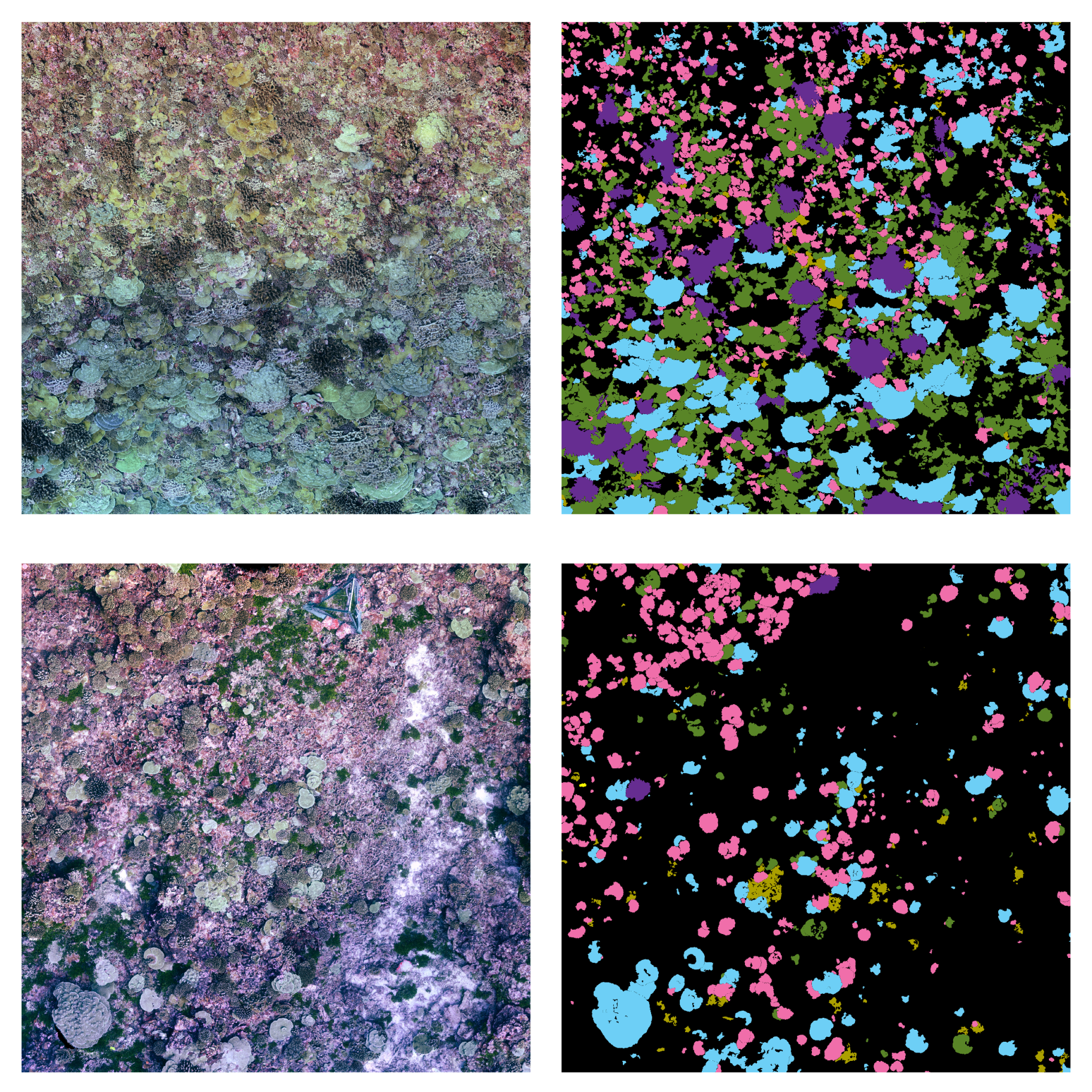
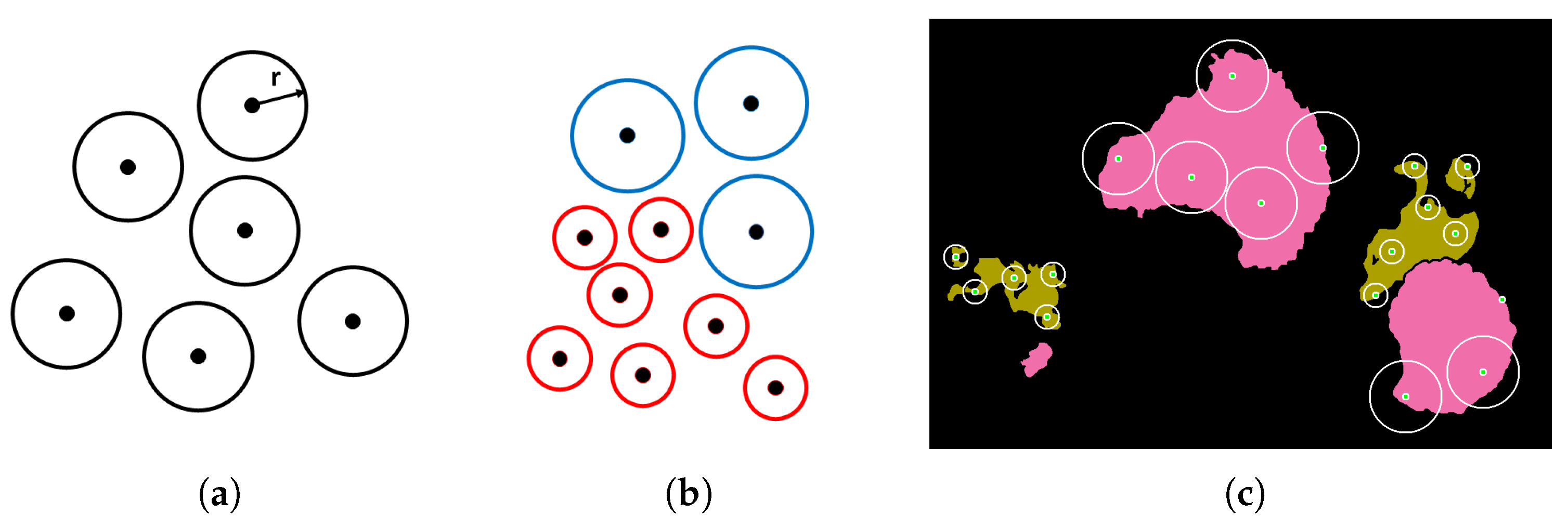
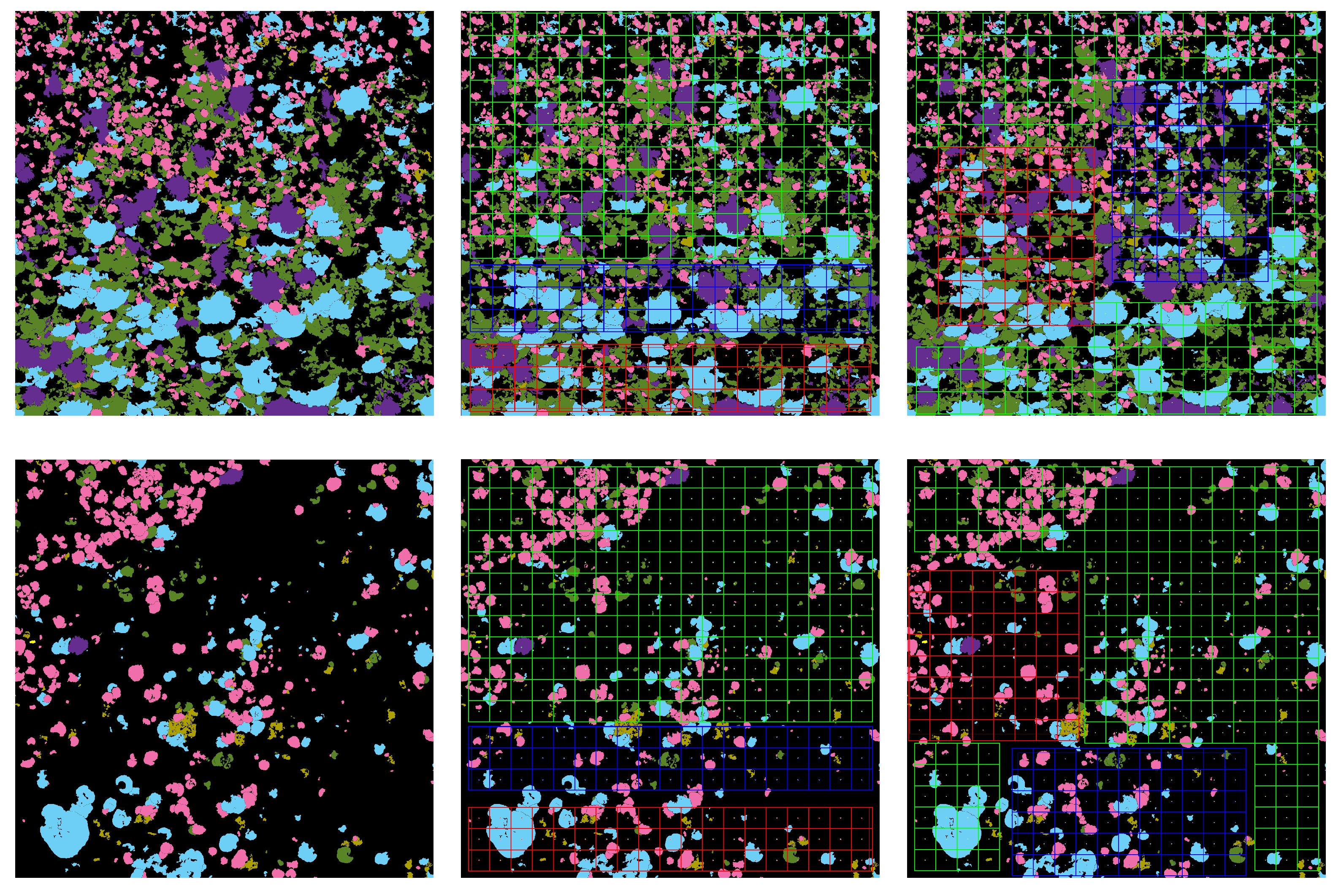
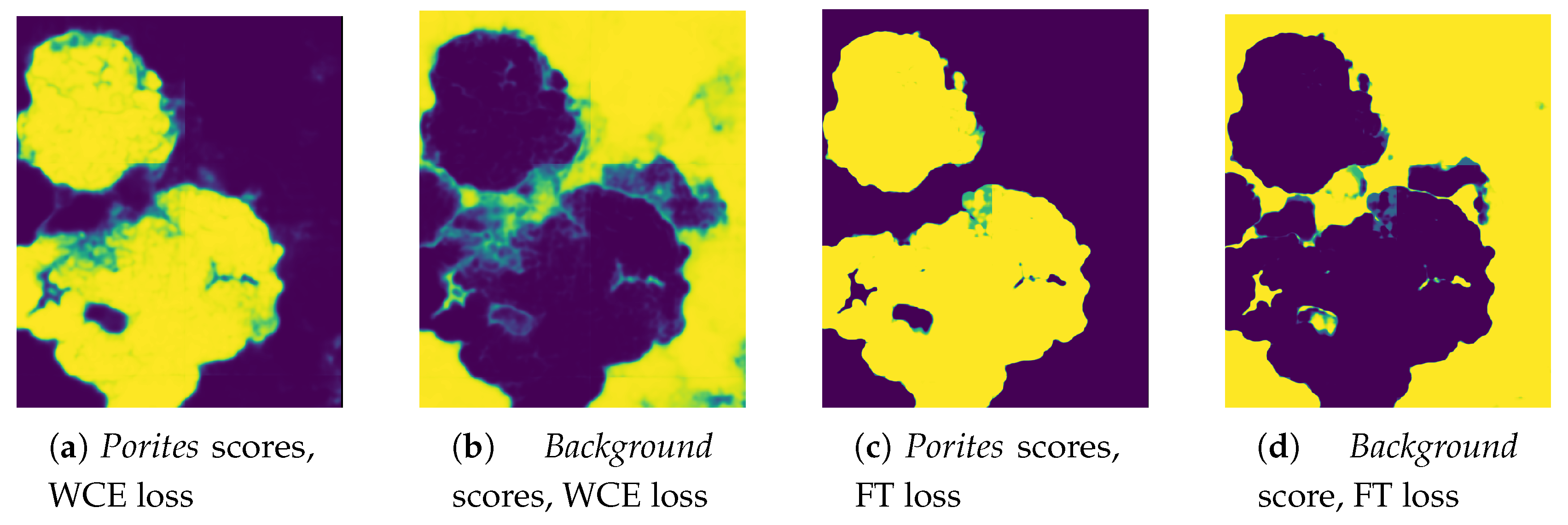
The Semantic Segmentation of Coral Reefs
Large-area imaging is an increasingly popular solution for the study of subtidal environments at scales of tens to hundreds of meters. In particular, orthographic imagery, e.g., orthophotomosaics, orthoprojections (for brevity,
orthos), is increasingly being used for spatio-temporal analysis of coral communities [
1,
2,
3,
4,
5]. Orthos enable the fine-scale mapping and accurate measurements of coral colony size and position that allow researchers to extract the information to better understand the demographic patterns and the spatial dynamics of benthic communities. Previously, such information could only be obtained through laborious in situ methods, which necessarily limits the scale of the monitoring campaigns. Importantly, as large-area imaging allows researchers to conduct ecological data extraction efforts digitally, researchers are now able to dramatically expand the spatial and temporal scales over which their work can be conducted. The challenge created, is thus in the efficient extraction of ecological information from large-area imagery.
