Stress Detection in New Zealand Kauri Canopies with WorldView-2 Satellite and LiDAR Data
Abstract
1. Introduction
1.1. Kauri Trees and Kauri Dieback Symptoms
1.2. Research Background
1.3. Objectives
- Test the performance of WV2 attributes for crown-based stress detection in kauri trees and define the recommended minimum crown size;
- Test a two-step method on WV2 attributes by first identifying dead and dying trees in a classification and then applying a regression for the remaining stress symptom levels;
- Test the performance of LiDAR attributes in combination with WV2 data for canopy stress detection.
2. Materials and Methods
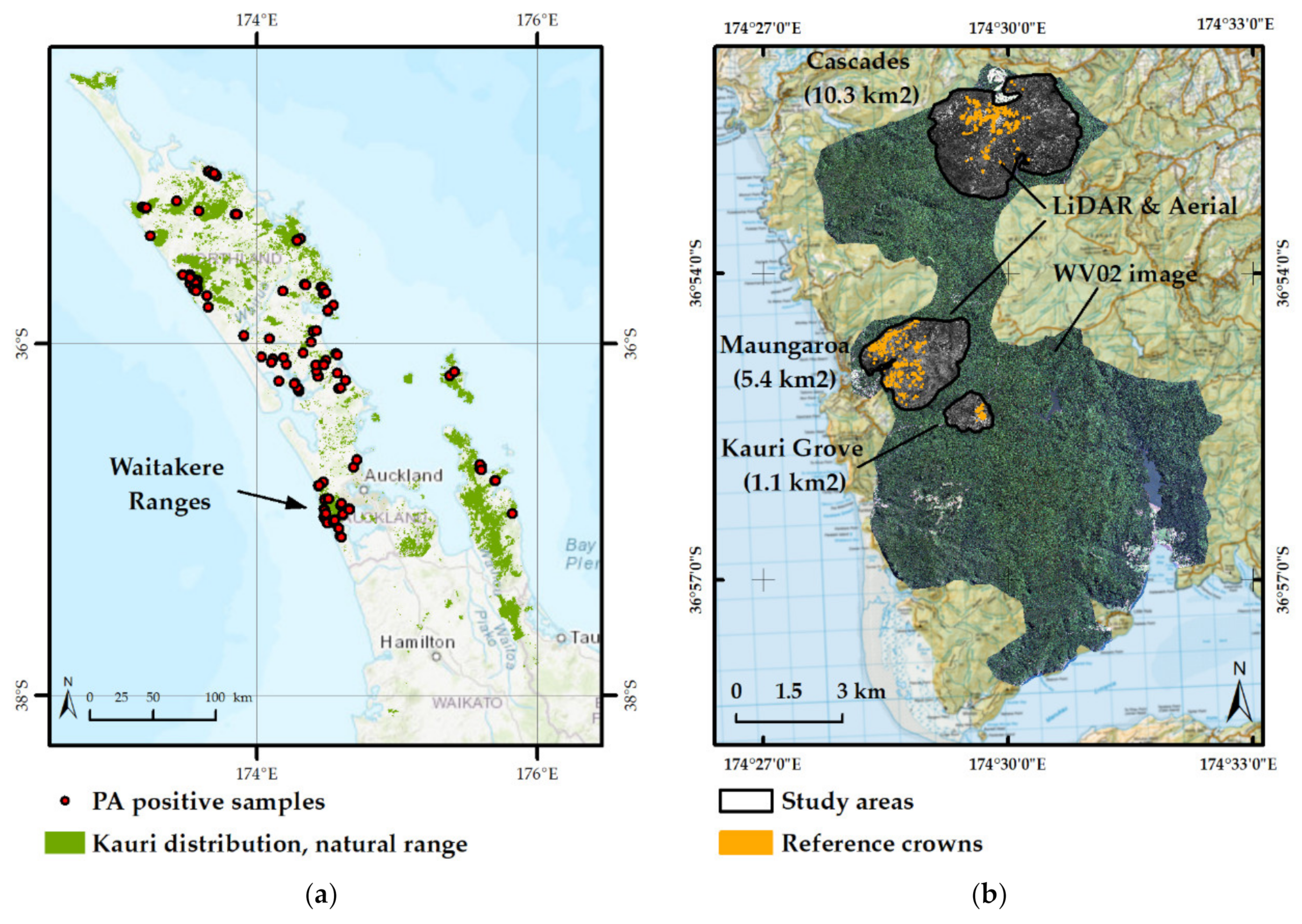
2.1. Study Area
2.2. LiDAR Data and Aerial Images
2.3. WorldView-2 Image
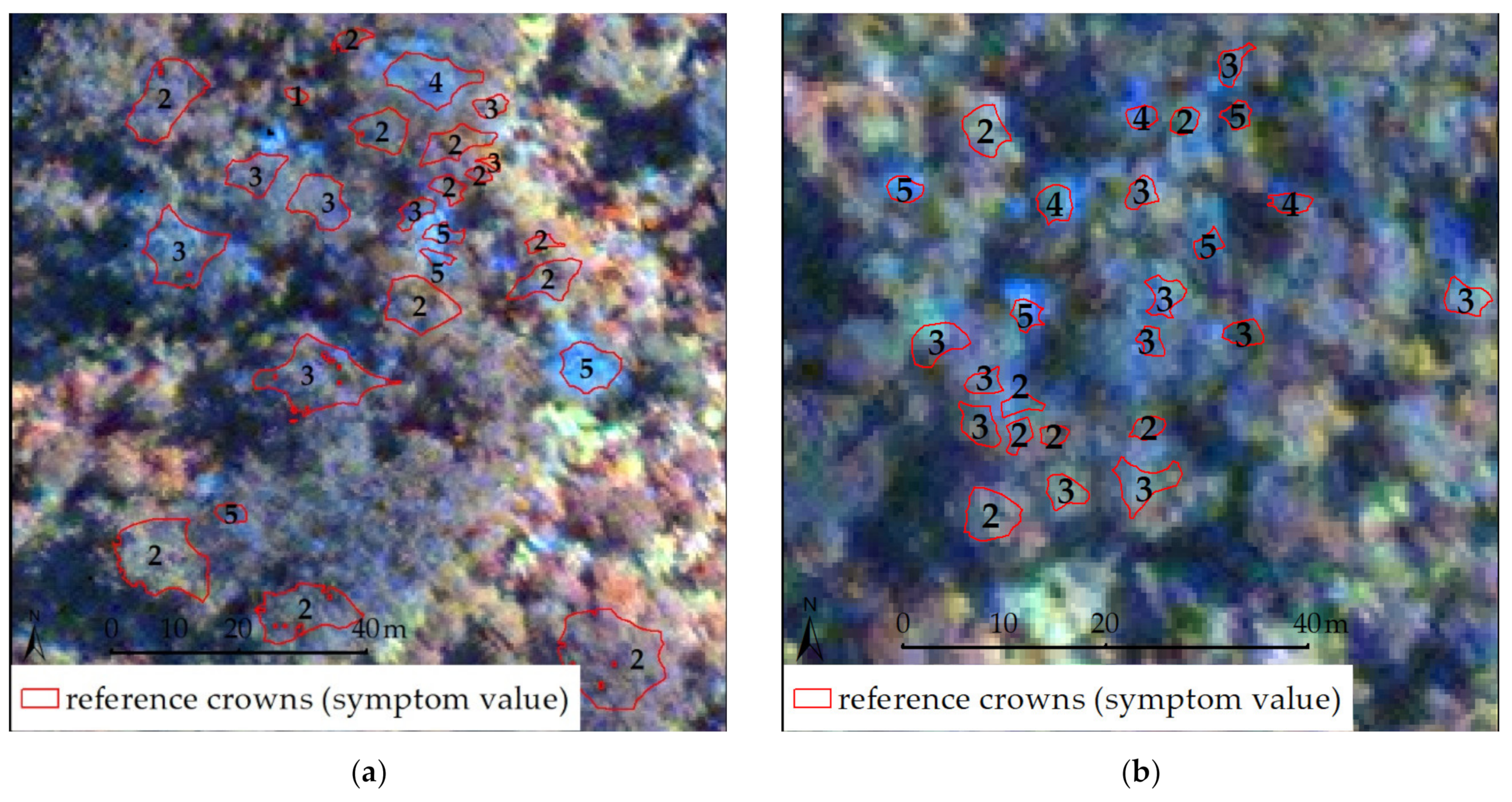
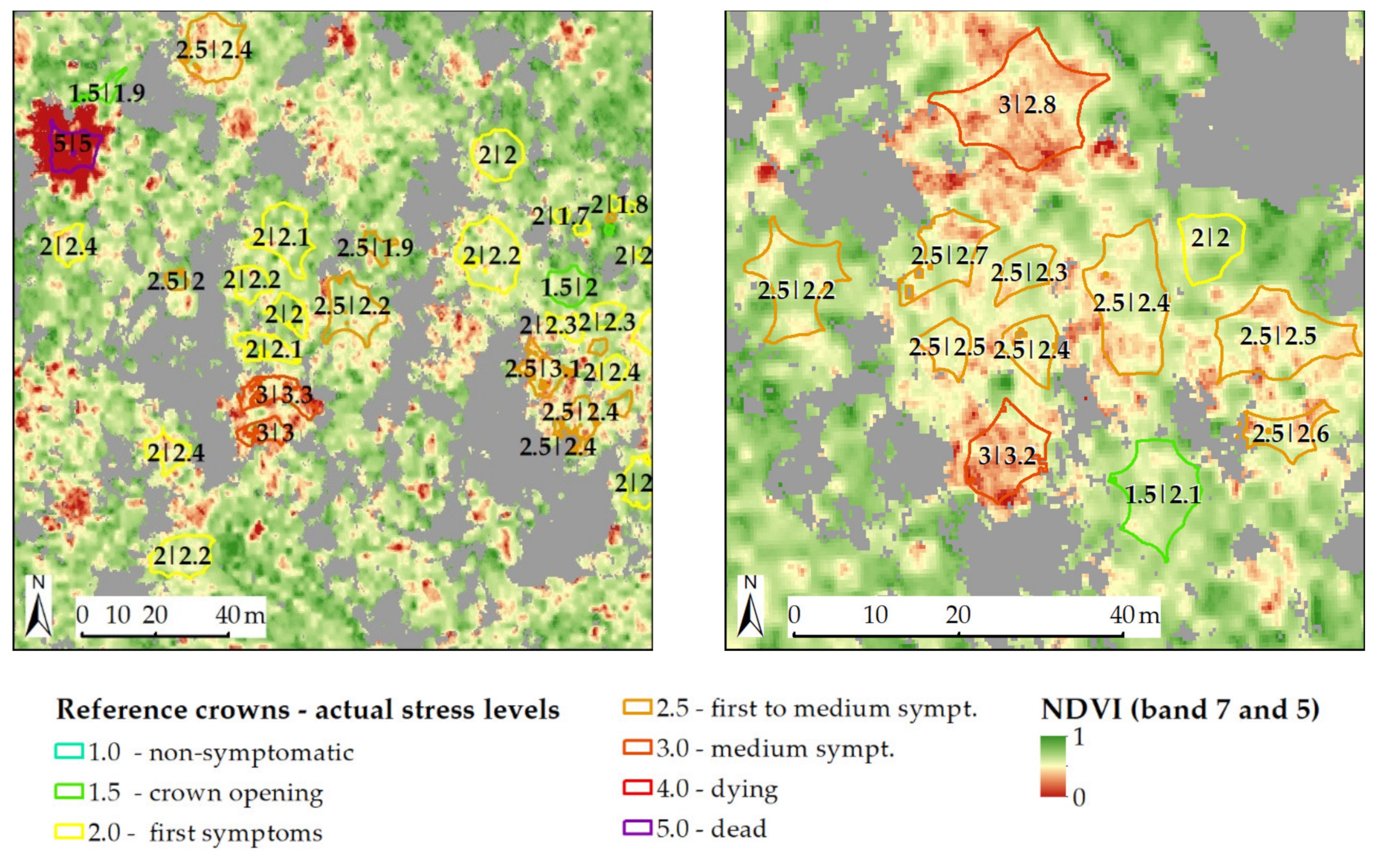
2.4. Reference Crown Set
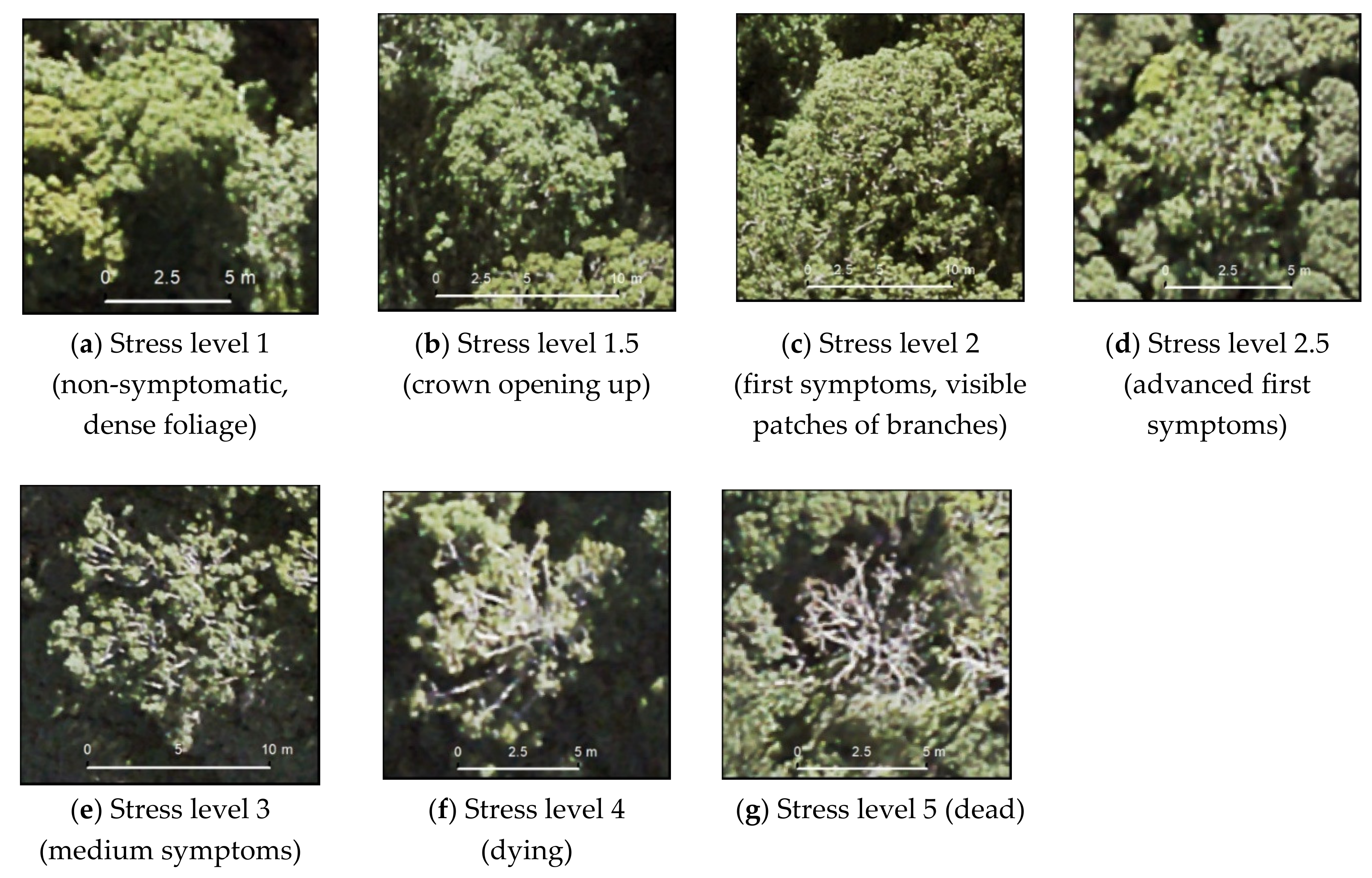
2.5. Reference Values for Stress Assessment
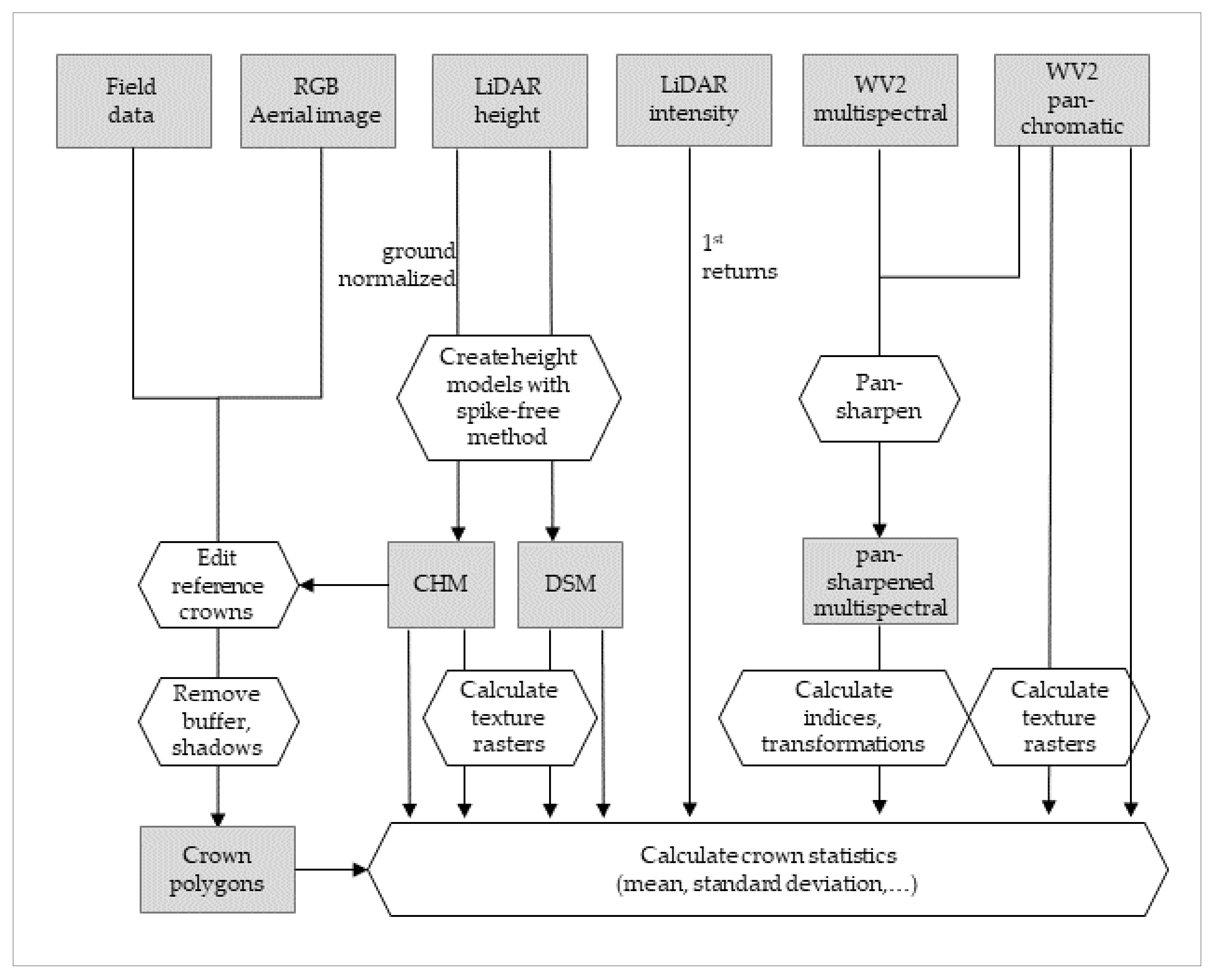
2.6. Attribute Calculation
2.7. Attribute Selection and Regression Approach
3. Results
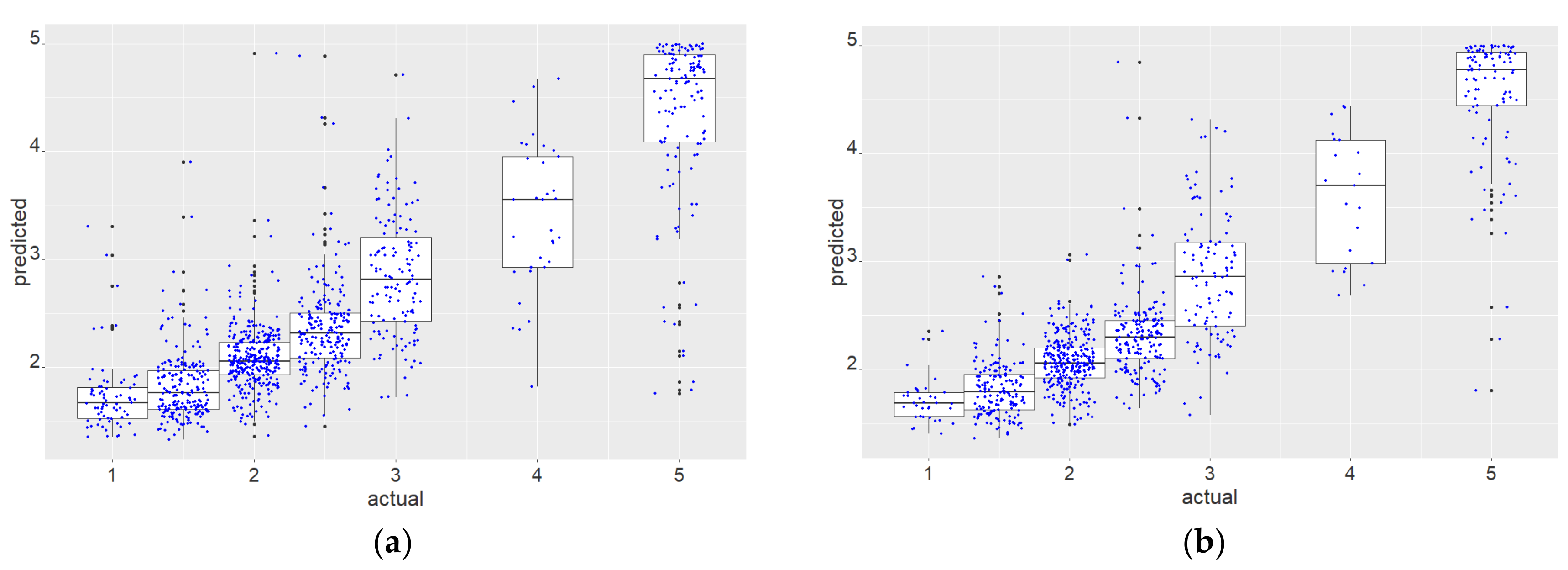
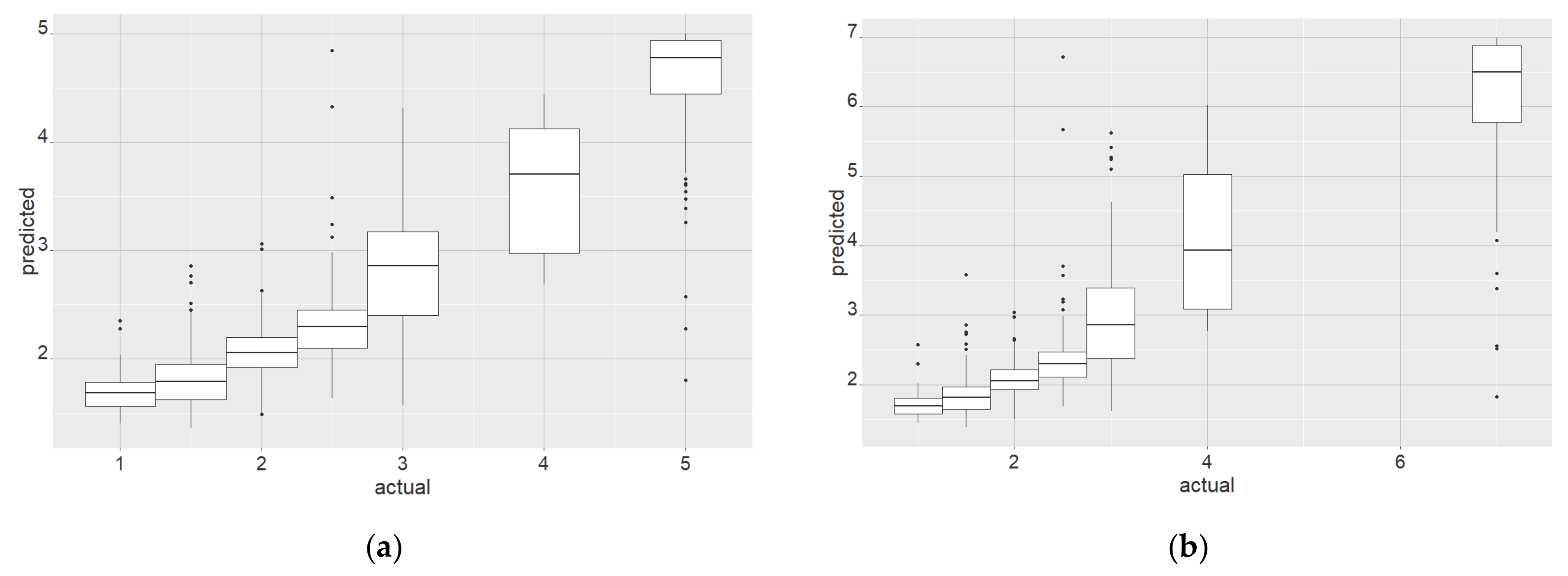
3.1. Results Objective 1: Test the Performance of WV2 Attributes for Crown-Based Stress Detection in Kauri Trees and Define the Recommended Minimum Crown Size
3.2. Results Objective 2: Classification to Identify Dead and Dying Trees
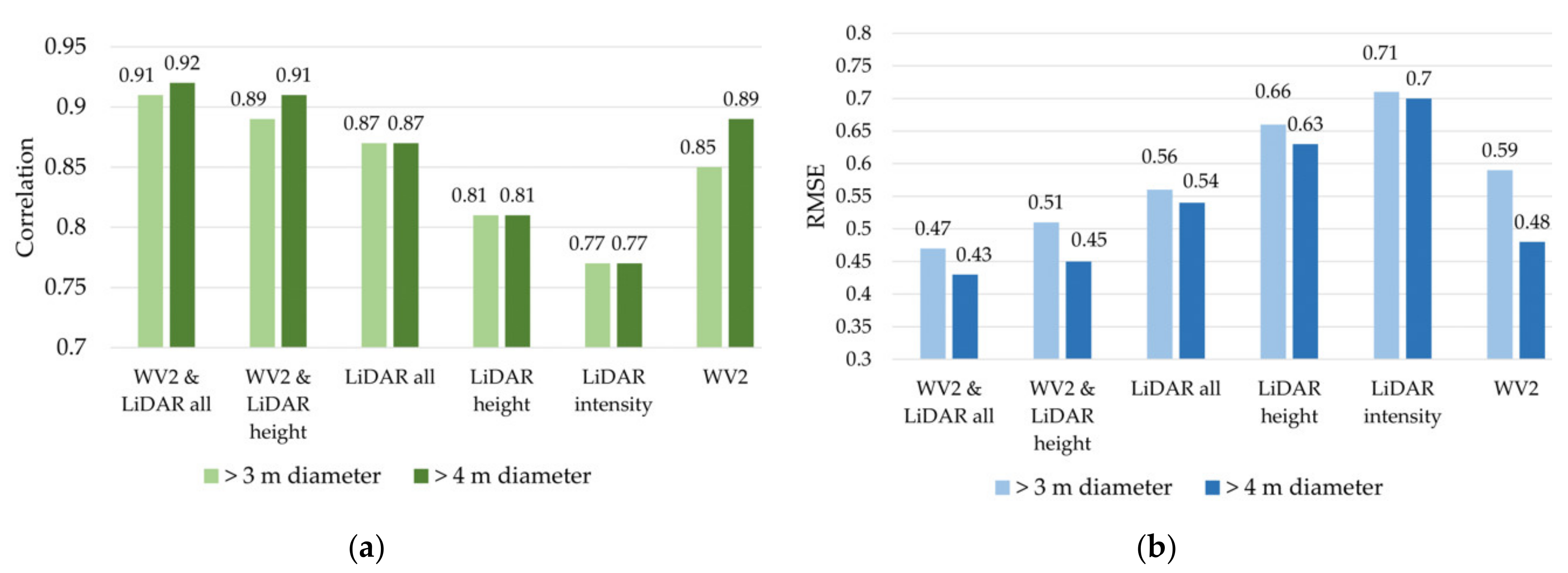
3.3. Results Objective 3: Test the Performance of LiDAR Attributes for Stress Detection
4. Discussion
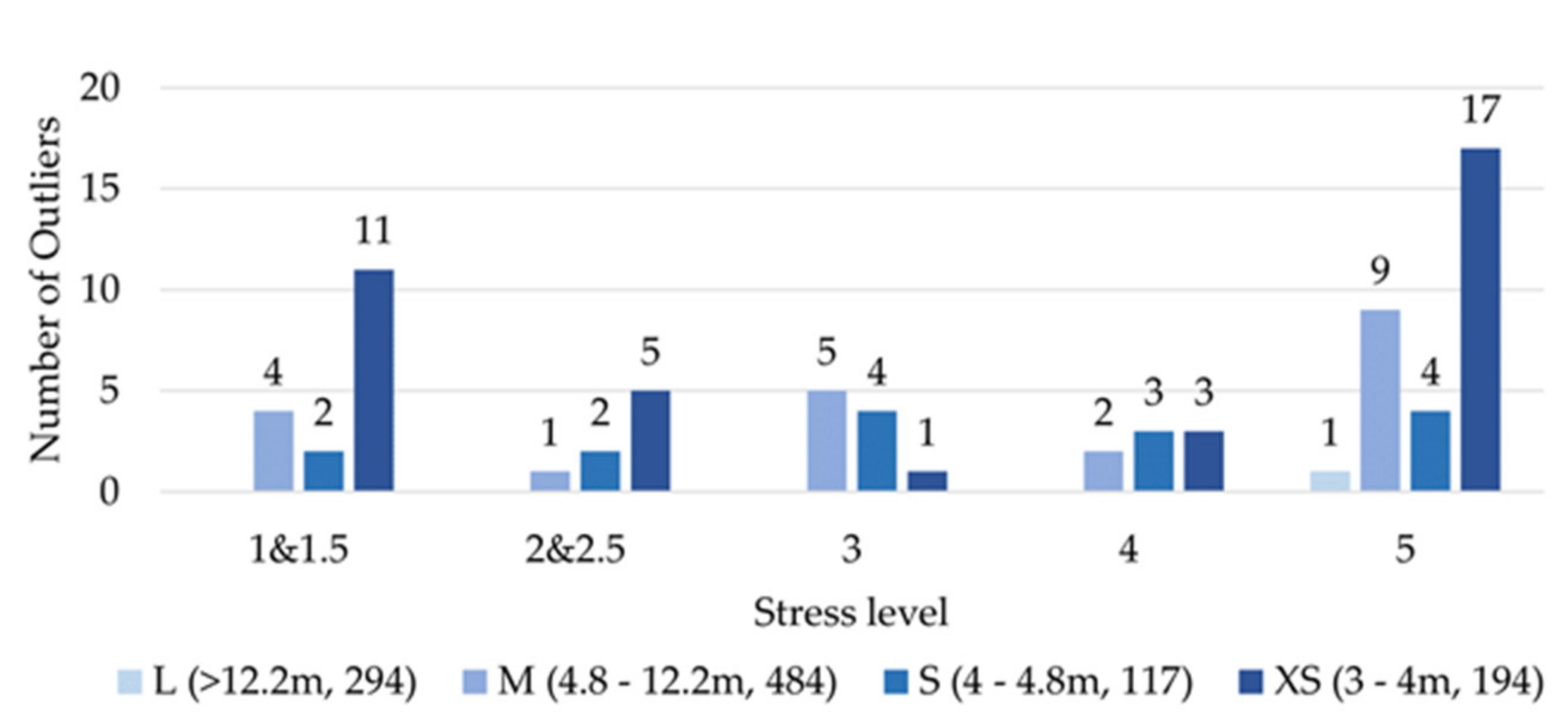
4.1. Discussion: Minimum Crown Size, Spatial Accuracy and Stratified Approach
4.2. Discussion: Attribute Performance
4.3. Discussion: Dealing with Non-Linearity: Rescaling and a Two-Step Approach
4.4. Discussion: Performance of Additional LiDAR Attributes for Stress Detection
4.5. Discussion: Recommendations for Further Studies
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Illustrations for the Orthorectification of the WV2 Image

Appendix B. Spatial offset of crown polygons to WV2 Image
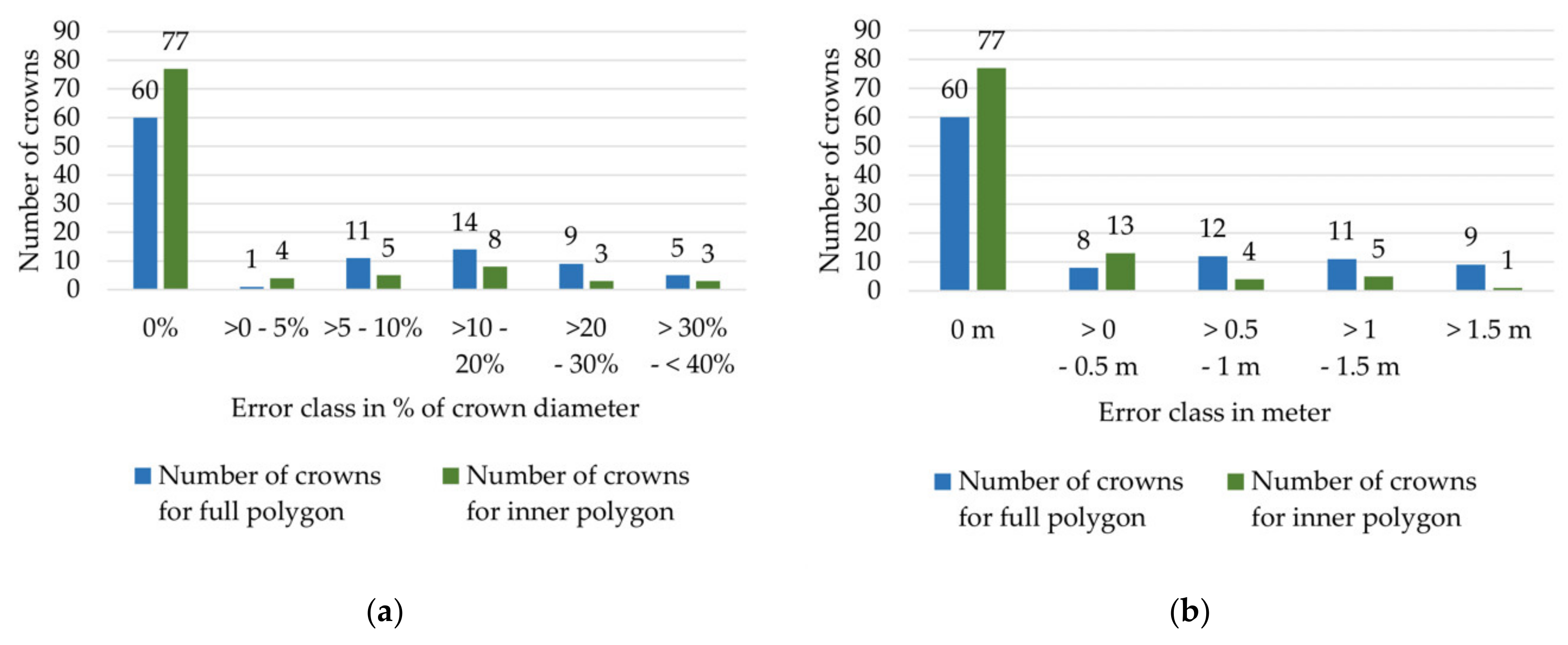
