How Do Methods Assimilating Sentinel-2-Derived LAI Combined with Two Different Sources of Soil Input Data Affect the Crop Model-Based Estimation of Wheat Biomass at Sub-Field Level?
Abstract
1. Introduction
- How does the soil texture information drawn from SoilGrids compare to detailed soil texture derived from laboratory-analyzed soil samples taken on 14 conventionally farmed fields across France, Germany, and the Netherlands?
- With regard to the average total biomass of winter wheat at harvest per field: How does the crop model prediction perform when using either analyzed soil texture (in the following: AS) or texture extracted from SoilGrids (in the following: SG) as model input in a standard run (i.e., no ensemble creation, no LAI assimilation), the ensemble mean (i.e., no LAI assimilation), and combined with the assimilation of S2 LAI information based on either of two different approaches (ensemble Kalman filter and weighted mean)?
- Which approach, in combination with AS or SG, reproduces the measured spatial variability of aboveground biomass at harvest within a field better?
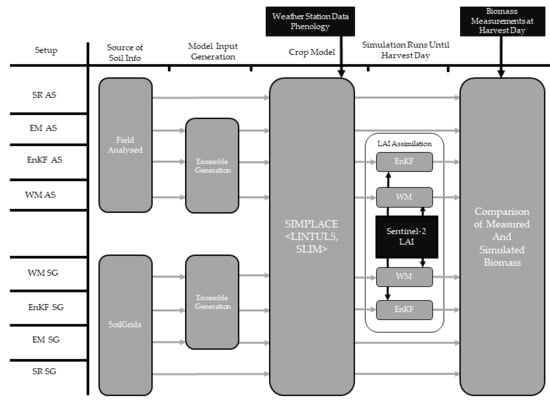
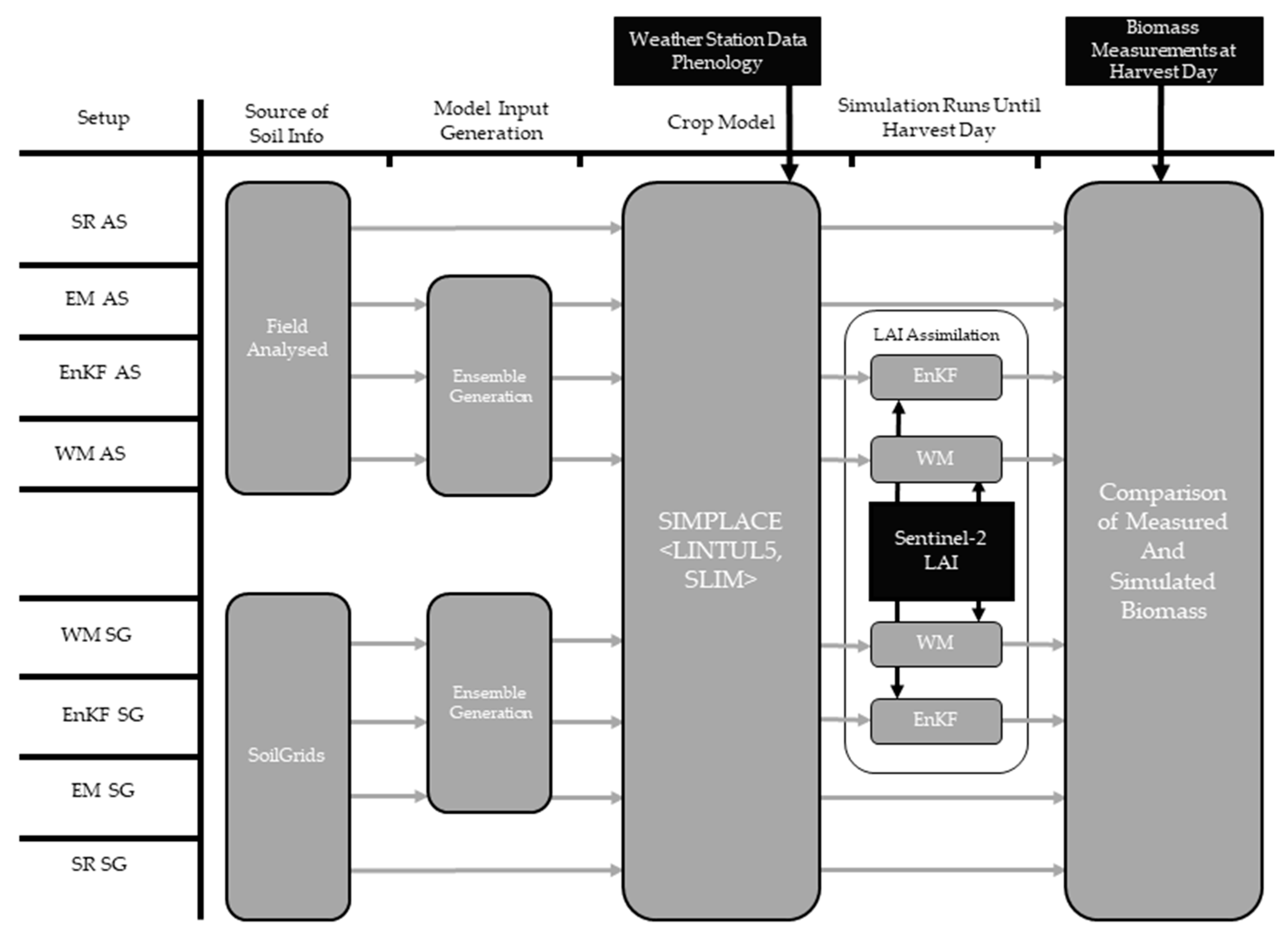
2. Materials and Methods
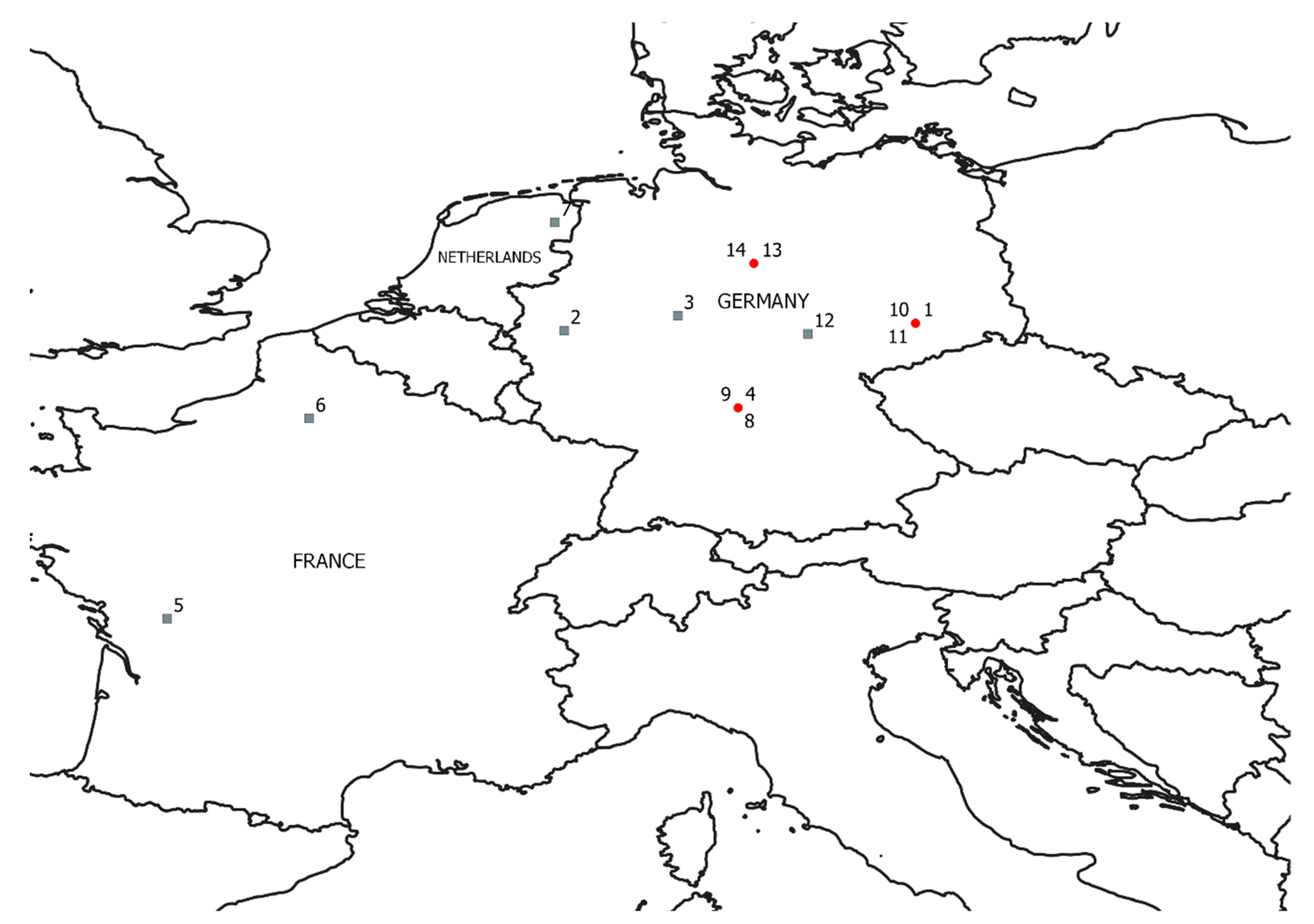
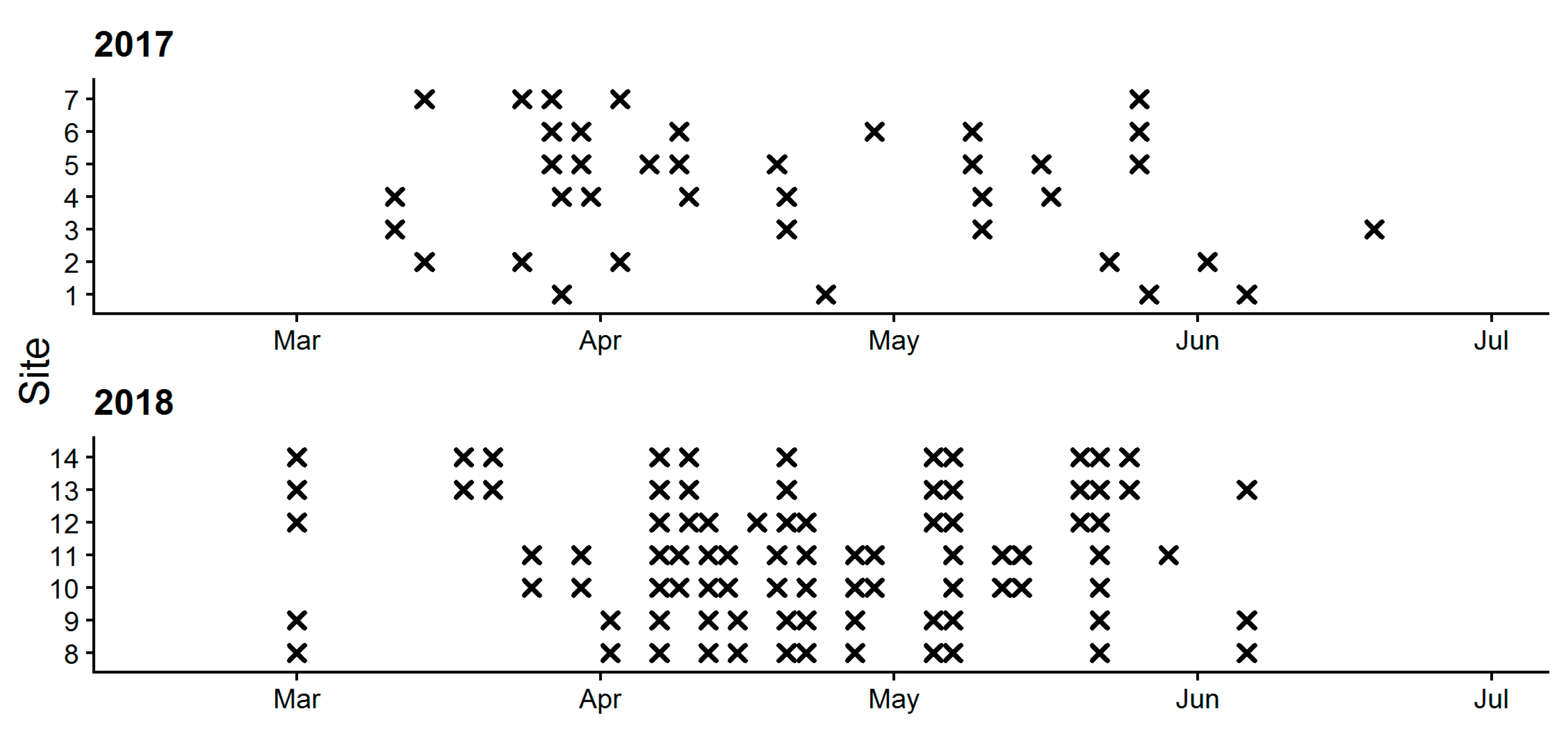
