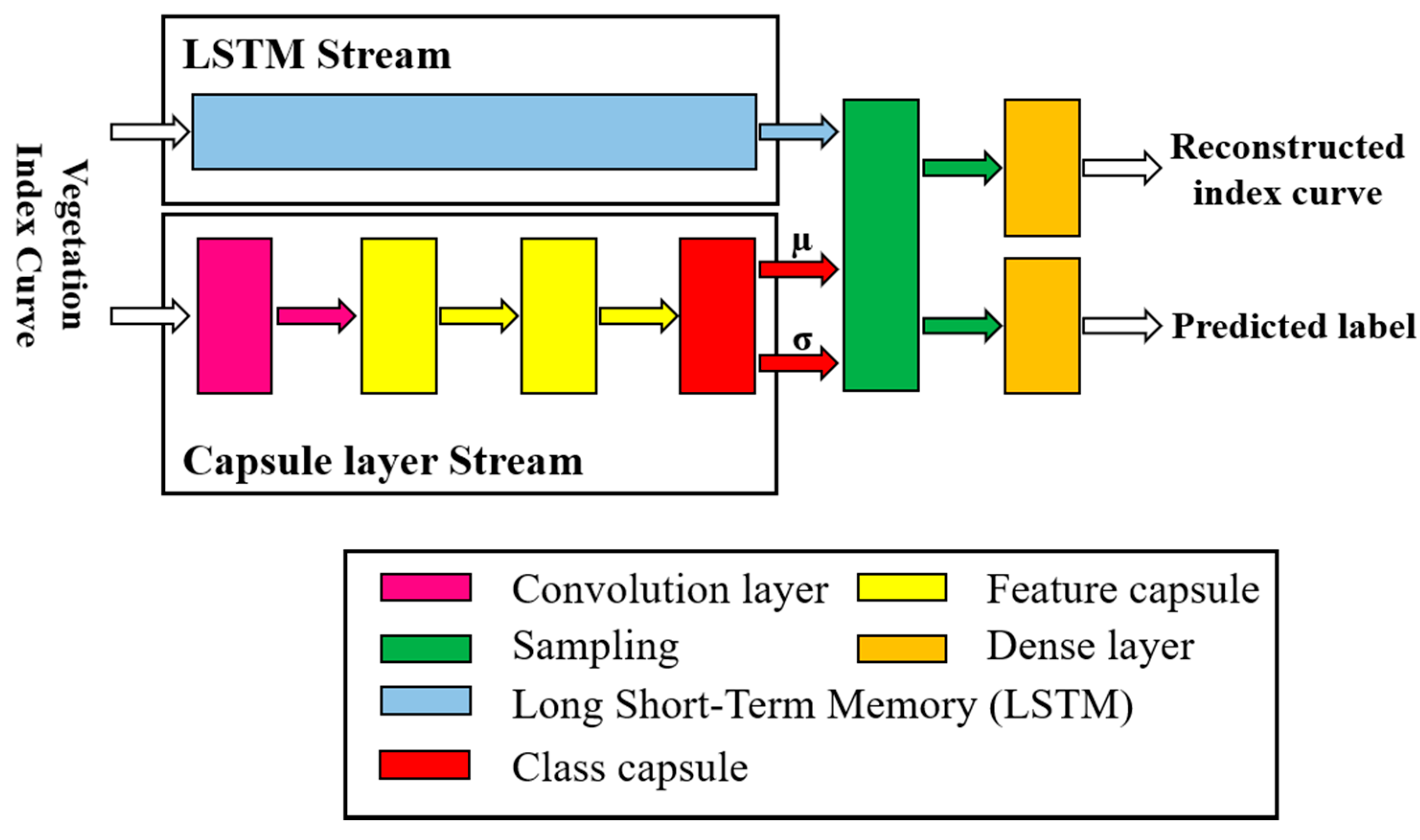
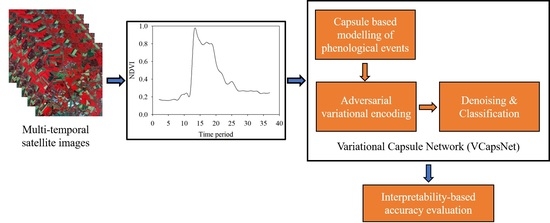
1. Introduction
Crops in a particular environment have specific phenological stages at defined time intervals in the season [
1,
2]. Modeling periodic events in the life cycle of crops is essential for distinguishing these crops. The derived information forms the basis for decision making in various irrigation scheduling activities to evaluate crop productivity [
2,
3]. Phenology-based analyses aim to track the change of phenological trajectories that vary from one crop to another in terms of the start, duration, and occurrences of crop events [
2,
3,
4,
5]. Although single-date satellite images have been widely employed for crop-type mapping, the tradeoff between spatial and spectral resolution of satellite sensors and the spectral similarities between the crops result in the misclassification of crop types [
6]. Hence, phenology-based metrics have been employed for crop type mapping tasks to overcome the issues related to conventional crop classification methods [
7]. A noticeable number of studies in agriculture focus on the extraction of phenological features using remotely sensed data [
2,
3,
8]. Most recent studies use vegetation index (VI) time-series derived from multi-temporal remote sensing data to determine specific phenological events [
9,
10].
