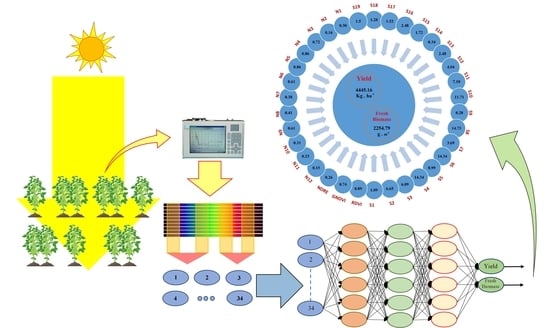
Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Test Sites and Experimental Designs
2.3. Data Acquisition
2.3.1. Field Data Collection
2.3.2. Hyperspectral Reflectance Data Collection
2.4. Data Pre-Processing and Statistical Analyses
2.5. Hyperspectral Vegetation Index (HVI) Extraction
2.6. Variable Selection
2.7. Yield and FBIO Prediction Model Calibration and Validation
2.7.1. Ensemble Method (Ensemble-Bagging Algorithm)
2.7.2. Deep Neural Network (DNN)
2.8. Optimization Process (SPEA2 Algorithm)
2.9. Quantification of Model Performance and Error Estimations
3. Results
3.1. Yield, FBIO and HVI Properties
3.2. Correlation Analysis of HVI vs. Soybean Yield and FBIO
3.3. Comparative Analysis of the EB and DNN Algorithms
3.4. Variable Selection
3.5. The DNN-SPEA2 Optimization Algorithm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colletti, A.; Attrovio, A.; Boffa, L.; Mantegna, S.; Cravotto, G. Valorisation of By-Products from Soybean (Glycine max (L.) Merr.) Processing. Molecules 2020, 25, 2129. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Kumar, A.; Abd_Allah, E.F.; Hashem, A.; Khan, M.L. Growing more with less: Breeding and developing drought resilient soybean to improve food security. Ecol. Indic. 2019, 105, 425–437. [Google Scholar] [CrossRef]
- De Pretto, C.; Giordano, R.d.L.C.; Tardioli, P.W.; Costa, C.B.B. Possibilities for producing energy, fuels, and chemicals from soybean: A biorefinery concept. Waste Biomass Valorization 2018, 9, 1703–1730. [Google Scholar] [CrossRef]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Reynolds, M.; Singh, R. Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 Genes Genomes Genet. 2016, 6, 2799–2808. [Google Scholar] [CrossRef] [Green Version]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [Green Version]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.L.; Mouazen, A.M. Wheat yield prediction using machine learning and advanced sensing techniques. Comput. Electron. Agric. 2016, 121, 57–65. [Google Scholar] [CrossRef]
- Schut, A.G.; Traore, P.C.S.; Blaes, X.; Rolf, A. Assessing yield and fertilizer response in heterogeneous smallholder fields with UAVs and satellites. Field Crop. Res. 2018, 221, 98–107. [Google Scholar] [CrossRef]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; de los Campos, G.; Alvarado, G.; Suchismita, M.; Rutkoski, J.; González-Pérez, L.; Burgueño, J. Predicting grain yield using canopy hyperspectral reflectance in wheat breeding data. Plant Methods 2017, 13, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, L.; Zhang, Z.; Ma, Y.; Du, Q.; Williams, P.; Drewry, J.; Luck, B. Alfalfa Yield Prediction Using UAV-Based Hyperspectral Imagery and Ensemble Learning. Remote Sens. 2020, 12, 2028. [Google Scholar] [CrossRef]
- Zha, H.; Miao, Y.; Wang, T.; Li, Y.; Zhang, J.; Sun, W.; Feng, Z.; Kusnierek, K. Improving unmanned aerial vehicle remote sensing-based rice nitrogen nutrition index prediction with machine learning. Remote Sens. 2020, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Lobell, D.B.; Di Tommaso, S.; You, C.; Yacoubou Djima, I.; Burke, M.; Kilic, T. Sight for sorghums: Comparisons of Satellite-and ground-based sorghum yield estimates in mali. Remote Sens. 2020, 12, 100. [Google Scholar] [CrossRef] [Green Version]
- Yoosefzadeh-Najafabadi, M.; Earl, H.J.; Tulpan, D.; Sulik, J.; Eskandari, M. Application of Machine Learning Algorithms in Plant Breeding: Predicting Yield From Hyperspectral Reflectance in Soybean. Front. Plant Sci. 2021, 11. [Google Scholar] [CrossRef]