2.1. Experimental Fields and Data Collection
2.2. SoilGrids Soil Information
2.3. Leaf Area Index Estimations
2.4. Crop Model
2.5. Data Assimilation
2.6. Model Run and Evaluation
3. Results
3.1. Comparison of Measured Soil Texture Data and Texture Data from SoilGrids
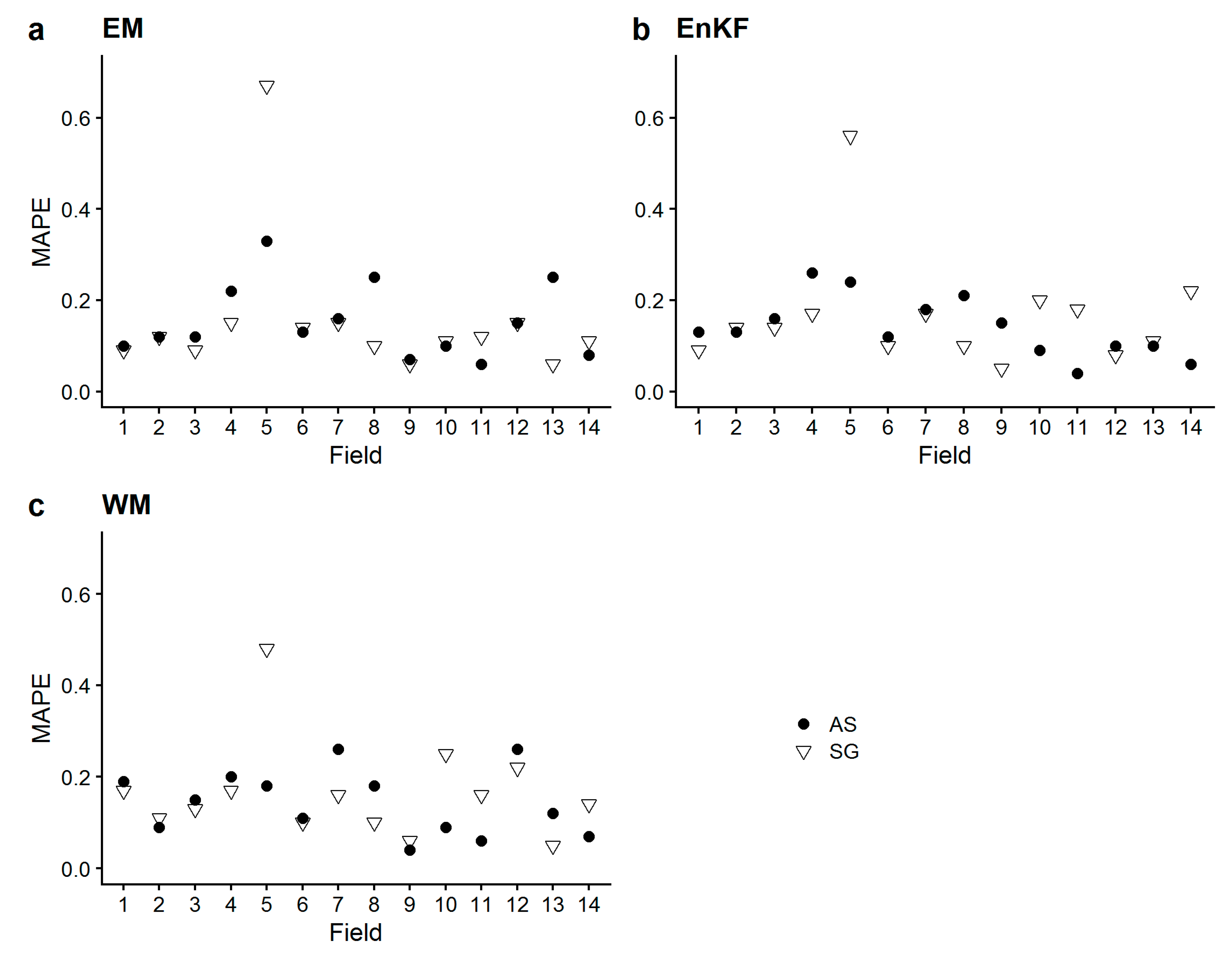
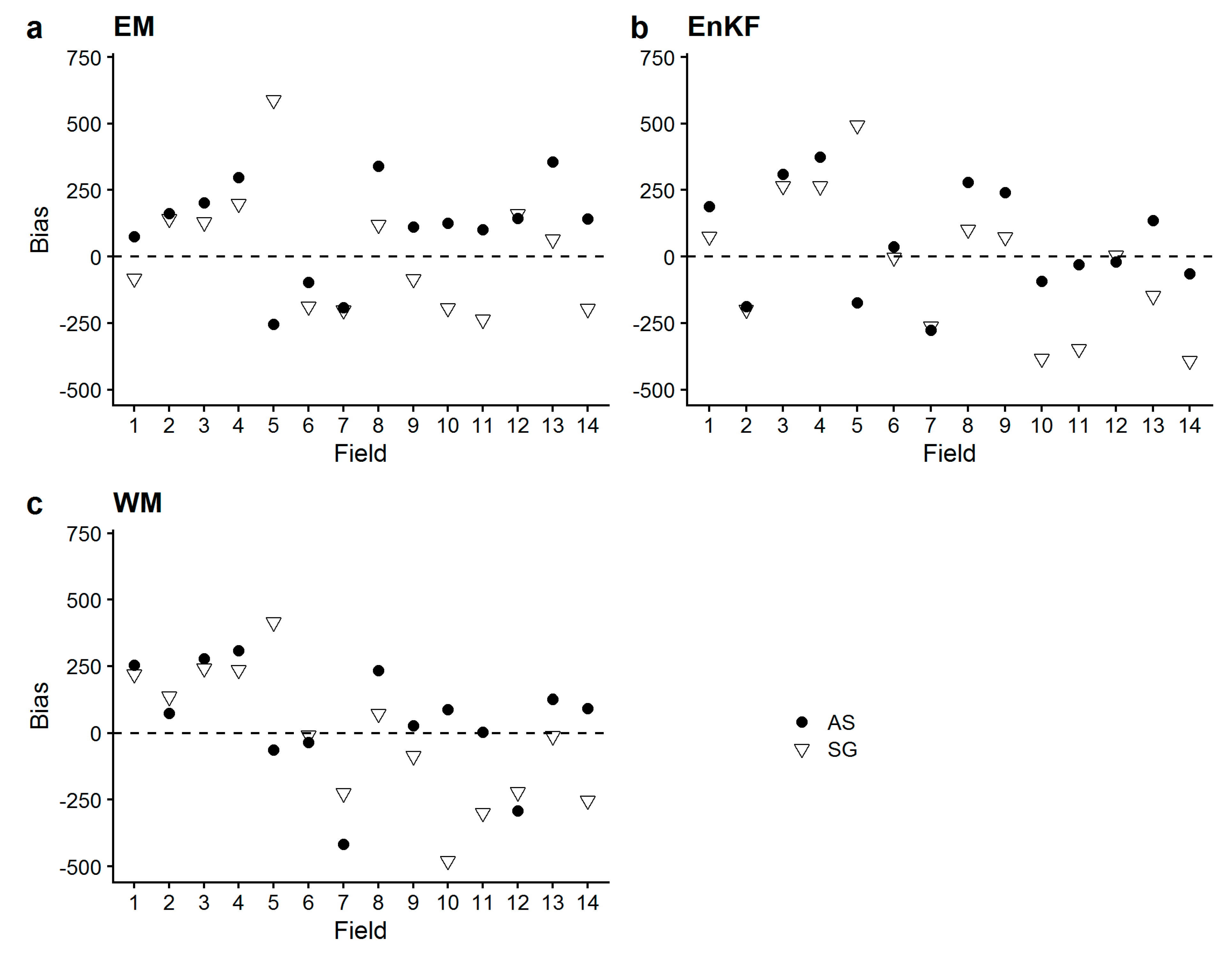
3.2. Comparison of Simulated vs. Measured Aboveground Biomass
3.3. Assessment of Reproduction of Sub-Field Variability
4. Discussion
4.1. Results of the SoilGrids vs. Measured Soil Texture Comparison
4.2. Results of the Crop Modeling Excersises
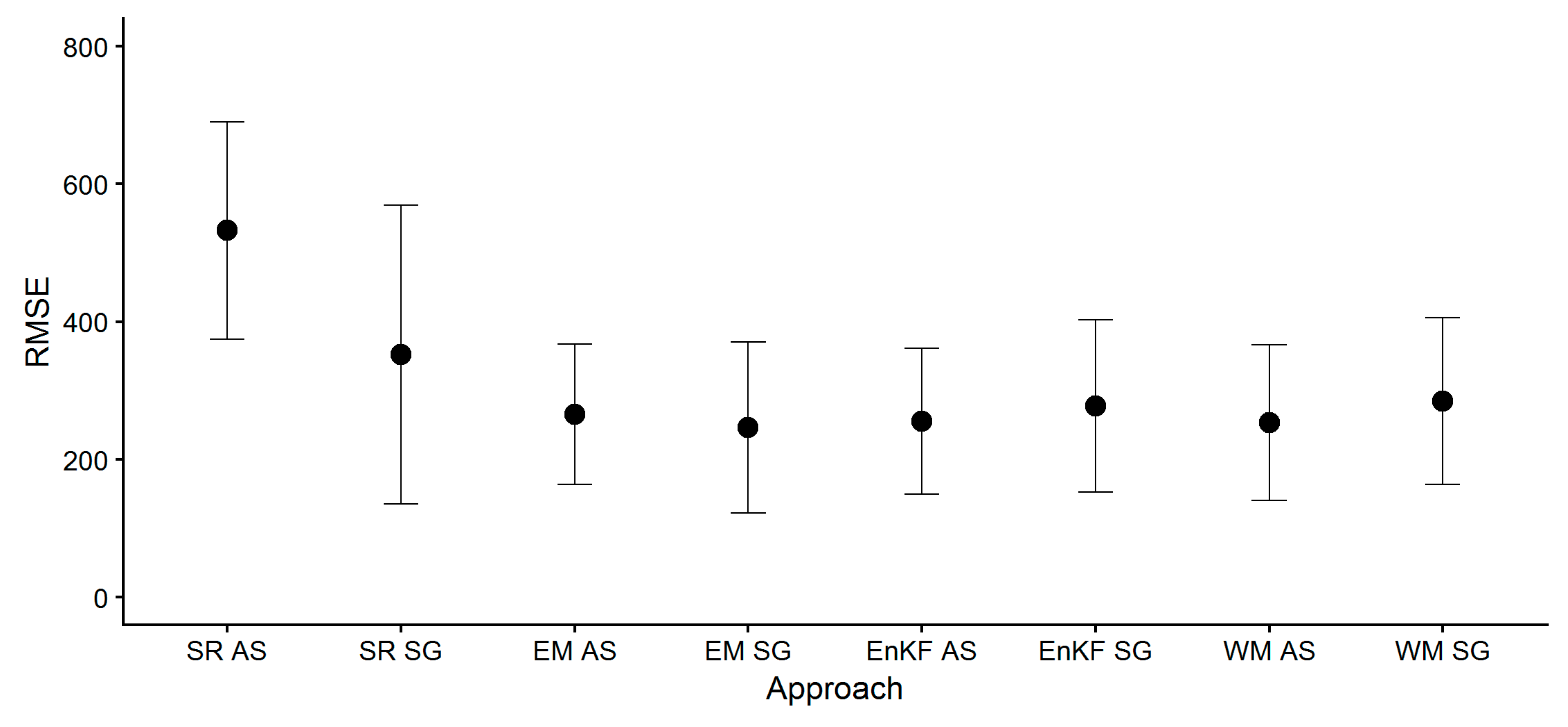
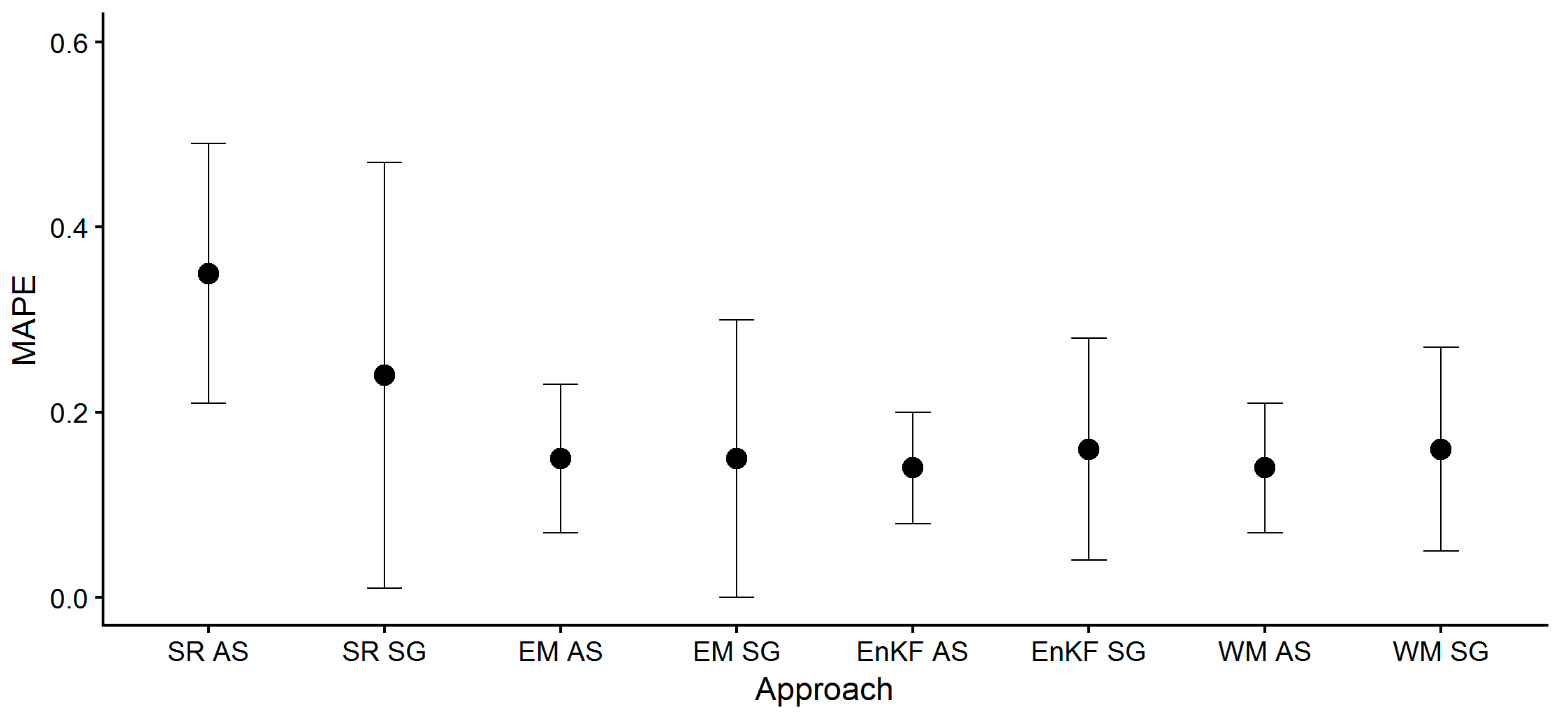