To date, these fine-scale ecological maps are generated following a manual workflow, using GIS drawing tools or image painting software, such as Adobe Photoshop, to digitize individual coral colonies. In image analysis, the accurate identification of colonies boundaries and assignment of a taxonomic class, i.e., classification, is known as semantic segmentation. Unfortunately, manual segmentation of the coral population is an extremely time-consuming process, and since corals have a slow growth (linearly, less than a 1 cm/yr), the detection of change demands that segmentation be conducted with a high degree of accuracy. The manual workflow to extract information from large-area imagery has required up to an hour of human effort for each square meter of imagery, representing a severe bottleneck between the data collection and ecological analysis.
Driven by the trending research on Convolutional Neural Networks (CNN), deep learning approaches have become increasingly used for the automatic recognition of marine organisms. The seminal work on the automatic classification of benthic coral reef communities [
6] led to the release of the Web-based platform for point annotations
CoralNet [
7]. However, point-based annotations of images are most valuable for determination of bulk metrics such as percent cover, and do not provide the data needed to quantify demographic change or to conduct spatial analyses: these investigations demand semantic segmentation. The optimization of a supervised model first requires the preparation of sizeable human-labeled datasets. In work by Alonso et al. 2017 [
8], the authors propose a method to propagate annotations from point-based to pixel-wise, by manipulating the fluorescence channel. The resulting labeled masks are then used to fine-tune a SegNet [
9] network. More recently, the same authors released the first extensive dataset of mask-labeled benthic images [
10], obtained by the propagation of sparse annotation with a multi-level superpixel method. A custom annotation tool for the creation of segmented orthos of the seafloor, based on SLIC and graph-cut, is described in King [
11]. The authors compared the performance of patch-based and semantic segmentation models, obtaining the best patch classification accuracy at
on ten coral taxa using a Resnet152 and the best pixel-wise accuracy of
with a Deeplab V2. In previous work [
12], we fine-tuned a Deeplab V3+ using a large training dataset composed of ten orthos generated from Structure-from-Motion generated dense point clouds. The model outputs labeled maps with an accuracy comparable with human annotators for the binary segmentation task. Despite this progress, the automatic segmentation of sessile organisms in underwater environments still presents many challenges.
Generalization. There are over 800 known species of reef-building corals,
(Scleractinia), worldwide with many locations having hundreds of common species [
13]. However, regional species pools vary widely [
14], and corals are well known for their phenotypic plasticity and capacity for morphological adaptation to local conditions. The ideal classifier would classify every taxon present in a given ortho, independent of locality, conditions during image collection, or the equipment and software used in the construction of orthos. However, a CNN trained on one dataset composed of orthos from one location, with a given regionally adapted taxonomic assemblage (source domain), might learn domain features specific to that collection of species or region, and perform worse on new orthos with another collection of species (target domain).