- Kycko, M.; Zagajewski, B.; Lavender, S.; Romanowska, E.; Zwijacz-Kozica, M. The Impact of Tourist Traffic on the Condition and Cell Structures of Alpine Swards. Remote Sens. 2018, 10, 220. [Google Scholar] [CrossRef] [Green Version]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Jin, X.; Xia, X.; Xiao, Y.; He, Z. Time-Series Multispectral Indices from Unmanned Aerial Vehicle Imagery Reveal Senescence Rate in Bread Wheat. Remote Sens. 2018, 10, 809. [Google Scholar] [CrossRef] [Green Version]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef] [Green Version]
- Duan, T.; Zheng, B.; Guo, W.; Ninomiya, S.; Guo, Y.; Chapman, S.C. Comparison of ground cover estimates from experiment plots in cotton, sorghum and sugarcane based on images and ortho-mosaics captured by UAV. Funct. Plant Biol. 2017, 44, 169–183. [Google Scholar] [CrossRef]
- Watanabe, K.; Guo, W.; Arai, K.; Takanashi, H.; Kajiya-Kanegae, H.; Kobayashi, M.; Yano, K.; Tokunaga, T.; Fujiwara, T.; Tsutsumi, N. High-throughput phenotyping of sorghum plant height using an unmanned aerial vehicle and its application to genomic prediction modeling. Front. Plant Sci. 2017, 8, 421. [Google Scholar] [CrossRef] [Green Version]
- Filippa, G.; Cremonese, E.; Migliavacca, M.; Galvagno, M.; Sonnentag, O.; Humphreys, E.; Hufkens, K.; Ryu, Y.; Verfaillie, J.; Morra di Cella, U.; et al. NDVI derived from near-infrared-enabled digital cameras: Applicability across different plant functional types. Agric. For. Meteorol. 2018, 249, 275–285. [Google Scholar] [CrossRef]
- Jolly, W.M.; Nemani, R.; Running, S.W. A generalized, bioclimatic index to predict foliar phenology in response to climate. Glob. Chang. Biol. 2005, 11, 619–632. [Google Scholar] [CrossRef]
- Lopez-Cruz, M.; Olson, E.; Rovere, G.; Crossa, J.; Dreisigacker, S.; Mondal, S.; Singh, R.; de los Campos, G. Regularized selection indices for breeding value prediction using hyper-spectral image data. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Development of support vector machine-based model and comparative analysis with artificial neural network for modeling the plant tissue culture procedures: Effect of plant growth regulators on somatic embryogenesis of chrysanthemum, as a case study. Plant Methods 2020, 16, 1–15. [Google Scholar] [CrossRef]
- Nezami-Alanagh, E.; Garoosi, G.-A.; Landín, M.; Gallego, P.P. Combining DOE with neurofuzzy logic for healthy mineral nutrition of pistachio rootstocks in vitro culture. Front. Plant Sci. 2018, 9, 1474. [Google Scholar] [CrossRef]
- Zhang, Z.; Jin, Y.; Chen, B.; Brown, P. California almond yield prediction at the orchard level with a machine learning approach. Front. Plant Sci. 2019, 10, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Galiano, V.; Sanchez-Castillo, M.; Chica-Olmo, M.; Chica-Rivas, M. Machine learning predictive models for mineral prospectivity: An evaluation of neural networks, random forest, regression trees and support vector machines. Ore Geol. Rev. 2015, 71, 804–818. [Google Scholar] [CrossRef]
- Gandhi, N.; Armstrong, L.J.; Petkar, O.; Tripathy, A.K. Rice crop yield prediction in India using support vector machines. In Proceedings of the 2016 13th International Joint Conference on Computer Science and Software Engineering (JCSSE), Kaen, Thailand, 13–15 July 2016; pp. 1–5. [Google Scholar]
- Bhojani, S.H.; Bhatt, N. Wheat crop yield prediction using new activation functions in neural network. Neural Comput. Appl. 2020, 32, 13941–13951. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Application of machine learning and genetic optimization algorithms for modeling and optimizing soybean yield using its component traits. PLoS ONE 2021, 16, e0250665. [Google Scholar] [CrossRef]