Appendix C. LiDAR and WorldView-2 Attributes
| Total | Based on | Attributes | Software/Tools |
|---|---|---|---|
| 14 | Intensity values | Avg, qav, std, ske, kur Percentiles (01, 05, 10, 25, 50, 75, 90, 95, 99) | LAStools, LAScanopy |
| 88 | DSM 1 | Stdev, var, rang, crosc, longc, mnc, mxc, slp, avgdev, kur, skw, on 3 × 3, 5 × 5, 7 × 7 kernel size | QGIS zonal statistic, GRASS: r.param.scale |
| 55 | CHM 1 | Mean, max, st. dev., rang, med, maj, var, vart, prc conv, sa, var, ent, asm, con, cor, idm, con, cor> all attributes were calculated on 3, 5, 7 kernel sizes | QGIS zonal statistic GRASS: r.statistics, r.texture, r.slope.aspect |
| 13 | Crown polygon | Ratio mdm/height; Ratio of max height with 95th, 75th, and 50th percentiles and average height of outer crown edge; difference of max height with 75 and 50 percentile, peak density, height, mdm, shp area and roundness (shp area/lengths), average height of outer crown edge | ArcGIS tools |
| 33 | LiDAR point cloud | Std, ske, kur, cov, dns (cut value 6 m and 8 m), min, max, avg, avg 1st return, percentiles (01, 05, 10, 25, 50, 75, 90, 95, 99), bincentiles (10, 20, 30, 40, 50, 60, 70, 80, 90) | LAStools LAScanopy |
| Total | Attributes 2 | Crown Statistics | Software/Tools |
|---|---|---|---|
| 8 | multispectral bands 1–8 | mean | ENVI |
| 1 | MNF transformation (1st band) | st. dev. | ENVI |
| 4 | HSVM transformation (3 bands), brightness layer (1 band) | mean | ENVI |
| 31 | 31 vegetation indices | mean | ENVI |
| 110 | textures on PAN channel and band 7 (NIR1) with 3 × 3, 5 × 5, and 7 × 7 kernels: rang, mean, var, ent, skw, hom, con, cor, sm, dis 2 | mean st. dev. | ENVI |
| asm | angular second m |
| avg | average |
| avgdev | average deviation |
| con | contrast |
| cor | correlation |
| cov | coverage |
| crosc | cross-sectional curvature |
| dis | dissimilarity |
| dns | density |
| ent | entropy |
| hom | homogeneity |
| idm | inverse difference |
| kur | kurtosis |
| longc | longitudinal curvature |
| maj | majority |
| mdm | mean diameter |
| med | median |
| mnc | mean curvature |
| mxc | max curvature |
| planc | plan curvature |