Differences between domains might be due to the considerable morphological variability within and among marine species, the presence of survey/reconstruction-related features which affect ortho quality, as well as the variability of class abundance on different plots. Machine learning models usually expect a similar class frequency between source and target domain. However, the abundance of coral populations can vary widely from location to location, even within a single reef or island. Although it seems convenient to assume equally probable distributions during the CNN optimization, this might bias predictions on heavily unbalanced real-world scenarios.
Class imbalance. Surveyed areas often contain only a small number of individuals of the taxa under consideration, the foreground classes. The background class represents the remainder of the benthos that is not actively targeted for monitoring, and includes everything from sand to less abundant coral taxa, and is often the predominant class in each input training tile. In our work to date, foreground class frequencies can vary from the 30 to of pixels, resulting in mild to severe class imbalance. When a dataset contains many more samples of one class than the others, the classifier penalizes under-represented classes by focusing on the more numerous ones. Moreover, there is also considerable variation in colony size, both within and among taxa, with colony area ranging from a few squared centimeters to several squared meters. These sizes, in many cases, are also correlated with the specific class. A class containing only small coral instances, each being represented by a handful of pixels, will result in a class-imbalance scenario.
Human errors. Previous work [
15] has shown that human recognition on photos reaches a variable accuracy on different benthic groups (hard coral, soft corals, algae): the average accuracy for hard corals is about
. In many cases, only in situ observations can disambiguate taxonomic classifications. Furthermore, humans do not recognize all coral classes equally well. To our experience, experts agree in identifying encrusting
Montipora less than half of the time with respect to the other classes. This uncertainty in the annotations of training datasets in turn affects model reliability.
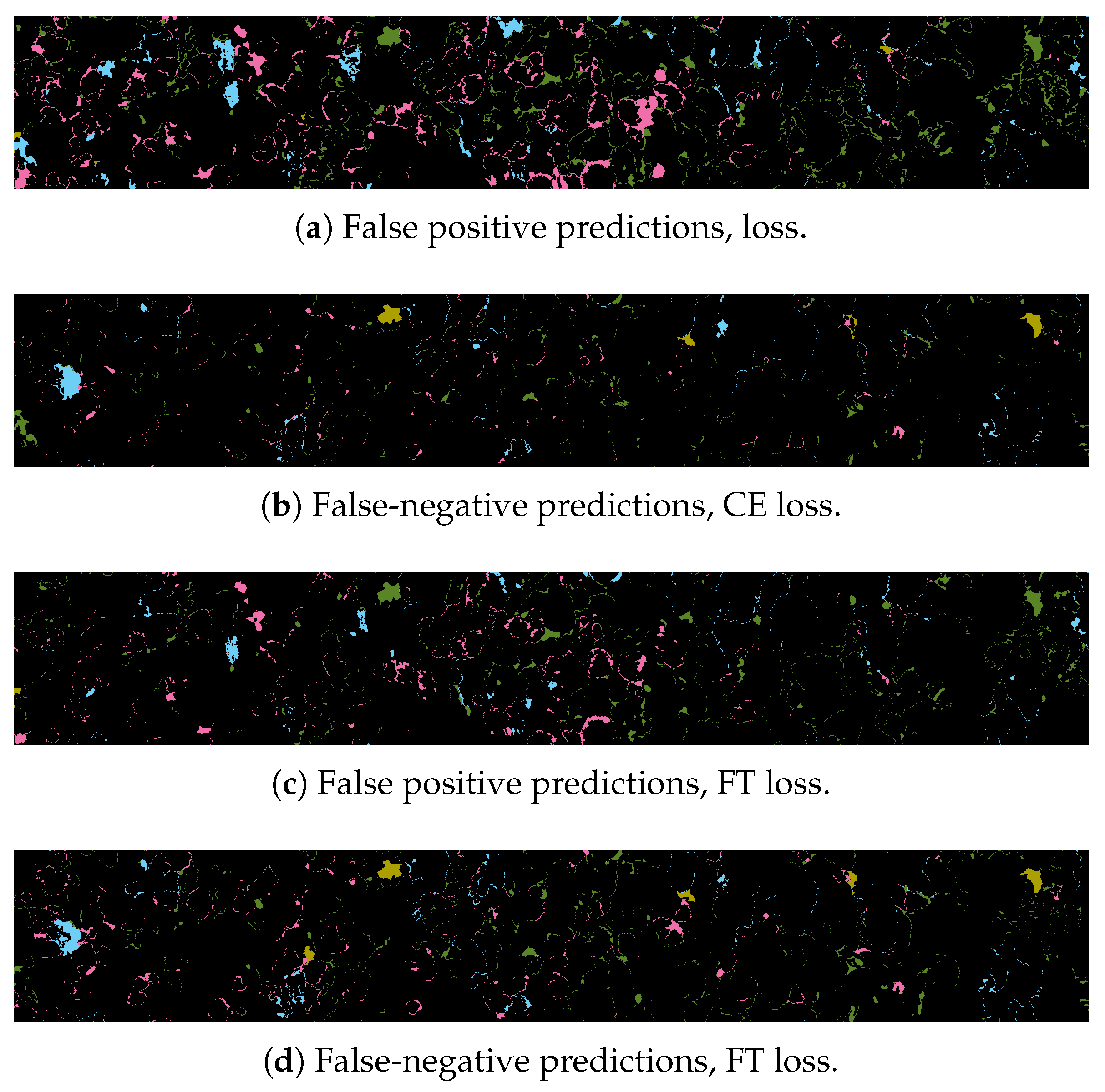
Complex Borders. The semantic segmentation of corals implies an accurate pixel-wise classification of the scene’s objects. In CNN-based segmentation, networks typically generate output objects with smooth contours that do not reflect the jagged and sharp boundaries of coral