- Jin, Z.; Zhuang, Q.; Tan, Z.; Dukes, J.S.; Zheng, B.; Melillo, J.M. Do maize models capture the impacts of heat and drought stresses on yield? Using algorithm ensembles to identify successful approaches. Glob. Chang. Biol. 2016, 22, 3112–3126. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S. Deep gaussian process for crop yield prediction based on remote sensing data. In Proceedings of the Thirty-First AAAI conference on artificial intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Benedetti, P.; Ienco, D.; Gaetano, R.; Ose, K.; Pensa, R.G.; Dupuy, S. M3Fusion: A Deep Learning Architecture for Multiscale Multimodal Multitemporal Satellite Data Fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 4939–4949. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Wu, G.; Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, D.; Wong, C.; Lo, B.; Yang, G.-Z. Deep learning for human activity recognition: A resource efficient implementation on low-power devices. In Proceedings of the 2016 IEEE 13th international conference on wearable and implantable body sensor networks (BSN), San Francisco, CA, USA, 14–17 June 2016; pp. 71–76. [Google Scholar]
- Yu, N.; Li, L.; Schmitz, N.; Tian, L.F.; Greenberg, J.A.; Diers, B.W. Development of methods to improve soybean yield estimation and predict plant maturity with an unmanned aerial vehicle based platform. Remote Sens. Environ. 2016, 187, 91–101. [Google Scholar] [CrossRef]
- Ali, I.; Cawkwell, F.; Green, S.; Dwyer, N. Application of statistical and machine learning models for grassland yield estimation based on a hypertemporal satellite remote sensing time series. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 5060–5063. [Google Scholar]
- Ribeiro, M.H.D.M.; dos Santos Coelho, L. Ensemble approach based on bagging, boosting and stacking for short-term prediction in agribusiness time series. Appl. Soft Comput. 2020, 86, 105837. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble methods in machine learning. In International workshop on multiple classifier systems; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–15. [Google Scholar]
- Divina, F.; Gilson, A.; Goméz-Vela, F.; García Torres, M.; Torres, J.F. Stacking ensemble learning for short-term electricity consumption forecasting. Energies 2018, 11, 949. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.L.; Ebecken, N.F.F.; Esquerdo, J.C.D.M. Sugarcane yield prediction in Brazil using NDVI time series and neural networks ensemble. Int. J. Remote Sens. 2017, 38, 4631–4644. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Galar, M.; Fernandez, A.; Barrenechea, E.; Bustince, H.; Herrera, F. A review on ensembles for the class imbalance problem: Bagging-, boosting-, and hybrid-based approaches. IEEE Trans. Syst. Manand Cybern. Part C (Appl. Rev.) 2011, 42, 463–484. [Google Scholar] [CrossRef]
- Schapire, R.E. A brief introduction to boosting. In Proceedings of the Ijcai, New York, NY, USA, 9–15 July 2016; pp. 1401–1406. [Google Scholar]
- Zhou, Z.-H. Ensemble Methods: Foundations and Algorithms; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 1–37. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Application of adaptive neuro-fuzzy inference system-non-dominated sorting genetic Algorithm-II (ANFIS-NSGAII) for modeling and optimizing somatic embryogenesis of Chrysanthemum. Front. Plant Sci. 2019, 10, 869. [Google Scholar] [CrossRef]
- Dao, D.-N.; Guo, L.-X. New hybrid SPEA/R-deep learning to predict optimization parameters of cascade FOPID controller according engine speed in powertrain mount system control of half-car dynamic model. J. Intell. Fuzzy Syst. 2020, 39, 53–68. [Google Scholar] [CrossRef]
- Zhao, F.; Lei, W.; Ma, W.; Liu, Y.; Zhang, C. An improved SPEA2 algorithm with adaptive selection of evolutionary operators scheme for multiobjective optimization problems. Math. Probl. Eng. 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Stroup, W.; Mulitze, D. Nearest neighbor adjusted best linear unbiased prediction. Am. Stat. 1991, 45, 194–200. [Google Scholar]
