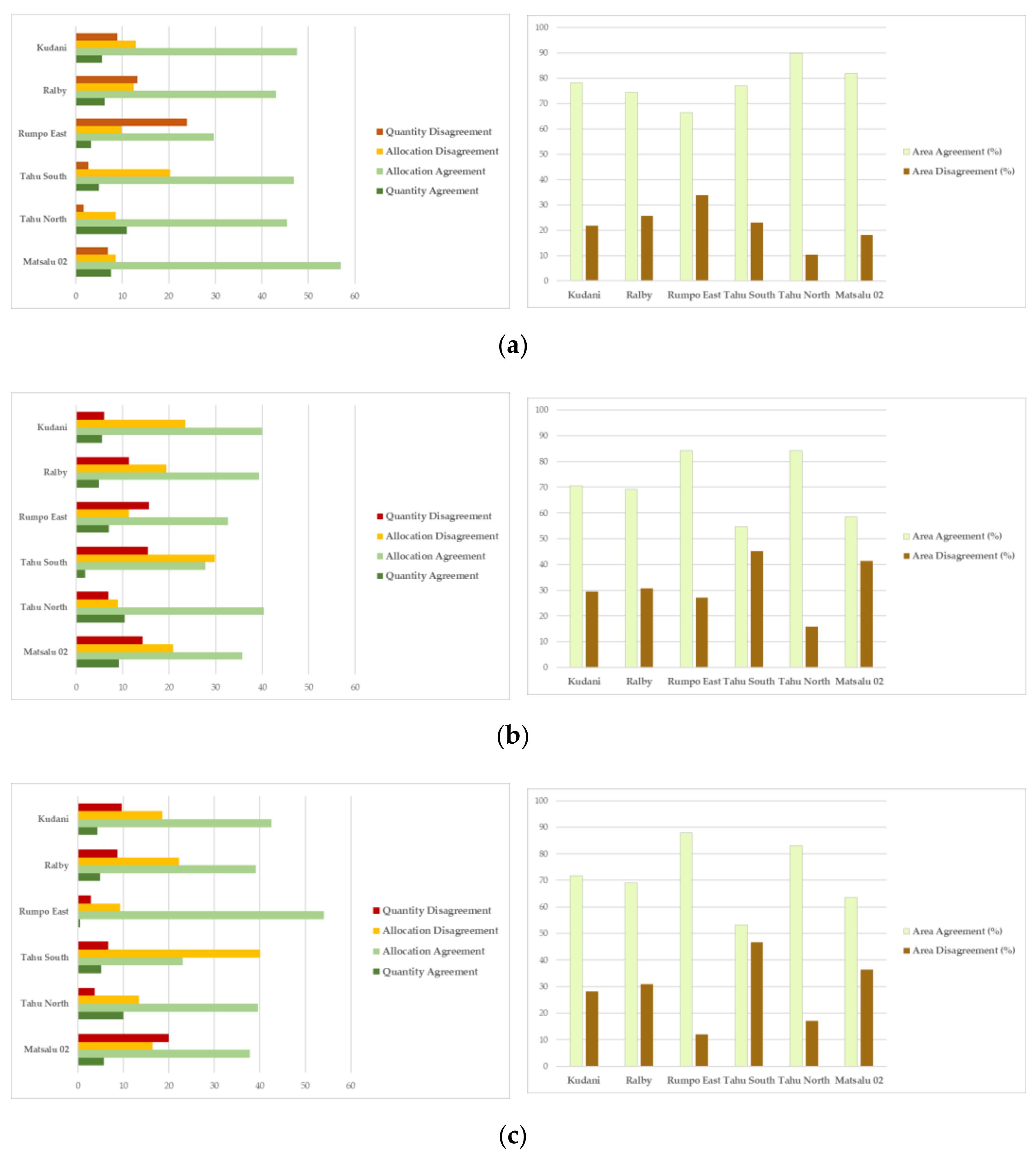
2.5. Map Comparisons
3. Results
3.1. Segmentation and Comparison between Training Areas
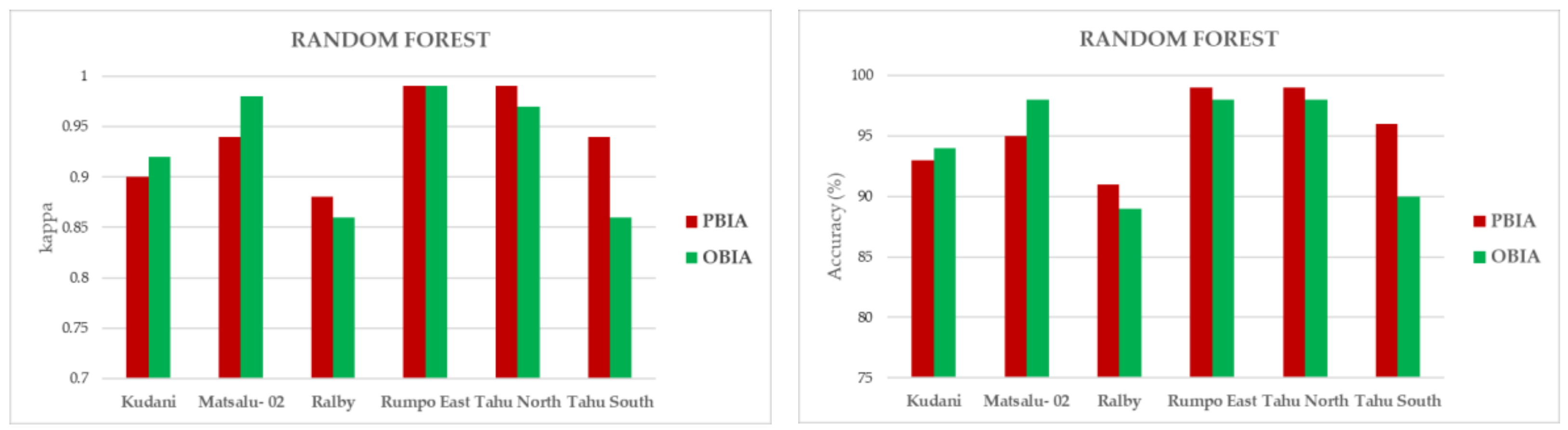
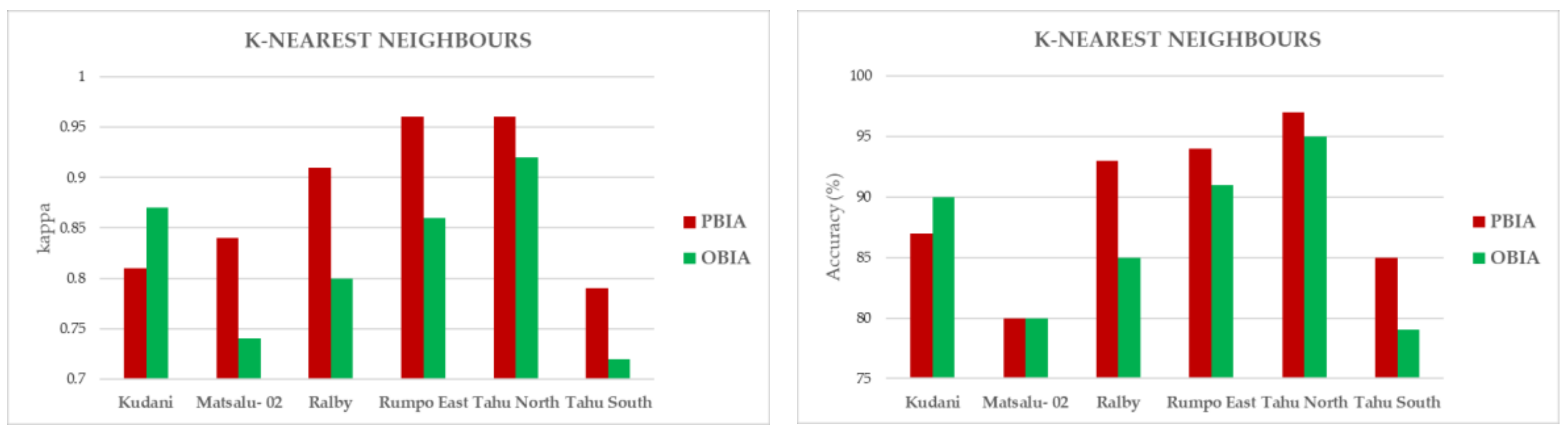
3.2. ML Accuracy Assesment
3.3. Map Comparisons
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A





References
- LaPaix, R.L.; Freedman, B.F.; Patriquin, D.P. Ground vegetation as an indicator of ecological integrity. Environ. Rev. 2009, 17, 249–265. [Google Scholar] [CrossRef]
- Berg, M.; Joyce, C.; Burnside, N. Differential responses of abandoned wet grassland plant communities to reinstated cutting management. Hydrobiologia 2012, 692, 83–97. [Google Scholar] [CrossRef]
- Pärtel, M.; Chiarucci, A.; Chytrý, M.; Pillar, V.D. Mapping plant community ecology. J. Veg. Sci. 2017, 28, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Van der Maarel, E. Vegetation Ecology—An Overview; van der Maarel, E., Ed.; Vegetation Ecology; Blackwell Publishing: Oxford, UK, 2015; pp. 1–3. [Google Scholar]
- Martínez-López, J.; Carreño, M.F.; Palazón-Ferrando, J.A.; Martínez-Fernández, J.; Esteve, M.A. Remote sensing of plant communities as a tool for assessing the condition of semiarid Mediterranean saline wetlands in agricultural catchments. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 193–204. [Google Scholar] [CrossRef]
- Pettorelli, N.; Laurance, W.F.; O’Brien, T.G.; Wegmann, M.; Nagendra, H.; Turner, W. Satellite remote sensing for applied ecologists: Opportunities and challenges. J. Appl. Ecol. 2014, 51, 839–848. [Google Scholar] [CrossRef]
- Kaplan, G.; Avdan, U. Mapping and monitoring wetlands using Sentinel-2 satellite imagery. ISPRS Ann. Photogramm Remote Sens. Spat. Inf. Sci. 2017, IV-4/W4, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Ceaușu, S.; Apaza-Quevedo, A.; Schmid, M.; Martín-López, B.; Cortés-Avizanda, A.; Maes, J.; Brotons, L.; Queiroz, C.; Pereira, H.M. Ecosystem service mapping needs to capture more effectively the biodiversity important for service supply. Ecosyst. Serv. 2021, 48, 101259. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Vanden Borre, J.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status—New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Díaz-Delgado, R.; Cazacu, C.; Adamescu, M. Rapid assessment of ecological integrity for LTER wetland sites by using UAV multispectral mapping. Drones 2019, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Baena, S.; Boyd, D.S.; Moat, J. UAVs in pursuit of plant conservation—Real world experiences. Ecol. Inform. 2018, 47, 2–9. [Google Scholar] [CrossRef]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Westoby, M.J.; Brasington, J.; Glasser, N.F.; Hambrey, M.J.; Reynolds, J.M. ‘Structure-from-Motion’ photogrammetry: A low-cost, effective tool for geoscience applications. Geomorphology 2012, 179, 300–314. [Google Scholar] [CrossRef] [Green Version]