frequently found in nature.The coarser resolution of predictions is partially due to the pyramidal structure of network architectures. Pooling layers combine high-level feature maps, downgrading the resolution and inducing a smoothing effect on outputs. As a result, most misclassification errors fall on instance boundaries. Further, the use of distribution-based loss functions, which do not take into account local properties, might aggravate this phenomenon. Finally, human annotation errors are also most likely to occur at colony boundaries, leading to inaccurate training datasets and inducing further ambiguity.
This paper is an extended version of our previous work [
16], presented at the Underwater 3D Recording and Modeling conference. In that study, we investigated several strategies to improve the performance of CNN-based semantic segmentation of
a single coral class on a
small, problematic dataset from a single location, exploiting the properties of orthos. However, research and monitoring campaigns generally have multiple target classes and will generate orthos from several geographic locations of interest. Here, we extend the previous methodologies to a multi-class scenario.
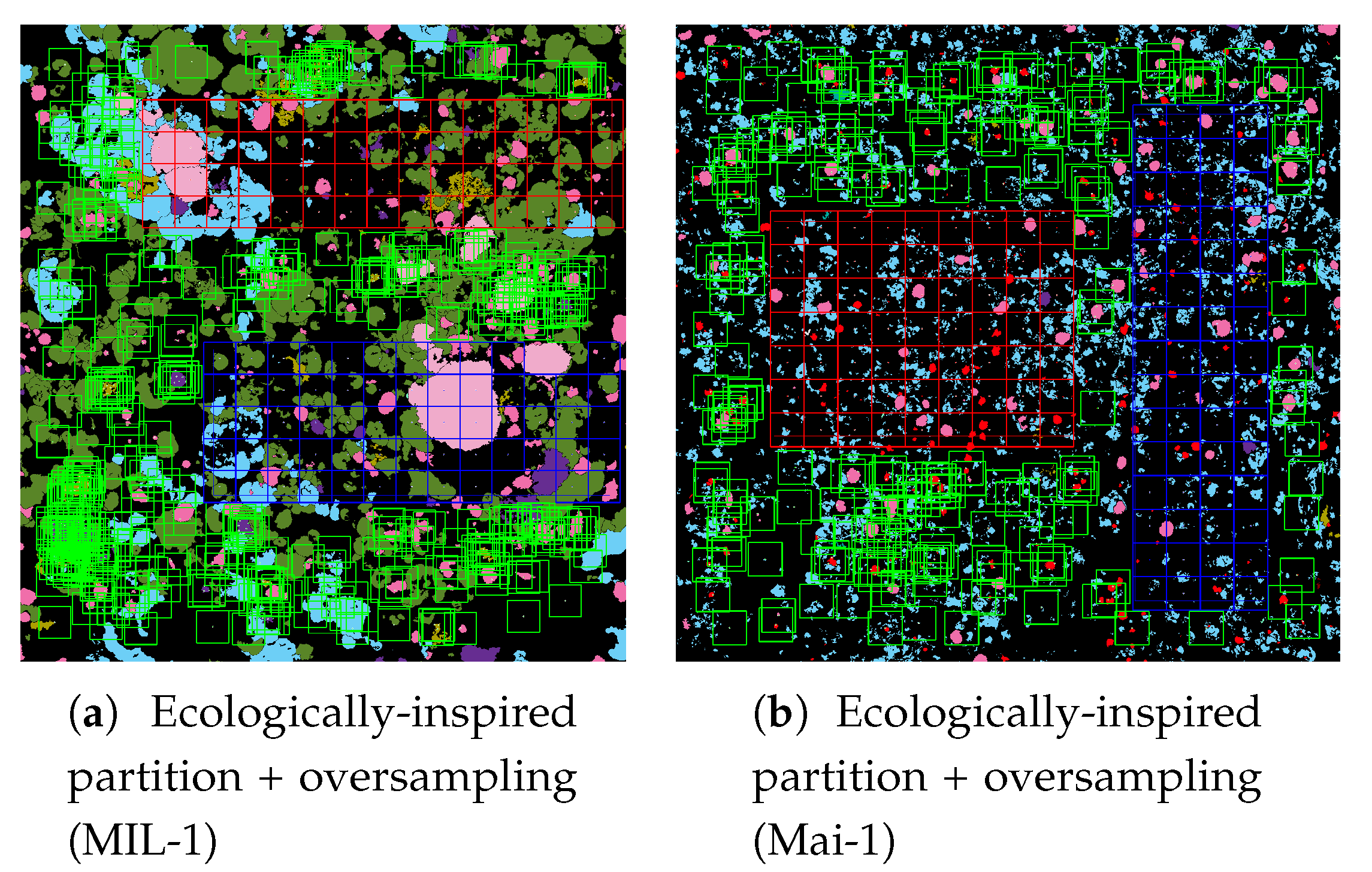