- Katsileros, A.; Drosou, K.; Koukouvinos, C. Evaluation of nearest neighbor methods in wheat genotype experiments. Commun. Biometry Crop Sci. 2015, 10, 115–123. [Google Scholar]
- Bowley, S. A Hitchhiker’s Guide to Statistics in Plant Biology; Any Old Subject Books: Guelph, ON, Canada, 1999. [Google Scholar]
- Fehr, W.; Caviness, C.; Burmood, D.; Pennington, J. Stage of development descriptions for soybeans, Glycine max (L.) Merrill 1. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Goldberger, A.S. Best linear unbiased prediction in the generalized linear regression model. J. Am. Stat. Assoc. 1962, 57, 369–375. [Google Scholar] [CrossRef]
- Ozaki, Y.; McClure, W.F.; Christy, A.A. Near-Infrared Spectroscopy in Food Science and Technology; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Rossel, R.A.V. ParLeS: Software for chemometric analysis of spectroscopic data. Chemom. Intell. Lab. Syst. 2008, 90, 72–83. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Huang, H.; Zeng, Q.; Pei, X.; Wong, S.; Xu, P. Predicting crash frequency using an optimised radial basis function neural network model. Transp. A: Transp. Sci. 2016, 12, 330–345. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Huang, H. A stable and optimized neural network model for crash injury severity prediction. Accid. Anal. Prev. 2014, 73, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Gal, T.; Greenberg, H.J. Advances in Sensitivity Analysis and Parametric Programming; Springer Science & Business Media: New York, NY, USA, 2012; Volume 6. [Google Scholar]
- Chen, X.-w.; Jeong, J.C. Enhanced recursive feature elimination. In Proceedings of the Sixth International Conference on Machine Learning and Applications (ICMLA 2007), Cincinnati, OH, USA, 13–15 December 2007; pp. 429–435. [Google Scholar]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meij, B.; Kooistra, L.; Suomalainen, J.; Barel, J.M.; De Deyn, G.B. Remote sensing of plant trait responses to field-based plant-soil feedback using UAV-based optical sensors. Biogeosciences 2017, 14, 733. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Ma, B.L.; Hu, Y.; Liu, J. Characterization of nitrogen and water status in oat leaves using optical sensing approach. J. Sci. Food Agric. 2015, 95, 367–378. [Google Scholar] [CrossRef]
- Yu, K.; Gnyp, M.; Gao, J.; Miao, Y.; Chen, X.; Bareth, G. Using Partial Least Squares (PLS) to Estimate Canopy Nitrogen and Biomass of Paddy Rice in China’s Sanjiang Plain. In Proceedings of the Workshop on UAV-based Remote Sensing Methods for Monitoring Vegetation, Cologne, Germany, 9–10 September 2013; Bendig, J., Bareth, G., Eds.; Kölner Geographische Arbeiten: Cologne, Germany; pp. 99–103. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-ausing reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W.; Rivard, B.; Feng, J. Remote estimation of gross primary production in wheat using chlorophyll-related vegetation indices. Agric. For. Meteorol. 2009, 149, 1015–1021. [Google Scholar] [CrossRef]
- Kooistra, L.; Suomalainen, J.; Iqbal, S.; Franke, J.; Wenting, P.; Bartholomeus, H.; Mücher, S.; Becker, R. Crop monitoring using a light-weight hyperspectral mapping system for unmanned aerial vehicles: First results for the 2013 season. In Proceedings of the 2013 Workshop on UAV-based Remote Sensing Methods for Monitoring Vegetation, Cologne, Germany, 9–10 September 2013; p. 5158. [Google Scholar]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Rodriguez, D.; Fitzgerald, G.; Belford, R.; Christensen, L. Detection of nitrogen deficiency in wheat from spectral reflectance indices and basic crop eco-physiological concepts. Aust. J. Agric. Res. 2006, 57, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Tian, Y.; Yao, X.; Yang, J.; Cao, W.; Hannaway, D.; Zhu, Y. Assessing newly developed and published vegetation indices for estimating rice leaf nitrogen concentration with ground-and space-based hyperspectral reflectance. Field Crop. Res. 2011, 120, 299–310. [Google Scholar] [CrossRef]