Different conventional classifiers have been employed to classify the time-series VI data [
11,
12,
13]. Most of these studies illustrate the need to consider the specific nature of the data. Maselli et al. [
14] employed a semi-empirical approach using multi-temporal meteorological data and normalized differential vegetation index (NDVI) images to estimate actual evapotranspiration. To address the issue of the effect of mixed pixels in crop area estimation, Pan et al. [
15] proposed a crop proportion phenology index to express the quantitative relationship between the VI time-series and winter wheat crop area. Zhang et al. [
8] integrated crop phenological information from the MODerate resolution Imaging Spectroradiometer (MODIS) to estimate the maize cultivated area over a large scale. Gumma et al. [
16] used MODIS data to map the spatial distribution of the seasonal rice crop extent and area in a related work. Similar work by Kontgis et al. [
16] highlights the importance of considering flooded and cloud-covered scenes within the dense time stacks of data to achieve effective mapping of seasonal rice cropland extents. Although the lengths and timings of different phenological events provide the distinguishable signatures for different crop types, the variations in these characteristics due to different plant-, environment-, and sensor-related constraints may affect the effectiveness of phenology-based crop fingerprint estimation [
2,
3,
17]. Hence, there is a need to derive the most important events from the data to distinguish different crops while resolving the issues of modeling errors. Some phenology-based classification approaches [
9,
18,
19,
20,
21,
22] have shown that mapping efficiency can be improved by adding important features that lead to better discrimination between the crop types. In this regard, phase and amplitude information derived using the Fourier transformation (FT) of the time-series data are employed to describe the vegetation status over time [
9,
19,
20,
21]. In addition to the Fourier-based harmonic analysis, thresholding and moving average of VI curves [
23,
24,
25,
26], slope and valley point analysis of the VI curves [
27,
28,
29], and curvature-change rate analysis of logistic vegetation growth models [
10,
24,
30,
31,
32] are applied to detect phenological events in the time-series remote sensing data [
9]. However, most of these approaches require manual fine-tuning, are either supervised or semi-supervised in nature, and are sensitive to noise.
Deep learning (DL) approaches, which learn abstract representations to transform inputs into intrinsic manifolds in an unsupervised manner, have reported better results than the conventional machine learning approaches for various Earth observation (EO) data applications [
33,
34]. Variational autoencoders (VAEs) [
35,
36] learn the latent space as composed of a mixture of distributions enabling latent variable disentanglement and facilitate interpretability. Wang et al. [
37] adopted an adversarial training process to adapt VAEs to model the inherent features of the spectral information effectively. Although convolutional neural networks (CNNs) have illustrated the capability to generate task-specific features, the handling of sensor limitations and acquisition errors requires these networks to have flexibility in defining the receptive fields [
38]. The generative adversarial network (GAN)-based approaches applicable for the classification of VI curves generally use a one-to-one correlation-based similarity measure and are prone to shifts and distortions prevalent in the VI curves [
33,
39,
40]. Long Short-Term Memory (LSTM)-based approaches adopt a recurrent guided architecture to model the sequential patterns. However, most of the existing DL classifiers, including LSTMs and GANs, consider the spectral curves as vectors that ignore the characteristics of physically significant features [
41]. Although dynamic time wrapping (DTW)-based approaches consider the shape similarity and shifting in VI curves, the parameter tuning requirements affect their effectiveness and generalizability [
42]. A recent advancement in CNN, called capsule networks, adopts capsules (a group of neurons) to address the issues of translation invariance prevalent in conventional CNNs [
41,
43,
44]. Shi et al. [
45] employed capsule blocks to model the spectral–spatial features to achieve high accuracy and interpretability in HSI-based classification tasks. Similar research has also been reported in [
41,
46,
47,
48,
49], in which capsule networks were explored for EO data classification. However, few studies have reported the use of capsules for time-series classification [
50,
51].