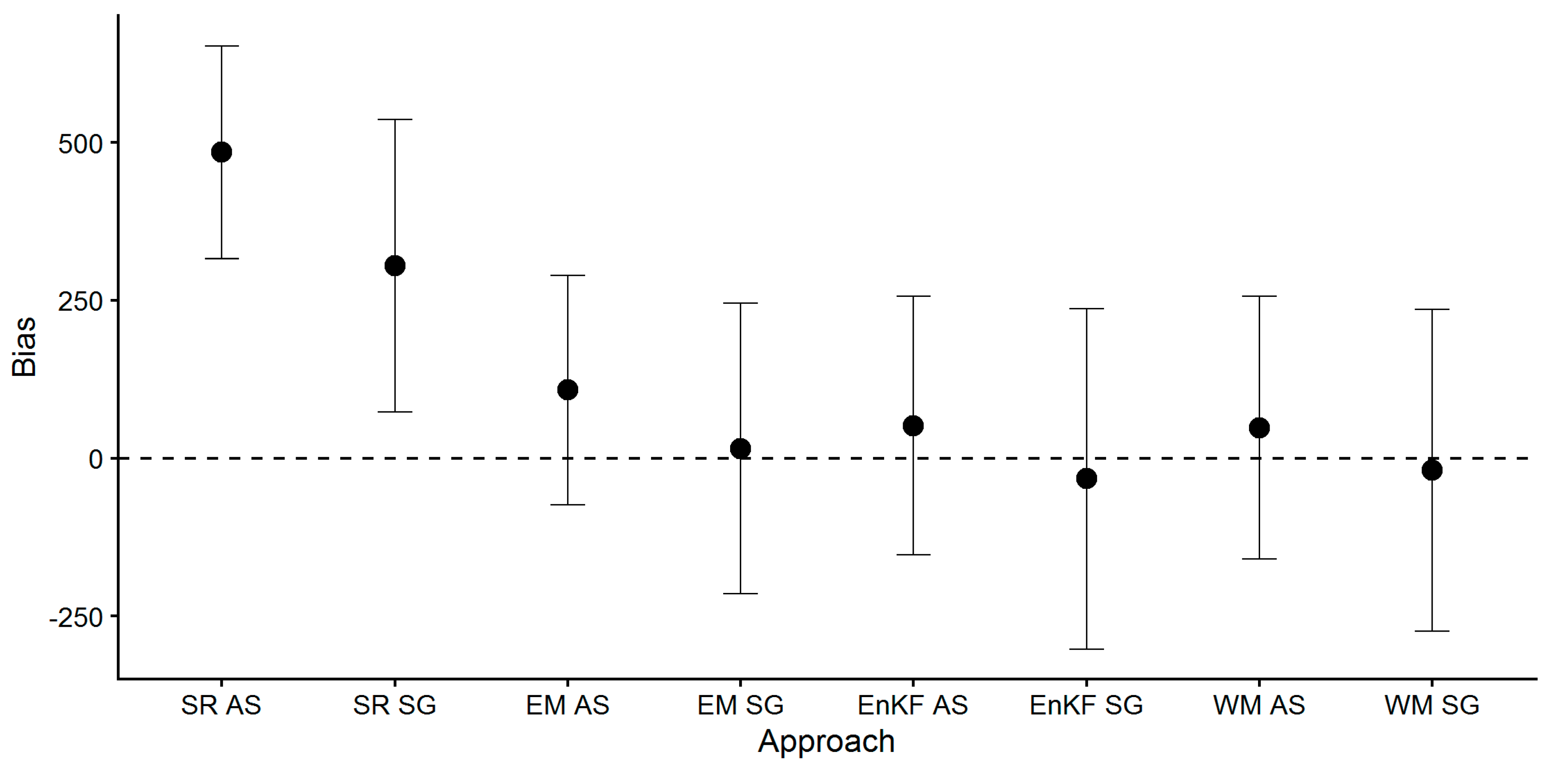
- Mean values of the calculated metrics (RMSE, MAPE, Bias) across all fields did not substantially differ between ensemble-based SG and AS approaches (i.e., EM, EnKF, WM), but standard deviations were greater for SG-based approaches. The model’s standard runs (SR, i.e., no assimilation, no ensemble generation), performed worse, and biomass yield was largely overestimated as indicated by the positive bias.These findings show that the average biomass prediction performance based on SG soil information was not inferior to AS soil information prediction performance, but came with a greater prediction uncertainty.