- Meza, J.; Marrugo, A.G.; Ospina, G.; Guerrero, M.; Romero, L.A. A Structure-from-motion pipeline for generating digital elevation models for surface-runoff analysis. J. Phys. Conf. Ser. 2019, 1247, 012039. [Google Scholar] [CrossRef]
- Cullum, C.; Rogers, K.H.; Brierley, G.; Witkowski, E.T.F. Ecological classification and mapping for landscape management and science: Foundations for the description of patterns and processes. Prog. Phys. Geogr. Earth Environ. 2016, 40, 38–65. [Google Scholar] [CrossRef] [Green Version]
- KopeĿ, D.; Michalska-Hejduk, D.; Berezowski, T.; Borowski, M.; Rosadziſski, S.; Chormaſski, J. Application of multisensoral remote sensing data in the mapping of alkaline fens natura 2000 habitat. Ecol. Indic. 2016, 70, 196–208. [Google Scholar] [CrossRef]
- Jensen, J.R. Introductory Digital Image Processing: A Remote Sensing Perspective, 4th ed.; Pearson Series in Geographic Information Science; Pearson: London, UK, 2015; ISBN 978-0-13-405816-0. [Google Scholar]
- Yu, L.; Liang, L.; Wang, J.; Zhao, Y.; Cheng, Q.; Hu, L.; Liu, S.; Yu, L.; Wang, X.; Zhu, P.; et al. Meta-discoveries from a synthesis of satellite-based land-cover mapping research. Int. J. Remote Sens. 2014, 35, 4573–4588. [Google Scholar] [CrossRef]
- Oddi, F.J.; Miguez, F.E.; Ghermandi, L.; Bianchi, L.O.; Garibaldi, L.A. A nonlinear mixed-effects modeling approach for ecological data: Using temporal dynamics of vegetation moisture as an example. Ecol. Evol. 2019, 9, 10225–10240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thessen, A.E. Adoption of machine learning techniques in ecology and earth science. One Ecosyst. 2016, 1, e8621. [Google Scholar] [CrossRef]
- Olden, J.D.; Lawler, J.J.; Poff, N.L. Machine learning methods without tears: A primer for ecologists. Q. Rev. Biol. 2008, 83, 171–193. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of machine-learning classification in remote sensing: An applied review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef] [Green Version]
- Brosofske, K.D.; Froese, R.E.; Falkowski, M.J.; Bans4kota, A. A review of methods for mapping and prediction of inventory attributes for operational forest management. For. Sci. 2013, 60, 733–756. [Google Scholar] [CrossRef]
- Chirici, G.; Mura, M.; McInerney, D.; Py, N.; Tomppo, E.O.; Waser, L.T.; Travaglini, D.; McRoberts, R.E. A meta-analysis and review of the literature on the k-Nearest Neighbors technique for forestry applications that use remotely sensed data. Remote Sens. Environ. 2016, 176, 282–294. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Dronova, I. Object-Based Image Analysis in Wetland Research: A Review. Remote Sens. 2015, 7, 6380–6413. [Google Scholar] [CrossRef] [Green Version]