- Datt, B. A new reflectance index for remote sensing of chlorophyll content in higher plants: Tests using Eucalyptus leaves. J. Plant Physiol. 1999, 154, 30–36. [Google Scholar] [CrossRef]
- Xue, L.; Cao, W.; Luo, W.; Dai, T.; Zhu, Y. Monitoring leaf nitrogen status in rice with canopy spectral reflectance. Agron. J. 2004, 96, 135–142. [Google Scholar] [CrossRef]
- Zhu, Y.; Yao, X.; Tian, Y.; Liu, X.; Cao, W. Analysis of common canopy vegetation indices for indicating leaf nitrogen accumulations in wheat and rice. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 1–10. [Google Scholar] [CrossRef]
- McMurtrey Iii, J.; Chappelle, E.; Kim, M.; Meisinger, J.; Corp, L. Distinguishing nitrogen fertilization levels in field corn (Zea mays L.) with actively induced fluorescence and passive reflectance measurements. Remote Sens. Environ. 1994, 47, 36–44. [Google Scholar] [CrossRef]
- Chen, P.; Haboudane, D.; Tremblay, N.; Wang, J.; Vigneault, P.; Li, B. New spectral indicator assessing the efficiency of crop nitrogen treatment in corn and wheat. Remote Sens. Environ. 2010, 114, 1987–1997. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey III, J.E. Ratio analysis of reflectance spectra (RARS): An algorithm for the remote estimation of the concentrations of chlorophyll a, chlorophyll b, and carotenoids in soybean leaves. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Gupta, R.; Vijayan, D.; Prasad, T. Comparative analysis of red-edge hyperspectral indices. Adv. Space Res. 2003, 32, 2217–2222. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R. Land cover mapping at BOREAS using red edge spectral parameters from CASI imagery. J. Geophys. Res. Atmos. 1999, 104, 27921–27933. [Google Scholar] [CrossRef]
- Orr, M.J. Introduction to Radial Basis Function Networks; Technical Report; Center for Cognitive Science, University of Edinburgh: Edinburgh, UK, 1996. [Google Scholar]
- Wilamowski, B.M.; Jaeger, R.C. Implementation of RBF type networks by MLP networks. In Proceedings of the International Conference on Neural Networks (ICNN’96), Washington, DC, USA, 3–6 June 1996; pp. 1670–1675. [Google Scholar]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 2000. [Google Scholar]
- Ye, Y.; Chen, L.; Wang, D.; Li, T.; Jiang, Q.; Zhao, M. SBMDS: An interpretable string based malware detection system using SVM ensemble with bagging. J. Comput. Virol. 2009, 5, 283. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. Acm Sigkdd Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Du, J.; Xu, Y. Hierarchical deep neural network for multivariate regression. Pattern Recognit. 2017, 63, 149–157. [Google Scholar] [CrossRef]
- Pal, S.K.; Mitra, S. Multilayer perceptron, fuzzy sets, classifiaction. IEEE Trans. Neural Netw. 1992, 3, 683–697. [Google Scholar] [CrossRef]
- Dao, D.-N.; Guo, L.-X. New hybrid between SPEA/R with deep neural network: Application to predicting the multi-objective optimization of the stiffness parameter for powertrain mount systems. J. Low Freq. Noisevibration Act. Control 2020, 39, 53–68. [Google Scholar] [CrossRef] [Green Version]
- Maheta, H.H.; Dabhi, V.K. An improved SPEA2 Multi objective algorithm with non dominated elitism and Generational Crossover. In Proceedings of the 2014 International Conference on Issues and Challenges in Intelligent Computing Techniques (ICICT), Ghaziabad, India, 7–8 February 2014; pp. 75–82. [Google Scholar]
- Zitzler, E.; Laumanns, M.; Thiele, L. SPEA2: Improving the strength Pareto evolutionary algorithm. TIK-Report 2001, 103. [Google Scholar] [CrossRef]