Phenology-based index curve classification requires that VI curves are denoised to produce a smooth time-series [
52]. Different algorithms, such as iterative weighted moving filters [
53,
54,
55,
56,
57], nonlinear curve fitting [
57,
58,
59,
60,
61,
62], filtering in the Fourier domain [
63,
64], and spline-based smoothing [
52,
65,
66], are being widely used for VI curve smoothing. However, most of these approaches either do not consider the phenological events, or require manual fine-tuning to avoid extraneous oscillations and to consider the specific nature of the phenological index curves. Although DL-based denoising approaches learn nonlinear feature spaces to avoid linear events, sparsity, and low-rank assumptions of the traditional interpolation methods [
67,
68,
69], they generally do not consider the irregular sampling of the data and phenological events [
70]. The DL-based approaches that have attained success in processing irregularly distributed point data [
38,
70,
71] are not directly applicable to denoising VI curves. Moreover, the existing phenology-based classification approaches consider denoising, data imputation, outlier elimination, and classification as independent problems.
In this research, we hypothesize that capsule-based feature learning can adequately model the characteristic features of the VI curves and the crop-specific phenological events, such as growth transitions, planting, heading, and harvesting. The DTW-based neural units and interpolated convolution are hypothesized to dynamically learn kernels for estimating feature-specific shape similarity correspondences of the VI curves. It is also proposed that the joint optimization of denoising, data imputation, outlier elimination, and classification stages yields better results than the conventional approach of independently optimizing them. In addition, variational encoding conditioned with time-series aggregation is hypothesized to facilitate the consideration of the intra-crop phenological event variations and resolution of outliers. The current study also verified that the high measurement accuracy results from an appropriate latent representation and not from the exploitation of artifacts in the data [
72,
73,
74]. The main contributions of the study are: (1) an interpretable VI curve classifier is proposed to consider the specific characteristic phenological events and the available prior knowledge; and (2) denoising, imputation, and classification stages are jointly optimized, considering the modeling errors and outlier effects, with a minimal number of training samples.
3. Results
To verify the effectiveness of VCapsNet, extensive experiments were conducted for phenological curve-based crop classification. The pixel-level NDVI curves derived from multi-date images were used to train VCapsNet to distinguish different types of crops. The ancillary data was used to assign labels to ground-truth phenological curves. Data augmentation, similar to that adopted in [
81,
82], was used to increase the number of training samples. In addition, random Gaussian noise was added in irregular intervals to evaluate the effect of denoising. An example of augmented patterns for the barley crop is presented in
Figure 4. VCapsNet was extensively analyzed using the data of three farms over three consecutive crop years. A total of 4600 samples were used for training and testing the model, among which 800 were augmented patterns. Hyper-parameter optimization, proposed in [
88], was employed to optimize the parameters of the different models analyzed in this study. It should be noted that an early stopping framework using k-fold validation forms the basis of the parameter selection. For all of the experiments adopted, k-fold validation was used with k set to 10. The confusion-matrix-based Kappa statistic and overall accuracy were used for evaluating the classification results. High values of Kappa statistic and overall accuracy indicate high accuracy. A Z-score-based test statistic (discussed in [
89,
90]) was employed to analyze the significance of the results presented in the current study. In addition to confusion-matrix-based measures, proposed interpretability techniques (
Section 2.2.4) were used to evaluate the physical significance and interpretability of the models. The peak signal-to-noise ratio (PSNR) was used to estimate the denoising accuracy of VCapsNet and other benchmark denoising approaches. The ablation analysis of VCapsNet is discussed in
Section 3.1. A comparative analysis of VCapsNet with the benchmark approaches is presented in
Section 3.2.