| prc conv | percent convex |
| profc | profile curvature |
| qav | quadratic average |
| rang | range |
| skw | skewness |
| slp | slope |
| sm | second moment |
| st. dev. | standard deviation |
| var | variance |
| vart | variety |
Appendix D. WV2 Attribute Importance for the Basic and Refined Symptom Levels
| Abbrev. | Band | 5 Level | First | 7 Level | Crown Statistic | Description | Algorithm | Ref. |
|---|---|---|---|---|---|---|---|---|
| Imp. % | Imp. % | Imp. % | ||||||
| NDVI | 5, 7 | 32.2 | 26.7 | 31.5 | mean | NDVI | (b7 − b5)/(b7 + b5) 4 | [99,100] |
| RGRI | 3, 5 | 29.5 | 20.3 | 24.2 | mean | Red-Green Ratio Index | b5/b3 4 | [101] |
| MNF1 | all | 15.2 | 9.9 | 12.3 | st. dev. | 1st band of an MNF transformation | [102] | |
| NDVIg | 3, 6 | 13.1 | 9 | mean | Green NDVI | (b6 − b3)/(b6 + b3)) 4 | [103] | |
| mNDWI | 7, 8 | 5.0 | 5.7 | mean | modified Normalized Difference Water Index | (b7 − b8)/(b7 + b8) 4 | [108] | |
| Bright | 2, 3, 5, 7 | 4.7 | 3.8 | mean | brightness band | (b2 + b3 + b5 + b7)/4 4 | [91] | |
| b6 | 6 | 3.6 | mean | mean of band 6 | ||||
| b7_3krg | 7 | 7.4 | 4.2 | mean | range of a 3 × 3 kernel on band 7 | [104] | ||
| b7_5krg | 7 | 8.8 | st. dev. | range of a 5 × 5 kernel on band 7 | [104] | |||
| Pn_k7sm | PAN | 3.7 | mean | second moment of a 3 × 3 kernel on the PAN band | [104] | |||
| Pn_k7mn | PAN | 3.6 | 8.0 | 2.7 | st. dev. | mean of a 7 × 7 kernel on the PAN band | [104] | |
| CHM | 6.2 | 5.7 | max | maximum height on CHM raster (spike-free threshold 1 m) | [84] |
Appendix E. WV2 and LiDAR Attributes to Identify Dead and Dying Trees
| Abbrev. | Crown | Description | WV2 | WV2 and LiDAR | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Statistic | 1089 Crowns >3 m | 895 Crowns >4 m | 1089 Crowns >3 m | 895 Crowns >4 m | |||||||
| imp. % | corr. | imp. % | corr. | imp. % | corr. | imp. % | corr. | ||||
| NDVI_75 | mean | NDVI (b7 − b5)/(b7 + b5) 1 | 25.8 | 0.75 | 15.7 | 0.75 | [99,100] | ||||
| MCARI1 | mean | Modified Chlorophyll Absorption in Reflectance Index 11.2*(2.5*[b7-b5]-1.3*[b7-b3]) 1 | 16.2 | 0.39 | [116] | ||||||