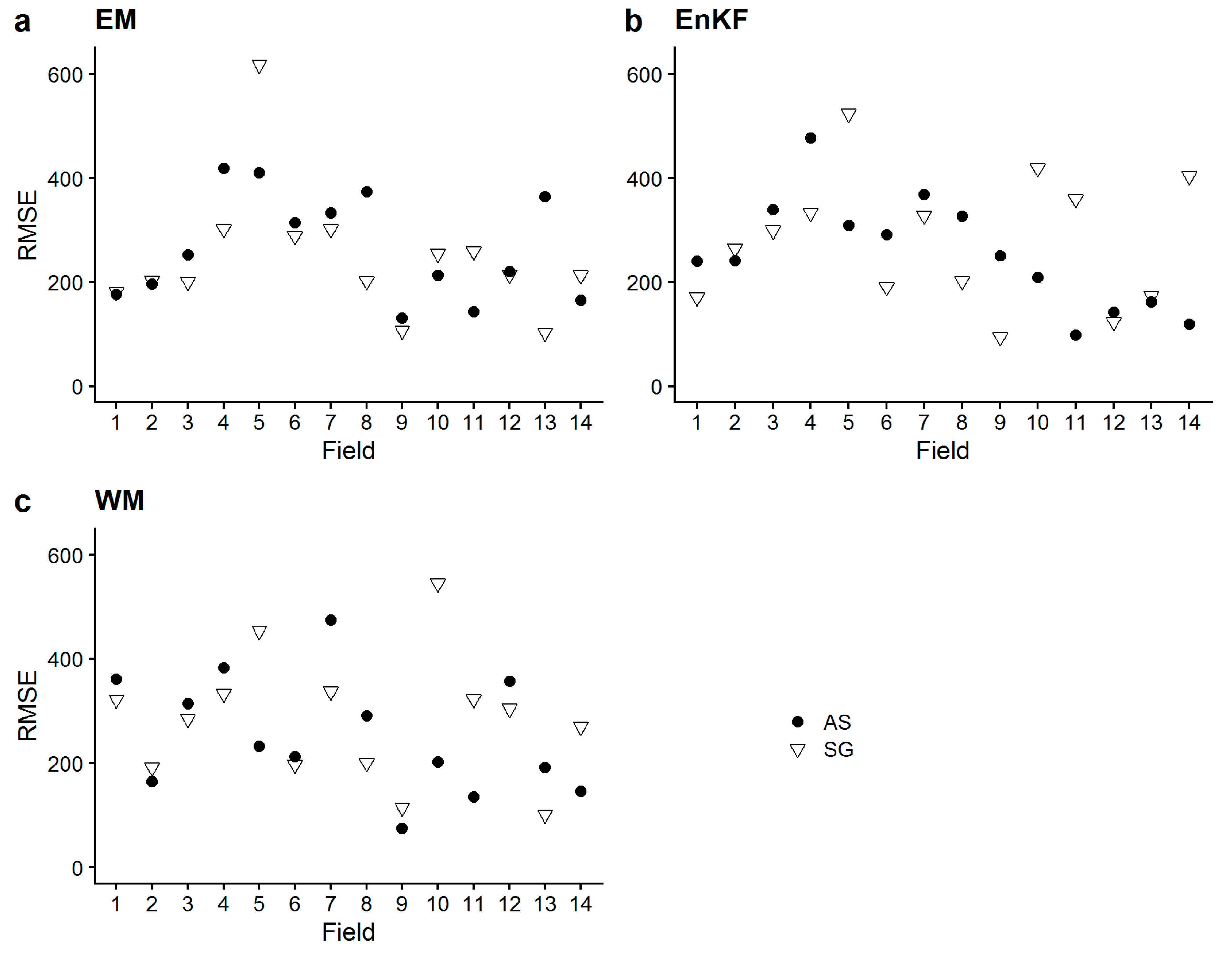
- The simulated sub-field heterogeneity of biomass at harvest did not accurately match the measured one in any of the sites, as no approach showed a congruent performance for any of the metrics. AS-based approaches did not outperform SG-based approaches concerning the reproduction of heterogeneity. The performance of the assimilation approaches were largely site-specific, as we saw the approaches performing differently in the different sites, with no clear approach standing out. To our surprise, EM did not perform substantially worse.
4.3. Limitations of Input Data
4.4. Sentinel-2 LAI Estimations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Country | Growing Season | Field | Location | Cultivar Grown | Planting Date | BMY | GY | HI |
|---|---|---|---|---|---|---|---|---|
| DE | 2016/2017 | 1 | Central Saxony | Benchmark | 2016-10-19 | 1645 | 912 | 0.55 |
| 2016-11-30 | ||||||||
| DE | 2016/2017 | 2 | Southern Northrine-Westphalia | Jonny | 2016-10-25 | 1455 | 597 | 0.41 |
| DE | 2016/2017 | 3 | Northern Hesse | Julius | 2016-10-04 | 1959 | 1023 | 0.52 |
| 2016-10-28 | ||||||||
| DE | 2016/2017 | 4 | Northern Bavaria | RGT Reform | 2016-10-05 | 1737 | 905 | 0.54 |
| 2016-10-19 | ||||||||
| FR | 2016/2017 | 5 | Northwestern Charente | Bologna | 2016-10-29 | 969 | 434 | 0.44 |
| 2016-11-15 | ||||||||
| FR | 2016/2017 | 6 | Northern Oise | Lyrik | 2016-10-03 | 1579 | 868 | 0.54 |
| 2016-10-18 | ||||||||
| NL | 2016/2017 | 7 | Eastern Drenthe | RGT Reform | 2016-10-19 | 1573 | 876 | 0.55 |
| 2016-11-03 | ||||||||
| 2016-11-14 | ||||||||
| DE | 2017/2018 | 8 | Northern Bavaria | RGT Reform | 2017-11-03 | 1451 | 773 | 0.53 |
| JB Asano | ||||||||
| DE | 2017/2018 | 9 | RGT Reform | 2017-11-16 | 1640 | 809 | 0.49 | |
| JB Asano | ||||||||
| DE | 2017/2018 | 10 | Central Saxony | RGT Reform | 2017-09-21 | 1889 | 1008 | 0.53 |
| JB Asano | ||||||||
| DE | 2017/2018 | 11 | RGT Reform | 2017-10-16 | 1882 | 954 | 0.50 | |
| JB Asano | ||||||||
| DE | 2017/2018 | 12 | Central Thuringia | JB Asano | 2017-09-19, 2017-10-19 | 1242 | 527 | 0.42 |
| RGT Reform | ||||||||
| DE | 2017/2018 | 13 | Central Lower Saxony | RGT Reform | 2017-11-03 | 1451 | 808 | 0.55 |
| JB Asano | ||||||||
| DE | 2017/2018 | 14 | RGT Reform | 2017-10-17 | 1795 | 889 | 0.49 | |
| JB Asano |
References
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Gilardelli, C.; Stella, T.; Confalonieri, R.; Ranghetti, L.; Campos-Taberner, M.; García-Haro, F.J.; Boschetti, M. Downscaling rice yield simulation at sub-field scale using remotely sensed LAI data. Eur. J. Agron. 2019, 103, 108–116. [Google Scholar] [CrossRef]
- Wallach, D.; Makowski, D.; Jones, J.W.; Brun, F. Chapter 8—Data Assimilation for Dynamic Models. In Working with Dynamic Crop Models, 2nd ed.; Wallach, D., Makowski, D., Jones, J.W., Brun, F., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 311–343. ISBN 978-0-12-397008-4. [Google Scholar]
- Chen, Y.; Zhang, Z.; Tao, F. Improving regional winter wheat yield estimation through assimilation of phenology and leaf area index from remote sensing data. Eur. J. Agron. 2018, 101, 163–173. [Google Scholar] [CrossRef]
- De Wit, A.J.W.; van Diepen, C.A. Crop model data assimilation with the Ensemble Kalman filter for improving regional crop yield forecasts. Agric. For. Meteorol. 2007, 146, 38–56. [Google Scholar] [CrossRef]
- Hu, S.; Shi, L.; Huang, K.; Zha, Y.; Hu, X.; Ye, H.; Yang, Q. Improvement of sugarcane crop simulation by SWAP-WOFOST model via data assimilation. Field Crop Res. 2019, 232, 49–61. [Google Scholar] [CrossRef]
- Pan, H.; Chen, Z.; de Wit, A.; Ren, J. Joint Assimilation of Leaf Area Index and Soil Moisture from Sentinel-1 and Sentinel-2 Data into the WOFOST Model for Winter Wheat Yield Estimation. Sensors 2019, 19, 3161. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, P.; Sun, H.; Zhang, S.; Li, L. Assimilation of Leaf Area Index and Surface Soil Moisture With the CERES-Wheat Model for Winter Wheat Yield Estimation Using a Particle Filter Algorithm. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 1303–1316. [Google Scholar] [CrossRef]
- Silvestro, P.C.; Pignatti, S.; Pascucci, S.; Yang, H.; Li, Z.; Yang, G.; Huang, W.; Casa, R. Estimating Wheat Yield in China at the Field and District Scale from the Assimilation of Satellite Data into the Aquacrop and Simple Algorithm for Yield (SAFY) Models. Remote Sens. 2017, 9, 509. [Google Scholar] [CrossRef]
- Huang, J.; Tian, L.; Liang, S.; Ma, H.; Becker-Reshef, I.; Huang, Y.; Su, W.; Zhang, X.; Zhu, D.; Wu, W. Improving winter wheat yield estimation by assimilation of the leaf area index from Landsat TM and MODIS data into the WOFOST model. Agric. For. Meteorol. 2015, 204, 106–121. [Google Scholar] [CrossRef]
- Ines, A.V.M.; Das, N.N.; Hansen, J.W.; Njoku, E.G. Assimilation of remotely sensed soil moisture and vegetation with a crop simulation model for maize yield prediction. Remote Sens. Environ. 2013, 138, 149–164. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Liu, G.; Jiang, Z.; Huang, C. Improving Winter Wheat Yield Estimation from the CERES-Wheat Model to Assimilate Leaf Area Index with Different Assimilation Methods and Spatio-Temporal Scales. Remote Sens. 2017, 9, 190. [Google Scholar] [CrossRef]