- Räsänen, A.; Virtanen, T. Data and resolution requirements in mapping vegetation in spatially heterogeneous landscapes. Remote Sens. Environ. 2019, 230, 111207. [Google Scholar] [CrossRef]
- Ward, R.D.; Burnside, N.G.; Joyce, C.B.; Sepp, K. Importance of microtopography in determining plant community distribution in baltic coastal Wetlands. J. Coast. Res. 2016, 32, 1062–1070. [Google Scholar] [CrossRef]
- Berhane, T.M.; Lane, C.R.; Wu, Q.; Anenkhonov, O.A.; Chepinoga, V.V.; Autrey, B.C.; Liu, H. Comparing Pixel- and Object-based approaches in effectively classifying wetland-dominated landscapes. Remote Sens. 2018, 10, 46. [Google Scholar] [CrossRef] [Green Version]
- Çiçekli, S.Y.; Sekertekin, A.; Arslan, N.; Donmez, C. Comparison of pixel and object-based classification methods in Wetlands using sentinel-2 Data. Int. J. Environ. Geoinf. 2018, 7, 213–220. [Google Scholar]
- Kimmel, K. Ecosystem Services of Estonian Wetlands. Ph.D. Thesis, Department of Geography, Institute of Ecology and Earth Sciences, Faculty of Science and Technology, University of Tartu, Tartu, Estonia, 2009. [Google Scholar]
- Rannap, R.; Briggs, L.; Lotman, K.; Lepik, I.; Rannap, V.; Põdra, P. Coastal Meadow Management—Best Practice Guidelines; Ministry of the Environment of the Republic of Estonia: Tallinn, Estonia, 2004; Volume 1, p. 100.
- Larkin, D.J.; Bruland, G.L.; Zedler, J.B. Heterogeneity theory and ecological restoration. In Foundations of Restoration Ecology; Palmer, M.A., Zedler, J.B., Falk, D.A., Eds.; Island Press/Center for Resource Economics: Washington, DC, USA, 2016; pp. 271–300. ISBN 978-1-61091-698-1. [Google Scholar]
- Ward, R.D.; Burnside, N.G.; Joyce, C.B.; Sepp, K.; Teasdale, P.A. Improved modelling of the impacts of sea level rise on coastal wetland plant communities. Hydrobiologia 2016, 774, 203–216. [Google Scholar] [CrossRef] [Green Version]
- Ward, R.D.; Burnside, N.G.; Joyce, C.B.; Sepp, K. The use of medium point density LiDAR elevation data to determine plant community types in Baltic coastal wetlands. Ecol. Indic. 2013, 33, 96–104. [Google Scholar] [CrossRef]
- Villoslada Peciña, M.; Bergamo, T.F.; Ward, R.D.; Joyce, C.B.; Sepp, K. A novel UAV-based approach for biomass prediction and grassland structure assessment in coastal meadows. Ecol. Indic. 2021, 122, 107227. [Google Scholar] [CrossRef]
- Burnside, N.G.; Joyce, C.B.; Puurmann, E.; Scott, D.M. Use of vegetation classification and plant indicators to assess grazing abandonment in Estonian coastal wetlands. J. Veg. Sci. 2007, 18, 645–654. [Google Scholar] [CrossRef]
- Kutser, T.; Paavel, B.; Verpoorter, C.; Ligi, M.; Soomets, T.; Toming, K.; Casal, G. Remote sensing of black lakes and using 810 nm reflectance peak for retrieving water quality parameters of optically complex waters. Remote Sens. 2016, 8, 497. [Google Scholar] [CrossRef]