| NDVIg | mean | green NDVI (b6 − b3)/(b6 + b3)) 1 | 26.7 | 0.55 | [103] | ||||||
| RDVI | mean | Renormalized Difference Vegetation Index (b7−b5) /sqrt(b7+b5) 1 | 22.2 | 0.51 | [116,117] | ||||||
| ARVI | mean | atmospherically resistant vegetation index (b7) - (2*b5-b2)/(b7) + (2*b5-b2) 1 | 18.5 | 0.70 | 16.5 | 0.70 | [118] | ||||
| MNF_1st | st. dev. | 1st band of an MNF transformation | 15.5 | 0.65 | [102] | ||||||
| Pan_k3sk | mean | skewness of a 3 × 3 kernel on the PAN band | 17.2 | 0.01 | [104] | ||||||
| Bright | mean | brightness band (b2 + b3 + b5 + b7)/4 1 | 20.2 | 0.06 | [91] | ||||||
| Pan_k3hm | mean | homogeneity of a 3 × 3 kernel on the PAN band | 21.4 | 0.10 | [104] | ||||||
| Pan_k5Cr | mean | correlation of a 5 × 5 kernel on the PAN band | 15.7 | 0.03 | [104] | ||||||
| b7_k3rg | mean | range of a 3 × 3 kernel on the band 7 | 15.5 | 0.02 | 16.9 | 0.07 | 18.2 | 0.02 | 16.4 | 0.07 | [104] |
| intensity | skewness | LiDAR intensity value | 14.7 | 0.55 | 12.1 | 0.52 | [83] | ||||
| D6_k5var | variance | variance of a 5 × 5 kernel on the DSM (spike-free threshold 0.6 m) | 13.8 | 0.17 | [104] | ||||||
| Cg6_k5var | median | variance of a 5 × 5 kernel on the CHM (spike-free threshold 0.6 m) | 12.3 | 0.51 | [104] | ||||||
| RatDMHght | ratio between the mean diameter and maximum height | 13.6 | 0.00 | ||||||||
| R_max_P50 | ratio between the maximum height and the 50th percentile | 11.4 | 0.58 | ||||||||
| CHM | maximum | maximum height on CHM raster (spike-free threshold 1 m) | 11.8 | 0.28 | 11.8 | 0.27 | [84] | ||||
References
- Beever, R.E.; Waipara, N.W.; Ramsfield, T.D.; Dick, M.A.; Horner, I.J. Kauri (Agathis australis) under threat from Phytophthora. Phytophthoras For. Nat. Ecosyst. 2009, 74, 74–85. [Google Scholar]
- Weir, B.S.; Paderes, E.; Anand, N.; Uchida, J.Y.; Pennycook, S.R.; Bellgard, S.E.; Beever, R.E. A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and P. cocois. Phytotaxa 2015, 205, 21–38. [Google Scholar] [CrossRef]
- MPI. Map “Kauri Dieback Distribution”. Available online: https://www.kauridieback.co.nz/media/2037/kauri-dieback-distribution_20190930_350dpi.jpg (accessed on 20 February 2020).