3.1. Ablation Analysis of VCapsNet
This Section evaluates the effect of the proposed architectural variations and loss functions on the results. The results are summarized in
Table 1. It is observed that the proposed strategies reduce the training sample requirement and significantly improve the results (in terms of Kappa and overall accuracy). As is evident from
Table 1, the implementation of VCapsNet without a capsule stream results in lower values of Kappa and overall accuracy. These results illustrate that the capsules model the phenological events and their characteristic features effectively to distinguish different classes. Moreover, the use of capsules significantly reduces the network depth. It may be noted from
Table 1 that the implementation of VCapsNet without a variational encoding strategy results in lower Kappa and overall accuracy values. The improvement in accuracy due to the variational encoding strategy can be attributed to the consideration of minor phenology variations in the same crop type and to the resolution of outliers. The use of DTW-based routing technique also improves the results by facilitating the consideration of shapes of the phenological events. In addition, as is observed from the Kappa and overall accuracy values in
Table 1, the use of the cell state learned using LSTM for conditioning the latent space sampling acts as a regularizer. This resolves the issues of vanishing gradient and convergence. The use of reconstruction loss as a means to regulate the classification loss facilitates denoising and data imputation of the index curves. Training of the proposed network for denoising and imputation ensures the classification is resilient to noise and other irregularities. Similarly, the use of piece-wise loss, interpolation-based convolution, and DTW-based neural units improves the classification accuracy. The proposed architectures and losses improve the modeling of phenological events that are evident from the concepts learned for different types of crops. The relevance analysis of input features indicates that VCapsNet focuses on features and phenological events that are physically significant to each crop type. Additionally, fine-tuning of the entanglement penalty facilitates the disentanglement of the latent codes concerning different crop classes.
A comparison of both of the proposed architectures indicates that, at a lower number of training samples, architecture-2 is preferred, whereas architecture-1 provides better results when enough training samples are available. In addition, architecture-2 yields acceptable results at much shallower depths compared to architecture-1.
The sensitivity analysis of the proposed VCapsNet models in terms of network parameters is presented in
Figure 5 and
Figure 6. For a given set of training samples, an increase in the network depth improves the Kappa value that, however, deteriorates as the number of layers increases beyond a limit. Empirically, for input VI curves having a length of 24–36, a 5–9 layered network yields the best results. The increase in the number and sizes of filters improves the accuracy, which slowly saturates and deteriorates following further blind increase. The increase in size and number of filters exponentially increases the computational complexity of the network. As the length of phenological features can vary, the use of multi-sized kernels significantly improves the results without significantly affecting the execution time. The reduction in the sensitivity of VCapsNet to network parameters can be attributed to the effective modeling of characteristic features of index curves.
3.2. Comparison of VCapsNet with the Commonly-Used DL Based Approaches
The commonly-used classifiers applicable to VI curve classification were compared with the proposed VCapsNet-based approach. The results are summarized in
Table 2. The significance of the results of VCapsNet (at a confidence level of 95%) in comparison with the other approaches is analyzed in
Table 3. In all of the experiments adopted in this study, the entire set of 4600 samples was split into disjoint training and testing subsets. For instance, when the percentage of training samples is 10%, 90% of the samples were used for testing the model. Based on the discussions in [
2,
3,
7,
12,
39,
83,
91], some main existing classification approaches were selected as the benchmark methods for comparison. In the experiments discussed in this study, a few of the benchmark approaches were modified to process the one-dimensional phenological curves. An analysis of the variation in accuracy of different approaches according to the variation in the percentage of training samples is presented in
Figure 7. Furthermore,
Figure 7 presents the variation in accuracies for different folds of k-fold validation in terms of the standard deviation. A total of 4600 training samples were used, and 10-fold validation was employed for each of the different sub-experiments (10%, 20%, 30%, etc.). As is evident from the results, VCapsNet better models phenological curves compared to other prominent approaches. The proper modeling of phenological events and features significantly improves the generalization capability of the network, resulting in improved classification accuracies even with a small number of training samples (
Figure 7). The learning of physically significant features and phenological events also resolves the issues of intra-crop variability of the phenological curves. In addition, the DTW-based convolutional units and interpolation-based convolutions, and the proposed losses and regularizations, facilitate the effective transformation of vectorized phenological curves to a latent space that is more discriminative than the original space.