- Karabulut, M. An examination of spectral reflectance properties of some wetland plants in Göksu Delta, Turkey. J. Int. Environ. Appl. Sci. 2018, 13, 194–203. [Google Scholar]
- Tadrowski, T. Accurate mapping using drones (UAV’s). GeoInformatics 2014, 17, 18. [Google Scholar]
- Villoslada, M.; Bergamo, T.F.; Ward, R.D.; Burnside, N.G.; Joyce, C.B.; Bunce, R.G.H.; Sepp, K. Fine scale plant community assessment in coastal meadows using UAV based multispectral data. Ecol. Indic. 2020, 111, 105979. [Google Scholar] [CrossRef]
- Strong, C.J.; Burnside, N.G.; Llewellyn, D. The potential of small-Unmanned Aircraft Systems for the rapid detection of threatened unimproved grassland communities using an enhanced normalized difference vegetation index. PLoS ONE 2017, 12, e0186193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.W.; Carrivick, J.L.; Quincey, D.J. Structure from motion photogrammetry in physical geography. Prog. Phys. Geogr. Earth Environ. 2016, 40, 247–275. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Zhang, W.; Liang, X.; Wan, P.; Qi, J.; Yu, S.; Yan, G.; Shao, J. Filtering airborne LiDAR data through complementary cloth simulation and progressive TIN densification filters. Remote Sens. 2019, 11, 1037. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, R.S. Using vegetation indices as input into random forest for soybean and weed classification. Am. J. Plant. Sci. 2016, 7, 2186–2198. [Google Scholar] [CrossRef] [Green Version]
- Filho, M.G.; Kuplich, T.M.; Quadros, F.L.F.D. Estimating natural grassland biomass by vegetation indices using Sentinel 2 remote sensing data. Int. J. Remote Sens. 2020, 41, 2861–2876. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, e1353691. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R Package Version 3.4-5. 2020. Available online: https://CRAN.R-project.org/package=raster (accessed on 10 September 2021).
- Bivand, R.; Keitt, T.; Barry, R. Rgdal: Bindings for the “Geospatial” Data Abstraction Library. R Package Version 1.5-18. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 10 September 2021).
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. Monit. Veg. Syst. Gt. Plains ERTS 1974, 351, 309–317. [Google Scholar]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Chen, P.-F.; Tremblay, N.; Wang, J.-H.; Vigneault, P.; Huang, W.-J.; Li, B.-G. New index for crop canopy fresh biomass estimation. Guang Pu Xue Yu Guang Pu Fen XiSpectroscopy Spectr. Anal. 2010, 30, 512–517. [Google Scholar]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T. Coincident detection of crop water stress, nitrogen status and canopy density using ground-based multispectral data. In Proceedings of the 5th International Conference on Precision Agriculture, Bloomingt, MN, USA, 16–19 July 2000. [Google Scholar]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus hippocastanum L. and Acer platanoides L. leaves. spectral features and relation to chlorophyll estimation. J. Plant. Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of Chlorophyll a, Chlorophyll b, Chlorophyll a+b, and total carotenoid content in eucalyptus leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Turpie, K.R. Explaining the spectral red-edge features of inundated marsh vegetation. J. Coast. Res. 2013, 29, 1111–1117. [Google Scholar] [CrossRef]
- Cheng, G.; Xie, X.; Han, J.; Guo, L.; Xia, G.-S. Remote sensing image scene classification meets deep learning: Challenges, methods, benchmarks, and opportunities. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 3735–3756. [Google Scholar] [CrossRef]
- Egorov, A.V.; Hansen, M.C.; Roy, D.P.; Kommareddy, A.; Potapov, P.V. Image interpretation-guided supervised classification using nested segmentation. Remote Sens. Environ. 2015, 165, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; ISBN 978-1-4614-6848-6. [Google Scholar]
- Lin, Z.; Zhang, G. Genetic algorithm-based parameter optimization for EO-1 Hyperion remote sensing image classification. Eur. J. Remote Sens. 2020, 53, 124–131. [Google Scholar] [CrossRef]
- Gonçalves, J.; Pôças, I.; Marcos, B.; Mücher, C.A.; Honrado, J.P. SegOptim—A new R package for optimizing object-based image analyses of high-spatial resolution remotely-sensed data. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 218–230. [Google Scholar] [CrossRef]
- Michel, J.; Youssefi, D.; Grizonnet, M. Stable mean-shift algorithm and its application to the segmentation of arbitrarily large remote sensing images. IEEE Trans. Geosci. Remote Sens. 2015, 53, 952–964. [Google Scholar] [CrossRef]
- Grizonnet, M.; Michel, J.; Poughon, V.; Inglada, J.; Savinaud, M.; Cresson, R. Orfeo toolbox: Open source processing of remote sensing images. Open Geospatial Data Softw. Stand. 2017, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Abu Alfeilat, H.A.; Hassanat, A.B.A.; Lasassmeh, O.; Tarawneh, A.S.; Alhasanat, M.B.; Eyal Salman, H.S.; Prasath, V.B.S. Effects of distance measure choice on K-nearest neighbor classifier performance: A review. Big Data 2019, 7, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Blower, J.D.; Bastin, L.; Lush, V.; Zabala, A.; Masó, J.; Cornford, D.; Díaz, P.; Lumsden, J. An integrated view of data quality in earth observation. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2013, 371, 20120072. [Google Scholar] [CrossRef]
- Foody, G.M. Thematic map comparison. Photogramm. Eng. Remote Sens. 2004, 70, 627–633. [Google Scholar] [CrossRef]
- Pontius, R.G. Quantification error versus location error in comparison of categorical maps. Photogramm. Eng. Amp Remote Sens. 2000, 66, 1011–1016. [Google Scholar]