- Ecroyd, C.E. Biological flora of New Zealand 8.Agathis australis (D. Don) Lindl. (Araucariaceae) Kauri. New Zealand J. Bot. 1982, 20, 17–36. [Google Scholar] [CrossRef]
- Steward, G.A.; Beveridge, A.E. A review of New Zealand kauri (Agathis australis (D. Don) Lindl.): Its ecology, history, growth and potential for management for timber. N. Z. J. For. Sci. 2010, 40, 33–59. [Google Scholar]
- Shortland, T.; Wood, W. Kia Toitu He Kauri, Kauri Dieback Tangata Whenua Roopu Cultural Impact Assessment. Report by Repo Consultancy Ltd., Comissioned by the Ministry of Primary Industries NZ/the Kauri Dieback Programme. Available online: https://www.kauridieback.co.nz/media/1813/shortland-wood-2011.pdf (accessed on 5 March 2020).
- Meiforth, J.J.; Buddenbaum, H.; Hill, J.; Shepherd, J.; Norton, D.A. Detection of New Zealand Kauri Trees with AISA Aerial Hyperspectral Data for Use in Multispectral Monitoring. Remote Sens. 2019, 11, 2865. [Google Scholar] [CrossRef]
- Meiforth, J.; Buddenbaum, H.; Hill, J.; Shepherd, J. Monitoring of Canopy Stress Symptoms in New Zealand Kauri Trees Analysed with AISA Hyperspectral Data. Remote Sens. 2020, 12, 926. [Google Scholar] [CrossRef]
- Bellgard, S.; Weir, B.; Pennycook, S.R.; Paderes, E.P.; Winks, C.; Beever, R.E.; Williams, S. Specialist Phytophthora Research: Biology, Pathology, Ecology and Detection of PTA; Final Report for the New Zealand Ministry for Primary Industries: Wellington, New Zealand, 2013. [Google Scholar]
- Macinnis-Ng, C.; Schwendenmann, L. Litterfall, carbon and nitrogen cycling in a southern hemisphere conifer forest dominated by kauri (Agathis australis) during drought. Plant. Ecol. 2015, 216, 247–262. [Google Scholar] [CrossRef]
- Ministry of Primary Industries. Airbone LiDAR and RGB Aerial Images in the Waitakere Ranges; Ministry of Primary Industries: Wellington, New Zealand, 2016.
- Auckland Council 0.075m Urban Aerial Photos RGB, Waitakere Ranges. Available online: https://data.linz.govt.nz/layer/95497-auckland-0075m-urban-aerial-photos-2017/ (accessed on 12 April 2019).