- Novelli, F.; Spiegel, H.; Sandén, T.; Vuolo, F. Assimilation of Sentinel-2 Leaf Area Index Data into a Physically-Based Crop Growth Model for Yield Estimation. Agronomy 2019, 9, 255. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, P.; Bai, X.; Khan, J.; Zhang, S.; Li, L.; Wang, L. Assimilation of the leaf area index and vegetation temperature condition index for winter wheat yield estimation using Landsat imagery and the CERES-Wheat model. Agric. For. Meteorol. 2017, 246, 194–206. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, S.; Shen, S. Assimilating remote sensing information with crop model using Ensemble Kalman Filter for improving LAI monitoring and yield estimation. Ecol. Model. 2013, 270, 30–42. [Google Scholar] [CrossRef]
- Jonckheere, I.; Fleck, S.; Nackaerts, K.; Muys, B.; Coppin, P.; Weiss, M.; Baret, F. Review of methods for in situ leaf area index determination: Part, I. Theories, sensors and hemispherical photography. Agric. For. Meteorol. 2004, 121, 19–35. [Google Scholar] [CrossRef]
- Dorigo, W.A.; Zurita-Milla, R.; de Wit, A.J.W.; Brazile, J.; Singh, R.; Schaepman, M.E. A review on reflective remote sensing and data assimilation techniques for enhanced agroecosystem modeling. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 165–193. [Google Scholar] [CrossRef]
- Huang, J.; Gómez-Dans, J.L.; Huang, H.; Ma, H.; Wu, Q.; Lewis, P.E.; Liang, S.; Chen, Z.; Xue, J.-H.; Wu, Y.; et al. Assimilation of remote sensing into crop growth models: Current status and perspectives. Agric. For. Meteorol. 2019, 276–277, 107609. [Google Scholar] [CrossRef]
- Jin, X.; Kumar, L.; Li, Z.; Feng, H.; Xu, X.; Yang, G.; Wang, J. A review of data assimilation of remote sensing and crop models. Eur. J. Agron. 2018, 92, 141–152. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, Z.; Chen, J.; Liu, J.; Ren, J.; Li, Z.; Sun, L.; Li, H. Application of Crop Model Data Assimilation With a Particle Filter for Estimating Regional Winter Wheat Yields. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 4422–4431. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, Z.; Chen, J.; Ren, J.; Li, Z.; Sun, L. The Estimation of Regional Crop Yield Using Ensemble-Based Four-Dimensional Variational Data Assimilation. Remote Sens. 2014, 6, 2664–2681. [Google Scholar] [CrossRef]
- Cheng, Z.; Meng, J.; Shang, J.; Liu, J.; Qiao, Y.; Qian, B.; Jing, Q.; Dong, T. Improving Soil Available Nutrient Estimation by Integrating Modified WOFOST Model and Time-Series Earth Observations. IEEE Trans. Geosci. Remote Sens. 2018, 2896–2908. [Google Scholar] [CrossRef]
- Li, X.; Du, H.; Mao, F.; Zhou, G.; Chen, L.; Xing, L.; Fan, W.; Xu, X.; Liu, Y.; Cui, L.; et al. Estimating bamboo forest aboveground biomass using EnKF-assimilated MODIS LAI spatiotemporal data and machine learning algorithms. Agric. For. Meteorol. 2018, 256–257, 445–457. [Google Scholar] [CrossRef]
- Zhuo, W.; Huang, J.; Li, L.; Zhang, X.; Ma, H.; Gao, X.; Huang, H.; Xu, B.; Xiao, X. Assimilating Soil Moisture Retrieved from Sentinel-1 and Sentinel-2 Data into WOFOST Model to Improve Winter Wheat Yield Estimation. Remote Sens. 2019, 11, 1618. [Google Scholar] [CrossRef]
- Huang, J.; Sedano, F.; Huang, Y.; Ma, H.; Li, X.; Liang, S.; Tian, L.; Zhang, X.; Fan, J.; Wu, W. Assimilating a synthetic Kalman filter leaf area index series into the WOFOST model to improve regional winter wheat yield estimation. Agric. For. Meteorol. 2016, 216, 188–202. [Google Scholar] [CrossRef]
- Tewes, A.; Hoffmann, H.; Schäfer, F.; Kerkhoff, C.; Krauss, G.; Gaiser, T. Assimilation of Leaf Area Index Measurements Into A Crop Model Framework: Performance Comparison of Two Assimilation Approaches. In Proceedings of the 12th European Conference on Precision Agriculture, SupAgro Montpellier, Montpellier, France, 8–11 July 2019; pp. 182–183, ISBN 978-2-900792-49-0. [Google Scholar]
- Hengl, T.; de Jesus, J.M.; Heuvelink, G.B.M.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef]
- Pätzold, S.; Mertens, F.M.; Bornemann, L.; Koleczek, B.; Franke, J.; Feilhauer, H.; Welp, G. Soil heterogeneity at the field scale: A challenge for precision crop protection. Precis. Agric. 2008, 9, 367–390. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Kooistra, L.; Van den Brande, M.M.M. Using Sentinel-2 Data for Retrieving LAI and Leaf and Canopy Chlorophyll Content of a Potato Crop. Remote Sens. 2017, 9, 405. [Google Scholar] [CrossRef]
- Pasqualotto, N.; Delegido, J.; Van Wittenberghe, S.; Rinaldi, M.; Moreno, J. Multi-Crop Green LAI Estimation with a New Simple Sentinel-2 LAI Index (SeLI). Sensors 2019, 19, 904. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; Hall, C.C.; Brown, L.; Shi, Y.; et al. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinformation 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Toscano, P.; Castrignanò, A.; Di Gennaro, S.F.; Vonella, A.V.; Ventrella, D.; Matese, A. A Precision Agriculture Approach for Durum Wheat Yield Assessment Using Remote Sensing Data and Yield Mapping. Agronomy 2019, 9, 437. [Google Scholar] [CrossRef]
- Vizzari, M.; Santaga, F.; Benincasa, P. Sentinel 2-Based Nitrogen VRT Fertilization in Wheat: Comparison between Traditional and Simple Precision Practices. Agronomy 2019, 9, 278. [Google Scholar] [CrossRef]
- Pause, M.; Raasch, F.; Marrs, C.; Csaplovics, E. Monitoring Glyphosate-Based Herbicide Treatment Using Sentinel-2 Time Series—A Proof-of-Principle. Remote Sens. 2019, 11, 2541. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. S2ToolBox Level 2 Products: LAI, FAPAR, FCOVER. Sentinel-2 Algorithm Theoretical Based Document. 2016. Available online: https://step.esa.int/docs/extra/ATBD_S2ToolBox_L2B_V1.1.pdf (accessed on 5 March 2020).