- Hagen-Zanker, A. Multi-method assessment of map similarity. In Proceedings of the 5th AGILE Conference on Geographic Information Science, Boulder, CO, USA, 25–28 September 2002. [Google Scholar]
- Pontius, R.G., Jr.; Millones, M. Death to Kappa: Birth of quantity disagreement and allocation disagreement for accuracy assessment. Int. J. Remote Sens. 2011, 32, 4407–4429. [Google Scholar] [CrossRef]
- Appelhans, T.; Otte, I.; Kuehnlein, M.; Meyer, H.; Forteva, S.; Nauss, T.; Detsch, F. Rsenal: Magic R Functions for Things Various; R Package Version 0.6.10; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Remmel, T.K. Investigating global and local categorical map configuration comparisons based on coincidence matrices. Geogr. Anal. 2009, 41, 144–157. [Google Scholar] [CrossRef]
- Dronova, I.; Gong, P.; Wang, L.; Zhong, L. Mapping dynamic cover types in a large seasonally flooded wetland using extended principal component analysis and object-based classification. Remote Sens. Environ. 2015, 158, 193–206. [Google Scholar] [CrossRef]
- Dribault, Y.; Chokmani, K.; Bernier, M. Monitoring seasonal hydrological dynamics of minerotrophic peatlands using multi-date GeoEye-1 very high resolution imagery and object-based classification. Remote Sens. 2012, 4, 1887–1912. [Google Scholar] [CrossRef] [Green Version]
- Lane, C.R.; Liu, H.; Autrey, B.C.; Anenkhonov, O.A.; Chepinoga, V.V.; Wu, Q. Improved wetland classification using eight-band high resolution satellite imagery and a hybrid approach. Remote Sens. 2014, 6, 12187–12216. [Google Scholar] [CrossRef] [Green Version]
- Doughty, C.L.; Ambrose, R.F.; Okin, G.S.; Cavanaugh, K.C. Characterizing spatial variability in coastal wetland biomass across multiple scales using UAV and satellite imagery. Remote Sens. Ecol. Conserv. 2021. [Google Scholar] [CrossRef]
- Carleer, A.P.; Debeir, O.; Wolff, E. Assessment of very high spatial resolution satellite image segmentations. Photogramm. Eng. Remote Sens. 2005, 71, 1285–1294. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Hayes, M.M.; Miller, S.N.; Murphy, M.A. High-resolution landcover classification using random forest. Remote Sens. Lett. 2014, 5, 112–121. [Google Scholar] [CrossRef]
- Maselli, F.; Chirici, G.; Bottai, L.; Corona, P.; Marchetti, M. Estimation of Mediterranean forest attributes by the application of k-NN procedures to multitemporal Landsat ETM+ images. Int. J. Remote Sens. 2005, 26, 3781–3796. [Google Scholar] [CrossRef] [Green Version]
- Wieland, M.; Pittore, M. Performance evaluation of machine learning algorithms for urban pattern recognition from multi-spectral satellite images. Remote Sens. 2014, 6, 2912–2939. [Google Scholar] [CrossRef] [Green Version]