- Wang, J.; Sammis, T.; Gutschick, V.P.; Gebremichael, M.; Dennis, S.O.; Harrison, R.E. Review of Satellite Remote Sensing Use in Forest Health Studies. Open Geogr. J. 2010, 3, 28–42. [Google Scholar] [CrossRef]
- Pause, M.; Schweitzer, C.; Rosenthal, M.; Keuck, V.; Bumberger, J.; Dietrich, P.; Heurich, M.; Jung, A.; Lausch, A. In Situ/Remote Sensing Integration to Assess Forest Health—A Review. Remote Sens. 2016, 8, 471. [Google Scholar] [CrossRef]
- Townsend, P.A.; Singh, A.; Foster, J.; Rehberg, N.J.; Kingdon, C.C.; Eshleman, K.; Seagle, S.W. A general Landsat model to predict canopy defoliation in broadleaf deciduous forests. Remote Sens. Environ. 2012, 119, 255–265. [Google Scholar] [CrossRef]
- Garrity, S.R.; Allen, C.D.; Brumby, S.P.; Gangodagamage, C.; McDowell, N.G.; Cai, D.M. Quantifying tree mortality in a mixed species woodland using multitemporal high spatial resolution satellite imagery. Remote Sens. Environ. 2013, 129, 54–65. [Google Scholar] [CrossRef]
- Ortiz, S.M.; Breidenbach, J.; Kändler, G. Early Detection of Bark Beetle Green Attack Using TerraSAR-X and RapidEye Data. Remote Sens. 2013, 5, 1912–1931. [Google Scholar] [CrossRef]
- Dotzler, S.; Hill, J.; Buddenbaum, H.; Stoffels, J. The Potential of EnMAP and Sentinel-2 Data for Detecting Drought Stress Phenomena in Deciduous Forest Communities. Remote Sens. 2015, 7, 14227–14258. [Google Scholar] [CrossRef]
- Meng, J.-H.; Li, S.; Wang, W.; Liu, Q.; Xie, S.; Ma, W. Mapping Forest Health Using Spectral and Textural Information Extracted from SPOT-5 Satellite Images. Remote Sens. 2016, 8, 719. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Brooks, D.; Reich, R.; Wheate, R.D. Detection of red attack stage mountain pine beetle infestation with high spatial resolution satellite imagery. Remote Sens. Environ. 2005, 96, 340–351. [Google Scholar] [CrossRef]
- Nitesh, P.; Ismail, R. Discriminating the occurrence of pitch canker fungus inPinus radiatatrees using QuickBird imagery and artificial neural networks. South. For. A J. For. Sci. 2013, 75, 29–40. [Google Scholar] [CrossRef]
- Waser, L.T.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the Potential of WorldView-2 Data to Classify Tree Species and Different Levels of Ash Mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef]
- Wang, H.; Pu, R.; Zhang, Z.; Zhao, Y. Mapping Robinia Pseudoacacia Forest Health Conditions by Using Combined Spectral, Spatial and Textureal Information Extracted from Ikonos Imagery. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 1425–1429. [Google Scholar] [CrossRef]
- Tuominen, J.; Lipping, T.; Kuosmanen, V.; Haapane, R. Remote Sensing of Forest Health. In Geoscience and Remote Sensing; IntechOpen: London, UK, 2009. [Google Scholar]
- Jones, H.G.; Vaughan, R.A. Remote Sensing of Vegetation: Principles, Techniques, and Applications; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Lausch, A.; Erasmi, S.; King, D.; Magdon, P.; Heurich, M. Understanding Forest Health with Remote Sensing -Part I—A Review of Spectral Traits, Processes and Remote-Sensing Characteristics. Remote Sens. 2016, 8, 1029. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Fundamentals, Sensor Systems, Spectral Libraries, and Data Mining for Vegetation; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Oumar, Z.; Mutanga, O. Integrating environmental variables and WorldView-2 image data to improve the prediction and mapping of Thaumastocoris peregrinus (bronze bug) damage in plantation forests. ISPRS J. Photogramm. Remote Sens. 2014, 87, 39–46. [Google Scholar] [CrossRef]
- Eitel, J.U.H.; Vierling, L.A.; Litvak, M.E.; Long, D.S.; Schulthess, U.; Ager, A.A.; Krofcheck, D.J.; Stoscheck, L. Broadband, red-edge information from satellites improves early stress detection in a New Mexico conifer woodland. Remote Sens. Environ. 2011, 115, 3640–3646. [Google Scholar] [CrossRef]
- Kayitakire, F.; Hamel, C.; Defourny, P. Retrieving forest structure variables based on image texture analysis and IKONOS-2 imagery. Remote Sens. Environ. 2006, 102, 390–401. [Google Scholar] [CrossRef]
- Ozdemir, I.; Karnieli, A. Predicting forest structural parameters using the image texture derived from WorldView-2 multispectral imagery in a dryland forest, Israel. Int. J. Appl. Earth Obs. Geoinformation 2011, 13, 701–710. [Google Scholar] [CrossRef]
- Eckert, S. Improved Forest Biomass and Carbon Estimations Using Texture Measures from WorldView-2 Satellite Data. Remote Sens. 2012, 4, 810–829. [Google Scholar] [CrossRef]
- Somers, B.; Verbesselt, J.; Ampe, E.; Sims, N.; Verstraeten, W.; Coppin, P. Spectral mixture analysis to monitor defoliation in mixed-aged Eucalyptus globulus Labill plantations in southern Australia using Landsat 5-TM and EO-1 Hyperion data. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 270–277. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Ghosh, A.; Joshi, P.K.; Koch, B. Assessing the potential of hyperspectral imagery to map bark beetle-induced tree mortality. Remote Sens. Environ. 2014, 140, 533–548. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S. A comparative analysis of high spatial resolution IKONOS and WorldView-2 imagery for mapping urban tree species. Remote Sens. Environ. 2012, 124, 516–533. [Google Scholar] [CrossRef]
- Meddens, A.J.H.; Hicke, J.A.; Vierling, L.A. Evaluating the potential of multispectral imagery to map multiple stages of tree mortality. Remote Sens. Environ. 2011, 115, 1632–1642. [Google Scholar] [CrossRef]
- Lottering, R.; Mutanga, O. Optimising the spatial resolution of WorldView-2 pan-sharpened imagery for predicting levels of Gonipterus scutellatus defoliation in KwaZulu-Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2016, 112, 13–22. [Google Scholar] [CrossRef]
- Ismail, R.; Mutanga, O.; Kumar, L.; Bob, U. Determining the optimal spatial resolution of remotely sensed data for the detection of sirex noctilio infestations in pine plantations in KwaZulu-Natal, South Africa. S. Afr. Geogr. J. 2008, 90, 22–31. [Google Scholar] [CrossRef]
- Aguilar, M.A.; del Mar Saldana, M.; Aguilar, F.J. Assessing geometric accuracy of the orthorectification process from GeoEye-1 and WorldView-2 panchromatic images. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 427–435. [Google Scholar] [CrossRef]
- Van de Voorde, T.; De Genst, W.; Canters, F.; Stephenne, N.; Wolff, E.; Binard, M. Extraction of land use/land cover related information from very high resolution data in urban and suburban areas. Remote Sensing in Transition. In Proceedings of the 23rd Symposium of the European Association of Remote Sensing Laboratories, Ghent, Belgium, 2–5 June 2003; pp. 237–244. [Google Scholar]
- Gao, Y.; Mas, J.F. A comparison of the performance of pixel-based and object-based classifications over images with various spatial resolutions. Online J. Earth Sci. 2008, 2, 27–35. [Google Scholar]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Boggs, G. Assessment of SPOT 5 and QuickBird remotely sensed imagery for mapping tree cover in savannas. Int. J. Appl. Earth Obs. Geoinformation 2010, 12, 217–224. [Google Scholar] [CrossRef]
- Yan, G.; Mas, J.F.; Maathuis, B.H.F.; Xiangmin, Z.; Van Dijk, P. Comparison of pixel-based and object-oriented image classification approaches—A case study in a coal fire area, Wuda, Inner Mongolia, China. Int. J. Remote Sens. 2006, 27, 4039–4055. [Google Scholar] [CrossRef]
- Weih, R.C.; Riggan, N.D. Object-based classification vs. pixel-based classification: Comparative importance of multi-resolution imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, C7. [Google Scholar]
- Whiteside, T.G.; Boggs, G.; Maier, S. Comparing object-based and pixel-based classifications for mapping savannas. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 884–893. [Google Scholar] [CrossRef]
- Desclée, B.; Bogaert, P.; Defourny, P. Forest change detection by statistical object-based method. Remote Sens. Environ. 2006, 102, 1–11. [Google Scholar] [CrossRef]
- Stagakis, S.; Gonzalez-Dugo, V.; Cid, P.; Guillén-Climent, M.; Zarco-Tejada, P.J. Monitoring water stress and fruit quality in an orange orchard under regulated deficit irrigation using narrow-band structural and physiological remote sensing indices. ISPRS J. Photogramm. Remote Sens. 2012, 71, 47–61. [Google Scholar] [CrossRef]