- Pasqualotto, N.; Bolognesi, S.F.; Belfiore, O.R.; Delegido, J.; D’Urso, G.; Moreno, J. Canopy chlorophyll content and LAI estimation from Sentine1-2: Vegetation indices and Sentine1-2 Leve1-2A automatic products comparison. In Proceedings of the 2019 IEEE International Workshop on Metrology for Agriculture and Forestry (MetroAgriFor), Portici, Italy, 24–26 October 2019; pp. 301–306. [Google Scholar] [CrossRef]
- Djamai, N.; Fernandes, R.; Weiss, M.; McNairn, H.; Goïta, K. Validation of the Sentinel Simplified Level 2 Product Prototype Processor (SL2P) for mapping cropland biophysical variables using Sentinel-2/MSI and Landsat-8/OLI data. Remote Sens. Environ. 2019, 225, 416–430. [Google Scholar] [CrossRef]
- Upreti, D.; Huang, W.; Kong, W.; Pascucci, S.; Pignatti, S.; Zhou, X.; Ye, H.; Casa, R. A Comparison of Hybrid Machine Learning Algorithms for the Retrieval of Wheat Biophysical Variables from Sentinel-2. Remote Sens. 2019, 11, 481. [Google Scholar] [CrossRef]
- Bodenkundliche Kartieranleitung. KA5; Bundesanstalt für Geowissenschaften und Rohstoffe, Ed.; Schweizerbart Science Publishers: Stuttgart, Germany, 2006; ISBN 978-3-510-95920-4. [Google Scholar]
- Main-Knorn, M.; Pflug, B.; Louis, J.; Debaecker, V.; Müller-Wilm, U.; Gascon, F. Sen2Cor for Sentinel-2. In Proceedings of the Image and Signal Processing for Remote Sensing XXIII, Warsaw, Poland, 11–14 September 2017; p. 3. [Google Scholar] [CrossRef]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, Version 2.6-7. 2018. Available online: https://cran.r-project.org/package=raster (accessed on 5 March 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wolf, J. User Guide for LINTUL5: Simple Generic Model for Simulation of Crop Growth under Potential, Water Limited and Nitrogen, Phosphorus and Potassium Limited Conditions; Wageningen UR: Wageningen, The Netherlands, 2012. [Google Scholar]
- Gabaldón-Leal, C.; Webber, H.; Otegui, M.E.; Slafer, G.A.; Ordóñez, R.A.; Gaiser, T.; Lorite, I.J.; Ruiz-Ramos, M.; Ewert, F. Modelling the impact of heat stress on maize yield formation. Field Crop Res. 2016, 198, 226–237. [Google Scholar] [CrossRef]
- Mboh, C.M.; Srivastava, A.K.; Gaiser, T.; Ewert, F. Including root architecture in a crop model improves predictions of spring wheat grain yield and above-ground biomass under water limitations. J. Agron. Crop Sci. 2019, 205, 109–128. [Google Scholar] [CrossRef]
- Webber, H.; Zhao, G.; Wolf, J.; Britz, W.; de Vries, W.; Gaiser, T.; Hoffmann, H.; Ewert, F. Climate change impacts on European crop yields: Do we need to consider nitrogen limitation? Eur. J. Agron. 2015, 71, 123–134. [Google Scholar] [CrossRef]
- Gaiser, T.; Perkons, U.; Küpper, P.M.; Kautz, T.; Uteau-Puschmann, D.; Ewert, F.; Enders, A.; Krauss, G. Modeling biopore effects on root growth and biomass production on soils with pronounced sub-soil clay accumulation. Ecol. Model. 2013, 256, 6–15. [Google Scholar] [CrossRef]
- Addiscott, T.M.; Whitmore, A.P. Simulation of solute leaching in soils of differing permeabilities. Soil Use Manag. 1991, 7, 94–102. [Google Scholar] [CrossRef]
- Wösten, J.H.M.; Lilly, A.; Nemes, A.; Le Bas, C. Development and use of a database of hydraulic properties of European soils. Geoderma 1999, 90, 169–185. [Google Scholar] [CrossRef]
- Wösten, J.H.M.; Pachepsky, Y.A.; Rawls, W.J. Pedotransfer functions: Bridging the gap between available basic soil data and missing soil hydraulic characteristics. J. Hydrol. 2001, 251, 123–150. [Google Scholar] [CrossRef]
- Evensen, G. The Ensemble Kalman Filter: Theoretical formulation and practical implementation. Ocean. Dyn. 2003, 53, 343–367. [Google Scholar] [CrossRef]
- Guo, C.; Tang, Y.; Lu, J.; Zhu, Y.; Cao, W.; Cheng, T.; Zhang, L.; Tian, Y. Predicting wheat productivity: Integrating time series of vegetation indices into crop modeling via sequential assimilation. Agric. For. Meteorol. 2019, 272–273, 69–80. [Google Scholar] [CrossRef]
- Krauss, G. Simplace: Interface to Use the Modelling Framework SIMPLACE, Version 4.2.4; Bonn, Germany, 2019. Available online: https://r-forge.r-project.org/projects/simplace/ (accessed on 5 March 2020).
- Tewes, A.; Hoffmann, H.; Krauss, G.; Schäfer, F.; Kerkhoff, C.; Gaiser, T. New approaches for the assimilation of LAI measurements into a crop model ensemble to improve wheat biomass estimations. Agronomy 2020. under review. [Google Scholar]
- Griffiths, R.I.; Thomson, B.C.; Plassart, P.; Gweon, H.S.; Stone, D.; Creamer, R.E.; Lemanceau, P.; Bailey, M.J. Mapping and validating predictions of soil bacterial biodiversity using European and national scale datasets. Appl. Soil Ecol. 2016, 97, 61–68. [Google Scholar] [CrossRef]
- Mulder, V.L.; Lacoste, M.; Richer-de-Forges, A.C.; Martin, M.P.; Arrouays, D. National versus global modelling the 3D distribution of soil organic carbon in mainland France. Geoderma 2016, 263, 16–34. [Google Scholar] [CrossRef]
- Tifafi, M.; Guenet, B.; Hatté, C. Large Differences in Global and Regional Total Soil Carbon Stock Estimates Based on SoilGrids, HWSD, and NCSCD: Intercomparison and Evaluation Based on Field Data From USA, England, Wales, and France. Glob. Biogeochem. Cycles 2018, 32, 42–56. [Google Scholar] [CrossRef]
- Dai, Y.; Shangguan, W.; Wei, N.; Xin, Q.; Yuan, H.; Zhang, S.; Liu, S.; Lu, X.; Wang, D.; Yan, F. A review of the global soil property maps for Earth system models. SOIL 2019, 5, 137–158. [Google Scholar] [CrossRef]
- Palosuo, T.; Kersebaum, K.C.; Angulo, C.; Hlavinka, P.; Moriondo, M.; Olesen, J.E.; Patil, R.H.; Ruget, F.; Rumbaur, C.; Takáč, J.; et al. Simulation of winter wheat yield and its variability in different climates of Europe: A comparison of eight crop growth models. Eur. J. Agron. 2011, 35, 103–114. [Google Scholar] [CrossRef]