In addition to the classification-based accuracy assessment, the models were also evaluated based on the prototypes learned for each crop, as discussed in
Section 2.2.4. For this purpose, the phenology curves that follow the correct timeline for each crop were selected based on the ancillary data and generalized using a variational autoencoder. The benchmark phenological curve for each crop, generated by sampling the mean of such a learned latent space, was compared with the learned concepts of the corresponding crops for each model. In this regard, cosine-based, DTW-based, and Fourier-based approaches were used as the similarity measures. The dates of crop-specific phenological events, such as growth transition, planting, heading, and harvesting, were adopted as characteristic features for comparison. A summary of these comparisons is presented in
Table 4. The harmonics of the Fourier transform of the VI curves properly capture the phenological events, whereas DTW approaches measure the shape-similarity irrespective of the minor shifts. The high values of similarity measures indicate that VCapsNet accurately learns the phenological events and physically significant features. A visual illustration of the predicted and actual NDVIs corresponding to the crop-specific phenological events for randomly selected wheat and barley fields is presented in
Figure 8. As is evident, VCapsNet provides accurate results.
To further explain the network and analyze the contribution of input features, the LRP approach (
Section 2.2.4) was adopted. The propagated relevance of VCapsNet for distinguishing different crop types indicates that the model places importance on the time frames related to phenological events.
To analyze the temporal effectiveness of VCapsNet, a leave-one-out validation strategy was adopted year-wise. The training samples of two crop years were used to classify the crop phenology from another crop year. The crop years of 2017–2018, 2018–2019, and 2019–2020 were considered for the analysis. A total of 3500 samples were used for training and 1500 samples for testing. The result of the experiment is presented in
Table 5 and the significance of the analysis in
Table 6. The performance and generalizability of the proposed approach can be attributed to the effective modeling of the characteristic phenological features of the crops.
3.3. Comparison of VCapsNet with the Commonly-Used Denoising Approaches
In addition to classification, VCapsNet reconstructs the phenological index curves, thereby facilitating denoising and data imputation. In this section, the commonly-used denoising approaches applicable to VI curve smoothing are compared with the proposed VCapsNet. In all of the experiments adopted in the study, the entire sample set was split into disjoint training and testing subsets. As the reconstruction is conditioned based on the crop type information, the proposed approach of joint classification and denoising provides better results than the existing denoising approaches. The selected benchmark approaches are improved versions of those that reported the state-of-the-art results in [
52,
54,
63,
64,
65,
96,
97]. The result of the comparative analysis is summarized in
Table 7 and
Figure 9.
Table 8 confirms the significance of the comparison at a confidence level of 95%. It is to be noted that
Figure 9 presents the bar graphs indicating the standard deviation during the k-fold validation. The local filtering methods (Savitzky–Golay filtering and locally weighted scatterplot smoothing) resulted in better performance with optimized parameter settings among the conventional smoothing approaches. However, fitting methods (asymmetric Gaussian function fitting and double logistic function fitting) are less sensitive to the parameters. As is evident from the results, VCapsNet yields higher PSNR (better reconstruction accuracy) than the benchmark approaches considered in this study. The results of VCapsNet can be attributed to its ability to model phenological events. In addition, the proposed variational encoding strategy, LSTM-based conditioning, and loss functions, significantly improve the generalizability. It should be noted that VCapsNet learns the smoothing parameters dynamically, whereas conventional smoothing approaches require manual fine-tuning. Additionally, the phenology event-based latent space resolves the issues of domain bias and inter-field variability of VI curves of the same crop.
4. Discussion
Experiments on VCapsNet (discussed in
Section 4) illustrate that the proposed approaches yield better results than the main existing approaches. A detailed analysis of the results is presented in the following subsections.