- Gärtner, P.; Förster, M.; Kurban, A.; Kleinschmit, B. Object based change detection of Central Asian Tugai vegetation with very high spatial resolution satellite imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 31, 110–121. [Google Scholar] [CrossRef]
- Sasaki, T.; Imanishi, J.; Ioki, K.; Morimoto, Y.; Kitada, K. Object-based classification of land cover and tree species by integrating airborne LiDAR and high spatial resolution imagery data. Landsc. Ecol. Eng. 2011, 8, 157–171. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X. WorldView-2 satellite imagery and airborne LiDAR data for object-based forest species classification in a cool temperate rainforest environment. In Developments in Multidimensional Spatial Data Models; Springer: Berlin, Germany, 2013; pp. 103–122. [Google Scholar]
- Machala, M. Forest Mapping Through Object-based Image Analysis of Multispectral and LiDAR Aerial Data. Eur. J. Remote Sens. 2014, 47, 117–131. [Google Scholar] [CrossRef]
- Wang, H.; Pu, R.; Zhu, Q.; Ren, L.; Zhang, Z. Mapping health levels of Robinia pseudoacacia forests in the Yellow River delta, China, using IKONOS and Landsat 8 OLI imagery. Int. J. Remote Sens. 2015, 36, 1114–1135. [Google Scholar] [CrossRef]
- Nikolakopoulos, K. Quality assessment of ten fusion techniques applied on Worldview-2. Eur. J. Remote Sens. 2015, 48, 141–167. [Google Scholar] [CrossRef]
- Jovanović, D.; Govedarica, M.; Sabo, F.; Važić, R.; Popović, D. Impact analysis of pansharpening Landsat ETM+, Landsat OLI, WorldView-2, and Ikonos images on vegetation indices. In Proceedings of the Fourth International Conference on Remote Sensing and Geoinformation of the Environment (RSCy2016), Paphos, Cyprus, 4–8 April 2016; p. 968814. [Google Scholar]
- Pontius, J.; Martin, M.; Plourde, L.; Hallett, R. Ash decline assessment in emerald ash borer-infested regions: A test of tree-level, hyperspectral technologies. Remote Sens. Environ. 2008, 112, 2665–2676. [Google Scholar] [CrossRef]
- Lazaridis, D.C.; Verbesselt, J.; Robinson, A.P. Penalised regression techniques for prediction: A case study for predicting tree mortality using remotely sensed vegetation indices. Can. J. For. Res. 2010, 41, 24–34. [Google Scholar] [CrossRef]
- Toomey, M.; Vierling, L.A. Multispectral remote sensing of landscape level foliar moisture: Techniques and applications for forest ecosystem monitoring. Can. J. For. Res. 2005, 35, 1087–1097. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation—Evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Barnes, C.; Balzter, H.; Barrett, K.; Eddy, J.; Milner, S.; Suárez, J.C. Individual Tree Crown Delineation from Airborne Laser Scanning for Diseased Larch Forest Stands. Remote Sens. 2017, 9, 231. [Google Scholar] [CrossRef]
- McMahon, C.A. Remote sensing pipeline for tree segmentation and classification in a mixed softwood and hardwood system. PeerJ 2019, 6, e5837. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.; Gloor, E.; Phillips, O.L.; Aragão, L.E. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171, 104–114. [Google Scholar] [CrossRef]
- Wilkes, P.; Jones, S.D.; Suarez, L.; Haywood, A.; Mellor, A.; Woodgate, W.; Soto-Berelov, M.; Skidmore, A.K. Using discrete-return airborne laser scanning to quantify number of canopy strata across diverse forest types. Methods Ecol. Evol. 2015, 7, 700–712. [Google Scholar] [CrossRef]
- Kim, S.; McGaughey, R.J.; Andersen, H.E.; Schreuder, G. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree species classification using airborne LiDAR–effects of stand and tree parameters, downsizing of training set, intensity normalisation, and sensor type. Silva. Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree-and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Solberg, S.; Næsset, E.; Hanssen, K.H.; Christiansen, E. Mapping defoliation during a severe insect attack on Scots pine using airborne laser scanning. Remote Sens. Environ. 2006, 102, 364–376. [Google Scholar] [CrossRef]
- Kantola, T.; Vastaranta, M.; Yu, X.; Lyytikainen-Saarenmaa, P.; Holopainen, M.; Talvitie, M.; Kaasalainen, S.; Solberg, S.; Hyyppa, J. Classification of defoliated trees using tree-level airborne laser scanning data combined with aerial images. Remote Sens. 2010, 2, 2665–2679. [Google Scholar] [CrossRef]
- Vastaranta, M.; Kantola, T.; Lyytikäinen-Saarenmaa, P.; Holopainen, M.; Kankare, V.; Wulder, M.; Hyyppä, J.; Hyyppä, H. Area-based mapping of defoliation of Scots pine stands using airborne scanning LiDAR. Remote Sens. 2013, 5, 1220–1234. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Stereńczak, K.; Kozak, J. Evaluation of digital terrain models generated in forest conditions from airborne laser scanning data acquired in two seasons. Scand. J. For. Res. 2011, 26, 374–384. [Google Scholar] [CrossRef]
- Balenović, I.; Gašparović, M.; Simic Milas, A.; Berta, A.; Seletković, A. Accuracy assessment of digital terrain models of lowland pedunculate oak forests derived from airborne laser scanning and photogrammetry. Croat. J. For. Eng. J. Theory Appl. For. Eng. 2018, 39, 117–128. [Google Scholar]
- Stereńczak, K.; Ciesielski, M.; Balazy, R.; Zawiła-Niedźwiecki, T. Comparison of various algorithms for DTM interpolation from LIDAR data in dense mountain forests. Eur. J. Remote Sens. 2016, 49, 599–621. [Google Scholar] [CrossRef]
- Jongkind, A.; Buurman, P. The effect of kauri (Agathis australis) on grain size distribution and clay mineralogy of andesitic soils in the Waitakere Ranges, New Zealand. Geoderma 2006, 134, 171–186. [Google Scholar] [CrossRef]
- MPI. Map Data (Shp Files) for PTA Positive Sampling Sites (updated 23 January 2019) and the Natural Range of Kauri Distribution. Regulations for Use and Liability are Stated on the Map. Available online: https://www.kauridieback.co.nz/kauri-maps/ (accessed on 13 February 2020).
- ESRI; World Topographic Map-WMTS Service; Map Data. Sources: Esri, HERE, Garmin, Intermap, INCREMENT P, GEBCO, USGS, FAO, NPS, NRCAN, GeoBase, IGN, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), © OpenStreetMap contributors, GIS User Community. 2020. Available online: https://services.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer (accessed on 2 February 2020).
- LINZ. NZ Topo50. Topographical Map for New Zealand. Imported on 14 April, 2019 from 445 GeoTIFF Sources in NZGD2000/New Zealand Transverse Mercator 2000. Available online: https://www.linz.govt.nz/land/maps/topographic-maps/topo50-maps (accessed on 20 July 2019).
- rapidlasso-GmbH. LAStools. Software Suite for LiDAR Processing. Developed by Martin Insenburg. Available online: https://rapidlasso.com/lastools/ (accessed on 5 May 2019).
- Khosravipoura, A.; Skidmore, A.K.; Isenburg, M. Generating spike-free digital surface models using LiDAR raw point clouds: A new approach for forestry applications. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 104–114. [Google Scholar] [CrossRef]
- Matthew, M.W.; Adler-Golden, S.M.; Berk, A.; Richtsmeier, S.C.; Levine, R.Y.; Bernstein, L.S.; Acharya, P.K.; Anderson, G.P.; Felde, G.W.; Hoke, M.L. Status of atmospheric correction using a MODTRAN4-based algorithm. In Proceedings of the Algorithms for Multispectral Hyperspectral, and Ultraspectral Imagery VI, Baltimore, MA, USA, 29 April–2 May 2013; pp. 199–207. [Google Scholar]
- Laben, C.A.; Brower, B.V. Process for Enhancing the Spatial Resolution of Multispectral Imagery Using Pan-Sharpening. U.S. Patent 6,011,875, 4 January 2000. [Google Scholar]
- Padwick, C.; Deskevich, M.; Pacifici, F.; Smallwood, S. WorldView-2 pan-sharpening. In Proceedings of the ASPRS 2010 Annual Conference, San Diego, CA, USA, 26–30 April 2010. [Google Scholar]
- Li, D.; Ke, Y.; Gong, H.; Li, X. Object-based urban tree species classification using bi-temporal WorldView-2 and WorldView-3 images. Remote Sens. 2015, 7, 16917–16937. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a WorldView-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]
- Auckland Council. Auckland LiDAR 1m DEM (2013). Available online: https://data.linz.govt.nz/x/QUmr7g (accessed on 3 May 2018).
- Adeline, K.; Chen, M.; Briottet, X.; Pang, S.; Paparoditis, N. Shadow detection in very high spatial resolution aerial images: A comparative study. ISPRS J. Photogramm. Remote Sens. 2013, 80, 21–38. [Google Scholar] [CrossRef]
- Globe, D. WorldView-2 Data Sheet. Available online: https://gbdxdocs.digitalglobe.com/docs/worldview-2 (accessed on 1 February 2020).