- Galli, G.; Horne, D.W.; Collins, S.D.; Jung, J.; Chang, A.; Fritsche-Neto, R.; Rooney, W.L. Optimization of UAS-based high-throughput phenotyping to estimate plant health and grain yield in sorghum. Plant Phenome J. 2020, 3, e20010. [Google Scholar] [CrossRef]
- Mladenova, I.E.; Bolten, J.D.; Crow, W.T.; Anderson, M.C.; Hain, C.R.; Johnson, D.M.; Mueller, R. Intercomparison of soil moisture, evaporative stress, and vegetation indices for estimating corn and soybean yields over the US. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 1328–1343. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.; Xu, X.; He, J.; Ge, X.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.; Tian, Y. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. Isprs J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Bala, S.; Islam, A. Correlation between potato yield and MODIS-derived vegetation indices. Int. J. Remote Sens. 2009, 30, 2491–2507. [Google Scholar] [CrossRef]
- Aparicio, N.; Villegas, D.; Casadesus, J.; Araus, J.L.; Royo, C. Spectral vegetation indices as nondestructive tools for determining durum wheat yield. Agron. J. 2000, 92, 83–91. [Google Scholar] [CrossRef]
- Kumar, R.; Silva, L. Light ray tracing through a leaf cross section. Appl. Opt. 1973, 12, 2950–2954. [Google Scholar] [CrossRef]
- Woolley, J.T. Reflectance and transmittance of light by leaves. Plant Physiol. 1971, 47, 656–662. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Scott, M.; Bhattacharyya, M. Light is Essential for Degradation of Ribulose-1, 5-Bisphosphate Carboxylase-Oxygenase Large Subunit During Sudden Death Syndrome Development in Soybean. Plant Biol. 2006, 8, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Bosquet, L.; Crossa, J.; von Zitzewitz, J.; Serret, M.D.; Luis Araus, J. High-throughput phenotyping and genomic selection: The frontiers of crop breeding converge. J. Integr. Plant Biol. 2012, 54, 312–320. [Google Scholar] [CrossRef] [Green Version]
- Kandel, Y.R.; Wise, K.A.; Bradley, C.A.; Tenuta, A.U.; Mueller, D.S. Effect of planting date, seed treatment, and cultivar on plant population, sudden death syndrome, and yield of soybean. Plant Dis. 2016, 100, 1735–1743. [Google Scholar] [CrossRef] [Green Version]
- Ford, D.M.; Shibles, R.; Green, D. Growth and Yield of Soybean Lines Selected for Divergent Leaf Photosynthetic Ability1. Crop Sci. 1983, 23, 517–520. [Google Scholar] [CrossRef]
- Karmakar, P.; Bhatnagar, P. Genetic improvement of soybean varieties released in India from 1969 to 1993. Euphytica 1996, 90, 95–103. [Google Scholar]
- Hennessy, A.; Clarke, K.; Lewis, M. Hyperspectral Classification of Plants: A Review of Waveband Selection Generalisability. Remote Sens. 2020, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Mariotto, I.; Thenkabail, P.S.; Huete, A.; Slonecker, E.T.; Platonov, A. Hyperspectral versus multispectral crop-productivity modeling and type discrimination for the HyspIRI mission. Remote Sens. Environ. 2013, 139, 291–305. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of hyperspectral narrowbands (HNBs) and composition of hyperspectral twoband vegetation indices (HVIs) for biophysical characterization and discrimination of crop types using field reflectance and Hyperion/EO-1 data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- G. Jones, T.; Coops, N.C.; Sharma, T. Employing ground-based spectroscopy for tree-species differentiation in the Gulf Islands National Park Reserve. Int. J. Remote Sens. 2010, 31, 1121–1127. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- CABRERA-BOSQUET, L.; Albrizio, R.; Nogues, S.; Araus, J.L. Dual Δ13C/δ18O response to water and nitrogen availability and its relationship with yield in field-grown durum wheat. Plantcell Environ. 2011, 34, 418–433. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Mišurec, J.; Kopačková, V.; Mielke, C.; Rogass, C. Assessment of red-edge position extraction techniques: A case study for norway spruce forests using hymap and simulated sentinel-2 data. Forests 2016, 7, 226. [Google Scholar] [CrossRef] [Green Version]