- Moffett, K.B.; Gorelick, S.M. Distinguishing wetland vegetation and channel features with object-based image segmentation. Int. J. Remote Sens. 2013, 34, 1332–1354. [Google Scholar] [CrossRef]
- van der Wel, F. Assessment and Visualisation of Uncertainty in Remote Sensing Land Cover Classifications; University of Utrecht: Utrecht, The Netherlands, 2000; ISBN 90-6266-181-5. [Google Scholar]

| Study Area | Plant Communities | Elevation Range (m.a.s.l) | Area (ha) |
|---|---|---|---|
| KUD | LS, OP, TG, US | 0.01–1.85 | 30 |
| MA2 | LS, OP, RS, TG, US | −0.74–2.98 | 41 |
| RAL | LS, OP, TG, US | 0.19–0.36 | 10 |
| RUE | LS, OP, US | −0.13–0.62 | 8 |
| TAN | LS, OP, US | −0.34–0.89 | 10 |
| TAS | LS, OP, TG, US | −0.64–2.66 | 12 |
| Study Area | Flight Dates |
|---|---|
| KUD | 30 June 2019 |
| MA2 | 29 June 2019 |
| RAL | 4 July 2019 |
| RUE | 2 July 2019 |
| TAN | 30 June 2019 |
| TAS | 23 July 2019 |
| Vegetation Index | Calculation | Reference |
|---|---|---|
| Normalized Difference Vegetation Index | [52] | |
| Green Normalized Difference Vegetation Index | [53] | |
| Chlorophyll Vegetation Index | [54] | |
| Modified Simple Ratio (red edge) | [55] | |
| Red edge triangular vegetation index (core only) | [56] | |
| Canopy Chlorophyll Content Index | [57] | |
| Chlorophyll Index (red edge) | [58] | |
| Red edge normalized difference vegetation index | [59] | |
| Datt4 | [60] | |
| Modified Green Red Vegetation Index | [61] |
| Study Area | Best RF | Best KNN | Mean Size RF | Mean Size KNN |
|---|---|---|---|---|
| KUD | 0.07, 0.09, 13 | 0.14, 0.09, 14 | 24 | 29 |
| MA2 | 0.1, 0.07, 10 | 0.18, 0.06, 8 | 16 | 14 |
| RAL | 0.2, 0.09, 5 | 0.07, 0.07, 9 | 11 | 20 |
| RUE | 0.2, 0.06, 8 | 0.09, 0.07, 7 | 16 | 18 |
| TAN | 0.2, 0.07, 7 | 0.07, 0.1, 10 | 12 | 21 |
| TAS | 0.18, 0.09, 9 | 0.15, 0.08, 11 | 16 | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez Prentice, R.; Villoslada Peciña, M.; Ward, R.D.; Bergamo, T.F.; Joyce, C.B.; Sepp, K. Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands. Remote Sens. 2021, 13, 3669. https://doi.org/10.3390/rs13183669
Martínez Prentice R, Villoslada Peciña M, Ward RD, Bergamo TF, Joyce CB, Sepp K. Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands. Remote Sensing. 2021; 13(18):3669. https://doi.org/10.3390/rs13183669
Chicago/Turabian StyleMartínez Prentice, Ricardo, Miguel Villoslada Peciña, Raymond D. Ward, Thaisa F. Bergamo, Chris B. Joyce, and Kalev Sepp. 2021. "Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands" Remote Sensing 13, no. 18: 3669. https://doi.org/10.3390/rs13183669
APA StyleMartínez Prentice, R., Villoslada Peciña, M., Ward, R. D., Bergamo, T. F., Joyce, C. B., & Sepp, K. (2021). Machine Learning Classification and Accuracy Assessment from High-Resolution Images of Coastal Wetlands. Remote Sensing, 13(18), 3669. https://doi.org/10.3390/rs13183669