| Field | Gravel AS | Gravel SG | Sand AS | Sand SG | Silt AS | Silt SG | Clay AS | Clay SG | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0–30 | 0.02 | 6.96 | 4.25 | 26.46 | 75.95 | 58.24 | 19.8 | 15.31 |
| 30–60 | 0.02 | 6.96 | 3.48 | 26.46 | 75.4 | 58.24 | 21.12 | 15.31 | |
| 60–90 | 0.15 | 7.67 | 3.26 | 25.19 | 76.01 | 58.10 | 20.73 | 16.71 | |
| 2 | 0–30 | 1.07 | 4.08 | 54.46 | 38.14 | 29.61 | 43.28 | 15.93 | 18.58 |
| 30–60 | 0.66 | 4.28 | 52.06 | 39.17 | 27.69 | 42.61 | 20.26 | 18.22 | |
| 60–90 | 0.47 | 5.08 | 56.64 | 38.17 | 25.59 | 43.14 | 17.77 | 18.69 | |
| 3 | 0–30 | 1.52 | 9.12 | 37.97 | 27.22 | 41.57 | 54.27 | 20.46 | 18.52 |
| 30–60 | 1.32 | 8.48 | 34.04 | 27.25 | 40.65 | 54.23 | 25.31 | 18.52 | |
| 60–90 | 3.34 | 9 | 36.18 | 25.22 | 39.79 | 54.78 | 24.02 | 20 | |
| 4 | 0–30 | 0.92 | 8.46 | 10 | 27.39 | 62.93 | 49.01 | 27.06 | 23.6 |
| 30–60 | 1.05 | 8.46 | 7.96 | 27.78 | 55.91 | 48.76 | 36.13 | 23.46 | |
| 60–90 | 1.41 | 9.94 | 8.11 | 26.67 | 59.06 | 48.47 | 32.82 | 24.86 | |
| 5 | 0–30 | 22.12 | 12.96 | 13.64 | 23.74 | 38.25 | 48.64 | 48.12 | 27.62 |
| 30–60 | 21.58 | 13.28 | 13.55 | 24.33 | 38.21 | 48.26 | 48.26 | 27.4 | |
| 6 | 0–30 | 0.23 | 9.89 | 4.75 | 16.94 | 72.3 | 62.18 | 22.96 | 20.88 |
| 30–60 | 0.05 | 10.29 | 2.65 | 17.71 | 67.15 | 61.36 | 30.2 | 20.93 | |
| 7 | 0–30 | 0.01 | 4.52 | 80.93 | 80.44 | 10.26 | 15.19 | 8.83 | 4.37 |
| 30–60 | 0.03 | 4.45 | 78.45 | 80.98 | 12.7 | 14.2 | 8.85 | 4.82 | |
| 8 | 0–30 | 0.76 | 7.37 | 7.06 | 22.24 | 69.22 | 55.96 | 26.41 | 21.8 |
| 30–60 | 0.72 | 7.37 | 8.79 | 21.51 | 70.97 | 57.06 | 25.11 | 21.43 | |
| 60–90 | 0.54 | 8.88 | 13.56 | 19.29 | 75.88 | 57.9 | 19.69 | 22.8 | |
| 9 | 0–30 | 0.03 | 7.41 | 6.26 | 20 | 65.02 | 58.48 | 28.72 | 21.52 |
| 30–60 | 0.54 | 8 | 4.82 | 20.3 | 67.7 | 58.48 | 27.48 | 21.23 | |
| 60–90 | 1.89 | 8.89 | 4.83 | 18.89 | 72.82 | 59.48 | 22.35 | 21.64 | |
| 10 | 0–30 | 0.41 | 5.83 | 3.64 | 26.78 | 80.7 | 56.1 | 15.66 | 17.12 |
| 30–60 | 0 | 5.83 | 3.33 | 26.97 | 79.53 | 56.31 | 17.15 | 16.72 | |
| 60–90 | 0 | 6.52 | 2.56 | 25 | 79.74 | 55.72 | 17.7 | 19.28 | |
| 11 | 0–30 | 0 | 6.05 | 4.63 | 20 | 84.32 | 62.33 | 11.05 | 17.67 |
| 30–60 | 0 | 6.33 | 3.02 | 20 | 85.07 | 62 | 11.91 | 18 | |
| 60–90 | 0 | 6.33 | 2.48 | 18 | 76.82 | 62.29 | 20.71 | 19.71 | |
| 12 | 0–30 | 2.34 | 6.83 | 12.16 | 21.15 | 42.85 | 52.11 | 44.99 | 26.74 |
| 30–60 | 1.94 | 7.02 | 10.93 | 21.64 | 48.1 | 51.89 | 40.97 | 26.47 | |
| 60–90 | 4.01 | 7.62 | 9.1 | 18.73 | 52.17 | 53.74 | 38.73 | 27.53 | |
| 13 | 0–30 | 0.2 | 6.24 | 2.8 | 18.34 | 82.3 | 63.95 | 14.91 | 17.71 |
| 30–60 | 0 | 6.13 | 2.06 | 19 | 81.82 | 63.37 | 16.12 | 17.63 | |
| 60–90 | 0 | 7.45 | 1.49 | 17.55 | 83.88 | 64.24 | 14.64 | 18.21 | |
| 14 | 0–30 | 0.25 | 6.19 | 4.47 | 17.26 | 84.2 | 61.91 | 11.33 | 20.84 |
| 30–60 | 0 | 6.19 | 2.9 | 18.56 | 83.76 | 61.05 | 13.33 | 20.4 | |
| 60–90 | 0 | 7.51 | 1.98 | 16.86 | 81.27 | 61.74 | 16.75 | 21.4 |
| Clay Content | Gravel Content | Plant-Available Water Content | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Field | R | RMSE | Bias | R | RMSE | Bias | R | RMSE | Bias | AWC AS (%) | AWC SG (%) |
| 1 | −0.05 | 5.14 | −2.27 | −0.01 | 9.58 | 9.39 | −0.21 | 0.03 | −0.03 | 0.17 | 0.14 |
| 2 | 0.49 | 5.27 | 0.81 | −0.22 | 5.16 | 4.95 | −0.22 | 0.01 | −0.01 | 0.124 | 0.133 |
| 3 | 0.45 | 5.67 | −2.59 | 0.16 | 12.05 | 10.42 | 0.53 | 0.01 | 0 | 0.133 | 0.13 |
| 4 | 0.24 | 11.65 | −5.93 | −0.02 | 10.70 | 10.37 | −0.12 | 0.02 | −0.01 | 0.135 | 0.122 |
| 5 | 0.00 | 17.61 | −16.25 | −0.05 | 8.91 | −7.28 | 0.22 | 0.05 | 0.05 | 0.184 | 0.112 |
| 6 | 0.67 | 6.17 | −4.87 | 0.05 | 9.94 | 9.92 | 0.27 | 0.02 | −0.02 | 0.15 | 0.134 |
| 7 | −0.17 | 5.13 | −4.07 | 0.03 | 4.30 | 4.28 | 0.09 | 0.04 | 0.04 | 0.106 | 0.145 |
| 8 | −0.60 | 4.75 | 0.67 | −0.21 | 10.26 | 10.04 | 0.34 | 0.03 | −0.03 | 0.157 | 0.13 |
| 9 | −0.53 | 6.08 | −2.38 | 0.55 | 10.13 | 9.99 | −0.11 | 0.02 | −0.02 | 0.152 | 0.133 |
| 10 | 0.31 | 4.44 | 3.77 | −0.53 | 8.39 | 8.18 | 0.27 | 0.04 | −0.04 | 0.177 | 0.138 |
| 11 | 0.84 | 7.40 | 6.66 | NA | 9.83 | 9.52 | 0.52 | 0.04 | −0.04 | 0.184 | 0.142 |
| 12 | −0.04 | 14.16 | −13.20 | 0.27 | 7.09 | 6.64 | 0.71 | 0.02 | 0.02 | 0.107 | 0.123 |
| 13 | 0.33 | 4.01 | 3.80 | −0.63 | 8.38 | 8.27 | 0.60 | 0.04 | −0.04 | 0.183 | 0.142 |
| 14 | 0.59 | 8.14 | 7.89 | −0.53 | 7.95 | 7.89 | −0.01 | 0.05 | −0.05 | 0.186 | 0.135 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tewes, A.; Hoffmann, H.; Nolte, M.; Krauss, G.; Schäfer, F.; Kerkhoff, C.; Gaiser, T. How Do Methods Assimilating Sentinel-2-Derived LAI Combined with Two Different Sources of Soil Input Data Affect the Crop Model-Based Estimation of Wheat Biomass at Sub-Field Level? Remote Sens. 2020, 12, 925. https://doi.org/10.3390/rs12060925
Tewes A, Hoffmann H, Nolte M, Krauss G, Schäfer F, Kerkhoff C, Gaiser T. How Do Methods Assimilating Sentinel-2-Derived LAI Combined with Two Different Sources of Soil Input Data Affect the Crop Model-Based Estimation of Wheat Biomass at Sub-Field Level? Remote Sensing. 2020; 12(6):925. https://doi.org/10.3390/rs12060925
Chicago/Turabian StyleTewes, Andreas, Holger Hoffmann, Manuel Nolte, Gunther Krauss, Fabian Schäfer, Christian Kerkhoff, and Thomas Gaiser. 2020. "How Do Methods Assimilating Sentinel-2-Derived LAI Combined with Two Different Sources of Soil Input Data Affect the Crop Model-Based Estimation of Wheat Biomass at Sub-Field Level?" Remote Sensing 12, no. 6: 925. https://doi.org/10.3390/rs12060925
APA StyleTewes, A., Hoffmann, H., Nolte, M., Krauss, G., Schäfer, F., Kerkhoff, C., & Gaiser, T. (2020). How Do Methods Assimilating Sentinel-2-Derived LAI Combined with Two Different Sources of Soil Input Data Affect the Crop Model-Based Estimation of Wheat Biomass at Sub-Field Level? Remote Sensing, 12(6), 925. https://doi.org/10.3390/rs12060925