4.1. Modeling Phenological Events
Learning and appropriate modeling of different phenological events and their characteristics, such as relative locations, length, shape, texture, and context, are essential for crop classification. The architecture of DL models is found to influence the capability in modeling the phenological curves. The use of capsules enables better modeling of the relative locations and features of the phenological events compared to the convolutional networks. The proposed DTW-based routing mechanism facilitates the consideration of the contexts of phenological events for the effective modeling of VI curves. Although VCapsNet yields better results than the existing approaches in terms of the Kappa statistic and overall accuracy, these accuracy measures do not ensure that the network models the phenological curves correctly. In this regard, interpretability and explanation-based analyses must be able to indicate if the network has correctly learned the concepts for each class and confirm if it is appropriately placing importance on the input features. Among the two proposed architectures of VCapsNet, architecture-2 yields better results when the samples are limited, whereas architecture-1 performs better when sufficient training samples are available. The use of variational encoding for fitting the latent distribution to a normally distributed space resolves the issues of crop phenological variability and the problem of outliers. However, additional sampling layers in variational autoencoders are found to adversely affect the modeling of VI curves, especially when training samples are limited. It is further observed that the use of a LSTM cell state to condition the latent space regulates the sampling process, resulting in a feature space that captures the characteristic phenology of each crop. It may be noted that convolutional networks rather than capsules do not yield acceptable results due to the need to consider the specific characteristics of the VI curves. The LSTM classifiers only consider the sequential nature of the curves and ignore the characteristic features of the phenological events and their relative locations. Moreover, as adopted in conventional capsules, the simple routing ignores the shape similarity of the phenological events.
This study illustrates that the joint optimization of denoising, data imputation, and classification yields better results than individually optimizing them. The proposed VCapsNet provides not only good classification results, but is also effective in denoising and data imputation. In VCapsNet, the reconstruction serves as a regularizer for classification, and class information is a regularizer for denoising and data imputation. The proposed constraints and losses use the input priors to improve the projected space, thereby resulting in meaningful and interpretable representations that capture the phenological events and features. Different experiments in the current study illustrate that using multi-sized kernels facilitates the modeling of phenological features having variable lengths.
VCapsNet yields good results even at shallower depths compared to the other conventional DL approaches. The proposed regularizations and data augmentations, in addition to capsule-based feature modeling, improve the generalizability of the network. VCapsNet also considers the inter-field variability of the crops through proper modeling of the phenological events and embedding the label information in the latent space. Moreover, the neural units, convolutions, and routing mechanisms of VCapsNet are modeled to consider the shape of the receptive fields.
The experiments indicate that the evaluation matrices based on classification or reconstruction accuracy are insufficient for DL models. The interpretation of the concepts learned and the relevance assigned to the input features for each crop provides insight into the meaningfulness and physical significance of the features learned by the network. VCapsNet fares well in terms of interpretability compared to the other DL models considered in this study. In addition to using the interpretability evaluation for mere quantitative comparison, the same approach can be used to refine the training data and select the most relevant features.
Due to its generalizability, VCapsNet is applicable to crop mapping at different scales. Sub-national and national level mapping requires the generalization of the phenological index curves. The pixel-level phenological VI curves are generalized to yield the crop fingerprints at the field level. The field level VI curves are then classified using VCapsNet to accomplish mapping at different scales. The VCapsNet is generic and can be extended to different applications.
4.2. Interpretability Based Comparison of the Benchmark DL Models
The analysis of phenological curves requires appropriate modeling of phenological events in terms of their characteristics such as depth, width, shape, and position. The interpretability-based evaluation strategies (
Section 2.2.4) were found to be useful in the understanding of the concepts learned by the models. Comparing the concepts learned for each of the crops with the ancillary data (related to the sowing, growth, and harvesting time) provides an understanding of the learning capability of the DL models. In addition to evaluating the learning capability of the network, the learned concepts also provide an indication of the suitability of the training data and the need to refine this data.
Although analysis of the prototypes learned by DL models provides insights into the learning capability of the network, analysis of the relevance of the different input features is required to interpret the manifold accordingly. As discussed in
Section 2.2.4, the LRP approach assigns relevance scores to the input features based on their contributions. Analysis of these relevance scores with respect to the ancillary data regarding the timing of crop cultivation and growth provides an insight into the physical significance of the features learned by the model.
Barley and wheat crops are often indistinguishable from each other and cause issues for crop mapping algorithms. As can be observed from the results, VCapsNet yields good results in classifying these crops. The performance of VCapsNet can be attributed to its ability to appropriately model the crop-specific characteristics of the index curves. The interpretability-based evaluation indicates that the crop-specific phenological events, such as growth transitions, planting, heading, and harvesting, are effectively identified using VCapsNet.