The ecologically inspired splitting is a dataset preparation strategy dealing with the non-random coral spatial patterns in orthos [
1,
4]. As the spatial dispersion of coral populations is more descriptive than the simple class distribution, balancing according to this information improved the model accuracy both in training and testing.
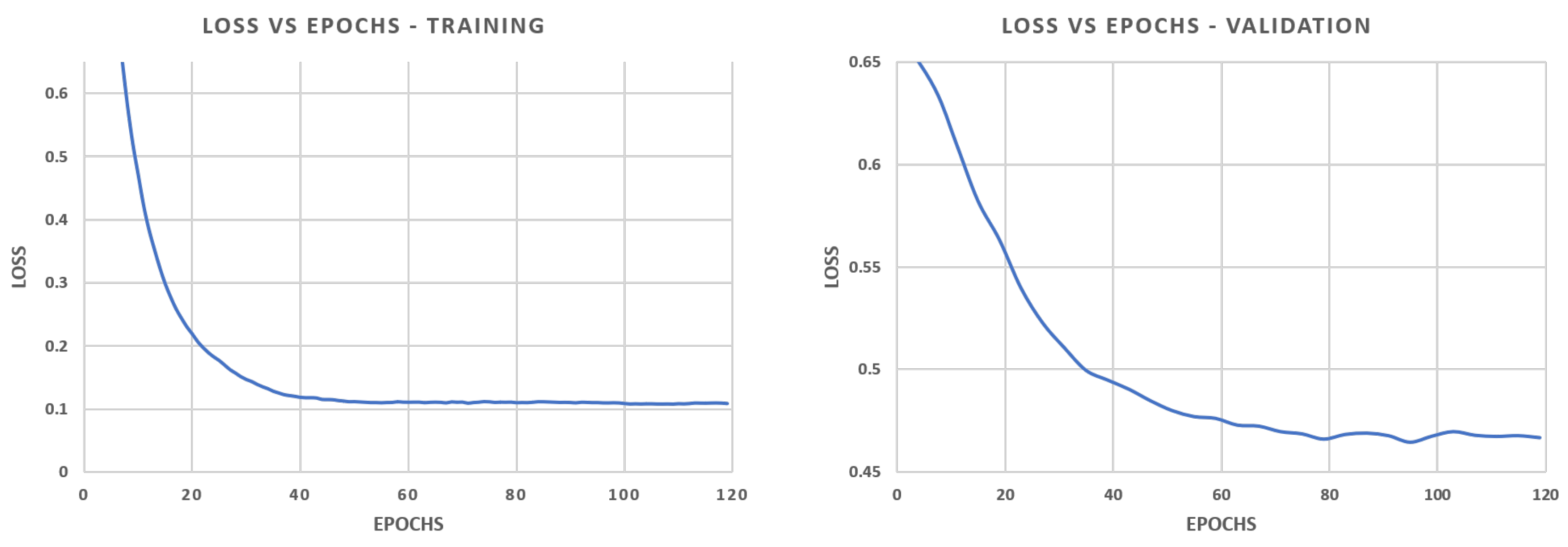
Traditional class-imbalance learning techniques increase the importance of deriving information from under-represented classes to reduce learning bias towards the predominant classes. These methods are divided into two main categories, (i) those that act at the data level, essentially through under- or over-sampling, and (ii) those that act at the algorithm level, such as the adoption of dedicated cost functions. Interested readers may refer to Dong et al. 2018 [
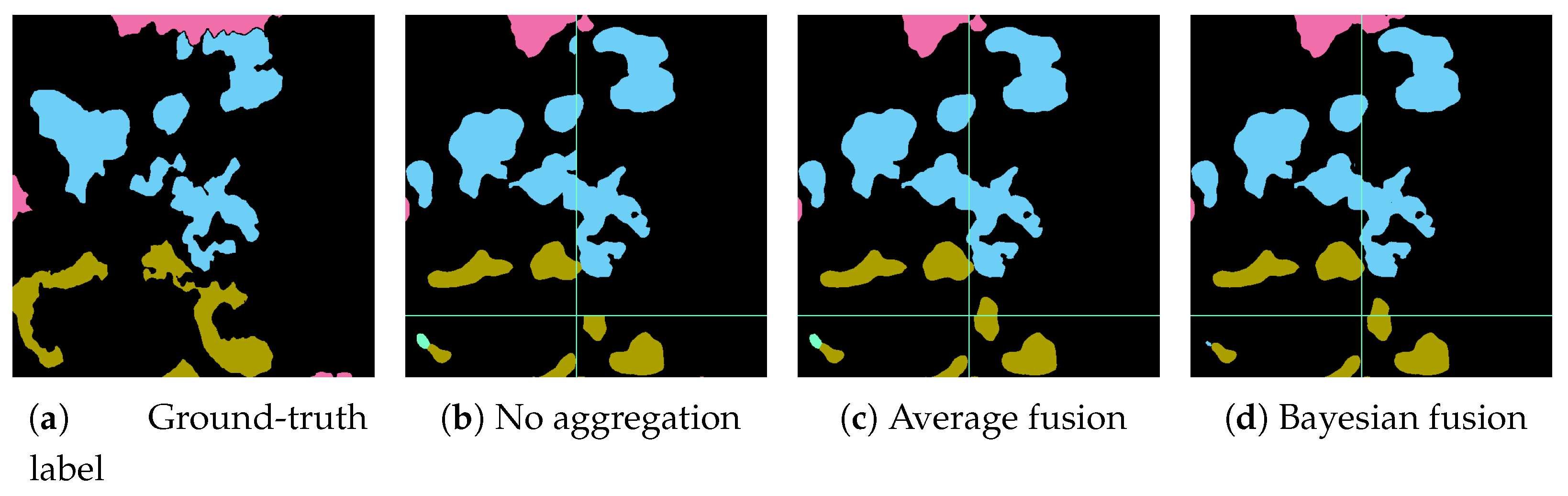
17] for an exhaustive overview of class-imbalance learning methodologies. Here, we tested different loss functions and then propose an over-sampling strategy based on Poisson-disk sampling. Some of the loss functions succeed in improving the accuracy on boundaries, while others better detect small coral instances. Finally, we re-propose a method to aggregate overlapping tile predictions using the prior information of the predicted coverage of the specimens on the surveyed area.