- Mehdaoui, R.; Anane, M. Exploitation of the red-edge bands of Sentinel 2 to improve the estimation of durum wheat yield in Grombalia region (Northeastern Tunisia). Int. J. Remote Sens. 2020, 41, 8986–9008. [Google Scholar] [CrossRef]
- Guerini Filho, M.; Kuplich, T.M.; Quadros, F.L.D. Estimating natural grassland biomass by vegetation indices using Sentinel 2 remote sensing data. Int. J. Remote Sens. 2020, 41, 2861–2876. [Google Scholar] [CrossRef]
- Fernández, C.I.; Leblon, B.; Haddadi, A.; Wang, K.; Wang, J. Potato late blight detection at the leaf and canopy levels based in the red and red-edge spectral regions. Remote Sens. 2020, 12, 1292. [Google Scholar] [CrossRef] [Green Version]
- Babar, M.; Reynolds, M.; Van Ginkel, M.; Klatt, A.; Raun, W.; Stone, M. Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci. 2006, 46, 1046–1057. [Google Scholar] [CrossRef]
- Rutherford, A. Introducing ANOVA and ANCOVA: A GLM Approach; Sage: London, UK, 2001. [Google Scholar]
- Homack, S.R. Understanding What ANOVA Post Hoc Tests Are, Really. In Proceedings of the Annual Meeting of the Southwest Educational Research Association, New Orleans, LA, USA, 1–3 February 2001. [Google Scholar]
- Yeom, S.; Giacomelli, I.; Menaged, A.; Fredrikson, M.; Jha, S. Overfitting, robustness, and malicious algorithms: A study of potential causes of privacy risk in machine learning. J. Comput. Secur. 2020, 28, 35–70. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Hwa Yang, Y.; B Zhou, B.; Y Zomaya, A. A review of ensemble methods in bioinformatics. Curr. Bioinform. 2010, 5, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Belayneh, A.; Adamowski, J.; Khalil, B.; Ozga-Zielinski, B. Long-term SPI drought forecasting in the Awash River Basin in Ethiopia using wavelet neural network and wavelet support vector regression models. J. Hydrol. 2014, 508, 418–429. [Google Scholar] [CrossRef]
- Mahalingam, T.; Subramoniam, M. ACO–MKFCM: An Optimized Object Detection and Tracking Using DNN and Gravitational Search Algorithm. Wirel. Pers. Commun. 2020, 110, 1567–1604. [Google Scholar] [CrossRef]
- Nigam, A.; Tiwari, A.K.; Pandey, A. Paddy leaf diseases recognition and classification using PCA and BFO-DNN algorithm by image processing. Mater. Today Proc. 2020, 33, 4856–4862. [Google Scholar] [CrossRef]
- Kukana, P. Hybrid Machine Learning Algorithm-Based Paddy Leave Disease Detection System. In Proceedings of the 2020 International Conference on Smart Electronics and Communication (ICOSEC), Trichy, India, 10–12 September 2020; pp. 512–519. [Google Scholar]
- Alam, M.; Mukhopadhyay, D. How secure are deep learning algorithms from side-channel based reverse engineering? In Proceedings of the 56th Annual Design Automation Conference 2019, Las Vegas, NV, USA, 2–6 June 2019; pp. 1–2. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Bisong, E. The Multilayer Perceptron (MLP). In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berkeley, CA, USA, 2019; pp. 401–405. [Google Scholar]
- Ge, Y.; Wang, Q.; Wang, L.; Wu, H.; Peng, C.; Wang, J.; Xu, Y.; Xiong, G.; Zhang, Y.; Yi, Y. Predicting post-stroke pneumonia using deep neural network approaches. Int. J. Med Inform. 2019, 132, 103986. [Google Scholar] [CrossRef] [PubMed]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4959–4962. [Google Scholar]
- Slinkard, A.; Solh, M.; Vandenberg, A. Breeding for yield: The direct approach. In Linking Research and Marketing Opportunities for Pulses in the 21st Century; Springer: Dordrecht, The Netherlands, 2000; pp. 183–190. [Google Scholar]
- Jin, J.; Liu, X.; Wang, G.; Mi, L.; Shen, Z.; Chen, X.; Herbert, S.J. Agronomic and physiological contributions to the yield improvement of soybean cultivars released from 1950 to 2006 in Northeast China. Field Crop. Res. 2010, 115, 116–123. [Google Scholar] [CrossRef]
- Ma, B.; Dwyer, L.M.; Costa, C.; Cober, E.R.; Morrison, M.J. Early prediction of soybean yield from canopy reflectance measurements. Agron. J. 2001, 93, 1227–1234. [Google Scholar] [CrossRef] [Green Version]
- Shigang, F.; Qian, A. Multi-objective reactive power optimization using SPEA2. High Volt. Eng. 2007, 33, 115–119. [Google Scholar]
- Zaloga, A.; Burakov, S.; Yakimov, I.; Gusev, K.; Dubinin, P. Multi-population evolutionary algorithm SPEA2 for crystal structure determination from X-ray powder diffraction data. In Proceedings of the IOP Conference Series: Materials Science and Engineering, Krasnoyarsk, Russia, 18–21 November 2019; p. 012102. [Google Scholar]
- King, R.A.; Deb, K.; Rughooputh, H. Comparison of NSGA-II and SPEA2 on the multiobjective environmental/economic dispatch problem. Univ. Maurit. Res. J. 2010, 16, 485–511. [Google Scholar]









| Index Category | Abbreviation | Formula | Reference |
|---|---|---|---|
| Normalized difference vegetation index (NDVI) | N1 | [584 nm − 471 nm]/[584 nm + 471 nm] | [65] |
| N2 | [689 nm − 521 nm]/[689 nm + 521 nm] | [65] | |
| N3 | [760 nm − 550 nm]/[760 nm + 550 nm] | [66] | |
| N4 | [740 nm − 667 nm]/[740 nm + 667 nm] | [67] | |
| N5 | [800 nm − 670 nm]/[800 nm + 670 nm] | [68] | |
| N6 | [750 nm − 705 nm]/[750 nm + 705 nm] | [69] | |
| N7 | [750 nm − 710 nm]/[750 nm + 710 nm] | [70] | |
| N8 | [780 nm − 710 nm]/[780 nm + 710 nm] | [71] | |
| N9 | [750 nm − 710 nm]/[750 nm + 710 nm] | [72] | |
| N10 | [732 nm − 717 nm]/[732 nm + 717 nm] | [72] | |
| N11 | [820 nm − 720 nm]/[820 nm + 720 nm] | [73] | |
| N12 | [750 nm − 735 nm]/[750 nm + 734 nm] | [72] | |
| Normalized difference red edge (NDRE) | NDRE | [790 nm − 720 nm]/[790 nm + 720 nm] | [74] |
| Green normalized difference vegetation index (GNDVI) | GNDVI | [750 nm − 550 nm]/[750 nm + 550 nm] | [75] |
| Renormalized difference vegetation index (RDVI) | RDVI | [800 nm − 670 nm]/[800 nm + 670 nm] | [76] |
| Simple ratio index (SRI) | S1 | [565 nm/533 nm] | [77] |
| S2 | [750 nm/550 nm] | [78] | |
| S3 | [760 nm/550 nm] | [66] | |
| S4 | [810 nm/560 nm] | [79] | |
| S5 | [734 nm/629 nm] | [67] | |
| S6 | [810 nm/660 nm] | [80] | |
| S7 | [700 nm/670 nm] | [81] | |
| S8 | [800 nm/670 nm] | [82] | |
| S9 | [675 nm/700 nm] | [83] | |
| S10 | [800 nm/680 nm] | [84] | |
| S11 | [752 nm/690 nm] | [78] | |
| S12 | [750 nm/700 nm] | [78] | |
| S13 | [750 nm/705 nm] | [69] | |
| S14 | [706 nm/755 nm] | [72] | |
| S15 | [747 nm/708 nm] | [85] | |
| S16 | [750 nm/710 nm] | [86] | |
| S17 | [741 nm/717 nm] | [85] | |
| S18 | [735 nm/720 nm] | [85] | |
| S19 | [738 nm/720 nm] | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices. Remote Sens. 2021, 13, 2555. https://doi.org/10.3390/rs13132555
Yoosefzadeh-Najafabadi M, Tulpan D, Eskandari M. Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices. Remote Sensing. 2021; 13(13):2555. https://doi.org/10.3390/rs13132555
Chicago/Turabian StyleYoosefzadeh-Najafabadi, Mohsen, Dan Tulpan, and Milad Eskandari. 2021. "Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices" Remote Sensing 13, no. 13: 2555. https://doi.org/10.3390/rs13132555
APA StyleYoosefzadeh-Najafabadi, M., Tulpan, D., & Eskandari, M. (2021). Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices. Remote Sensing, 13(13), 2555. https://doi.org/10.3390/rs13132555