- DOC. The Foliar Browse Index field manual. In An Update of a Method for Monitoring Possum (Trichosurus vulpecula) Damage to Forest Communities; Department of Conservation: Wellington, New Zealand, 2014. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Gutlein, M.; Frank, E.; Hall, M.; Karwath, A. Large-scale attribute selection using wrappers. In Proceedings of the 2009 IEEE Symposium on Computational Intelligence and Data Mining, Nashville, TN, USA, 30 March–2 April 2009; pp. 332–339. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of chlorophyll a, chlorophyll b, chlorophyll a+ b, and total carotenoid content in eucalyptus leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.; Deering, D. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation, NASA Gsfct Type Report; Texas A&M University: College Station, TX, USA, 1973. [Google Scholar]
- Gamon, J.; Surfus, J. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Metternicht, G. Vegetation indices derived from high-resolution airborne videography for precision crop management. Int. J. Remote Sens. 2003, 24, 2855–2877. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 610–621. [Google Scholar] [CrossRef]
- Kruse, F.; Lefkoff, A.; Boardman, J.; Heidebrecht, K.; Shapiro, A.; Barloon, P.; Goetz, A. The Spectral Image Processing System (SIPS): Software for Integrated Analysis of AVIRIS Data; NASA: Washington, DC, USA, 1992. [Google Scholar]
- Khosravipour, A.; Skidmore, A.K.; Wang, T.; Isenburg, M.; Khoshelham, K. Effect of slope on treetop detection using a LiDAR Canopy Height Model. ISPRS J. Photogramm. Remote Sens. 2015, 104, 44–52. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Gao, B.C. NDWI—A normalised difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Meigs, G.W.; Kennedy, R.E.; Gray, A.N.; Gregory, M.J. Spatiotemporal dynamics of recent mountain pine beetle and western spruce budworm outbreaks across the Pacific Northwest Region, USA. For. Ecol. Manag. 2015, 339, 71–86. [Google Scholar] [CrossRef]
- Verbesselt, J.; Hyndman, R.; Newnham, G.; Culvenor, D. Detecting trend and seasonal changes in satellite image time series. Remote Sens. Environ. 2010, 114, 106–115. [Google Scholar] [CrossRef]
- Zörner, J.; Dymond, J.R.; Shepherd, J.D.; Wiser, S.K.; Jolly, B. LiDAR-based regional inventory of tall trees—Wellington, New Zealand. Forests 2018, 9, 702. [Google Scholar] [CrossRef]
- Goodwin, N.; Coops, N.C.; Stone, C. Assessing plantation canopy condition from airborne imagery using spectral mixture analysis and fractional abundances. Int. J. Appl. Earth Obs. Geoinf. 2005, 7, 11–28. [Google Scholar] [CrossRef]
- Delalieux, S.; Zarco-Tejada, P.J.; Tits, L.; Bello, M.Á.J.; Intrigliolo, D.S.; Somers, B. Unmixing-based fusion of hyperspatial and hyperspectral airborne imagery for early detection of vegetation stress. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2571–2582. [Google Scholar] [CrossRef]
- Maack, J.; Kattenborn, T.; Fassnacht, F.E.; Enßle, F.; Hernández, J.; Corvalán, P.; Koch, B. Modeling forest biomass using Very-High-Resolution data—Combining textural, spectral and photogrammetric predictors derived from spaceborne stereo images. Eur. J. Remote Sens. 2015, 48, 245–261. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, Y.; Zhu, X.; Bai, Y.; Zhang, Y. Micro/Nano-Satellites: Opportunities and Challenges. Aerosp. China. 2016, 17, 6. [Google Scholar]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]

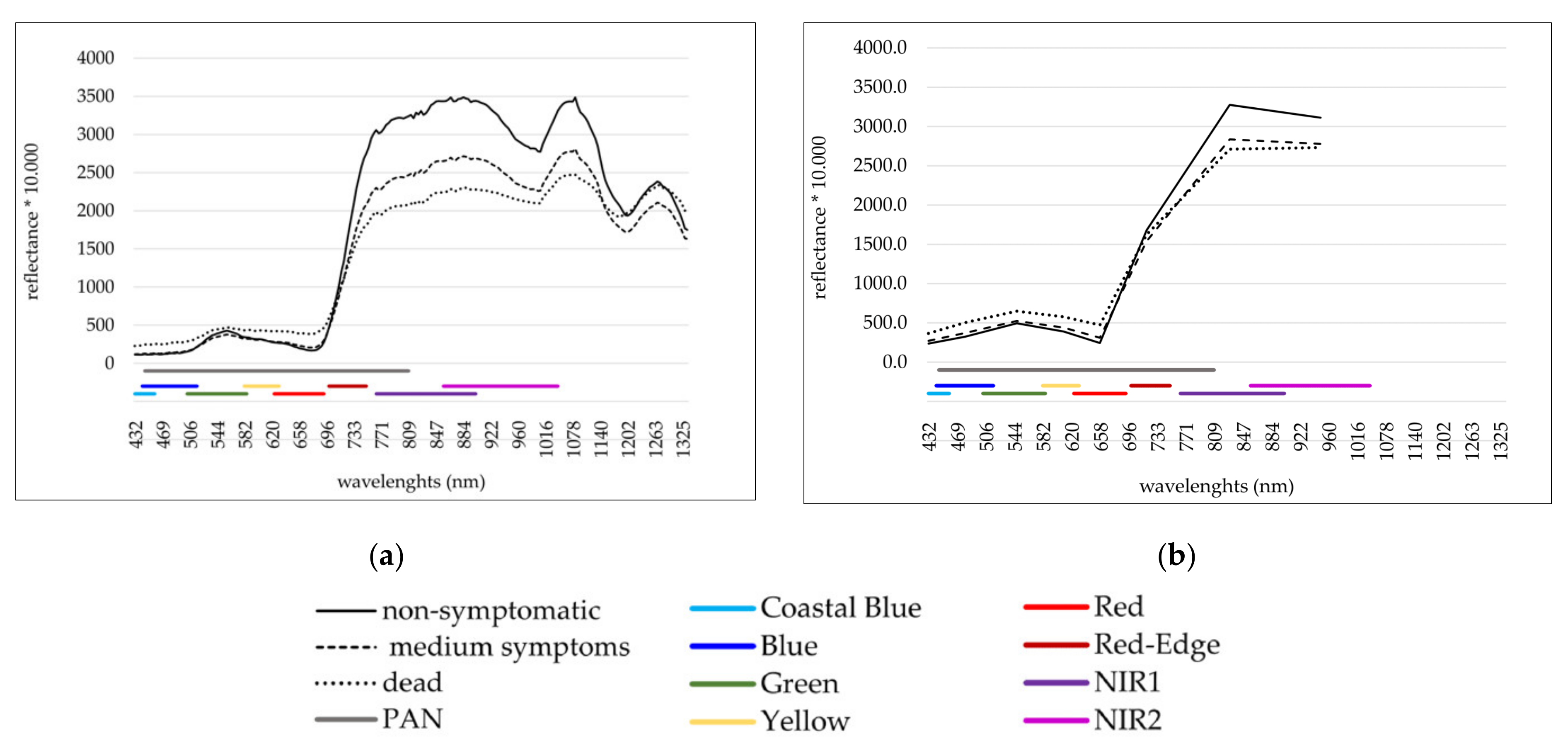

| Nr. | Band Name | Centre Wavelength (nm) | Lower Band Edge (nm) | Upper Band Edge (nm) | Spatial Resolution (m) |
|---|---|---|---|---|---|
| Panchromatic | 627 | 447 | 808 | 0.45 | |
| 1 | Coastal Blue | 427 | 396 | 458 | 1.8 |
| 2 | Blue | 478 | 442 | 515 | 1.8 |
| 3 | Green | 546 | 506 | 586 | 1.8 |
| 4 | Yellow | 608 | 584 | 632 | 1.8 |
| 5 | Red | 659 | 624 | 694 | 1.8 |
| 6 | Red-Edge | 724 | 699 | 749 | 1.8 |
| 7 | NIR1 | 833 | 765 | 901 | 1.8 |
| 8 | NIR2 | 949 | 856 | 1043 | 1.8 |
| Size Class | Crown Diameter | Stress Level | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Level 1 | Level 1.5 | Level 2 | Level 2.5 | Level 3 | Level 4 | Level 5 | |||
| Very Small | 3–4 m | 35 | 39 | 29 | 24 | 25 | 12 | 30 | 194 |
| Small | 4–4.8 m | 15 | 21 | 18 | 14 | 18 | 7 | 24 | 117 |
| Medium | 4.8–12.2 m | 20 | 117 | 126 | 87 | 61 | 10 | 63 | 484 |
| Large | >12.2 m | 29 | 144 | 78 | 26 | 3 | 14 | 294 | |
| Total | 70 | 206 | 317 | 203 | 130 | 32 | 131 | 1089 | |
| Crown Diameter | No. of Crowns | Correlation | RMSE | MAE |
|---|---|---|---|---|
| >3.0 m | 1089 | 0.85 | 0.59 | 0.4 |
| >3.5 m | 1000 | 0.86 | 0.56 | 0.38 |
| >4.0 m | 895 | 0.89 | 0.48 | 0.34 |
| >4.5 m | 825 | 0.9 | 0.45 | 0.32 |
| >5.0 m | 753 | 0.9 | 0.44 | 0.31 |
| Abbr. | Att. Imp. % | Correlation | Crown Statistic | Description | Algorithm | Reference |
|---|---|---|---|---|---|---|
| NDVI | 32.5 | −0.83 | mean | Normalized Difference Vegetation Index | (b7 − b5)/(b7 + b5) 1 | [99,100] |
| RGRI | 25.0 | 0.79 | mean | Red-Green Ratio Index | b5/b3 1 | [101] |
| MNF1 | 12.6 | 0.68 | st. dev. | 1st band of a minimum noise fraction (MNF) transformation | [102] | |
| NDVIg | 9.2 | 0.70 | mean | green NDVI | (b6 − b3)/(b6 + b3)) 1 | [103] |
| b7O3rg | 4.3 | −0.05 | mean | range of a 3 × 3 kernel of band 7 | [104] | |
| Bright | 3.9 | −0.14 | mean | brightness band | (b2 + b3 + b5 + b7)/4 1 | [91] |
| b06 | 3.7 | −0.15 | mean | mean of band 6 (red-edge) | ||
| PO7mn | 2.8 | −0.21 | st. dev. | mean of a 7 × 7 kernel of the panchromatic (PAN) band | [104] | |
| CHM | 5.8 | −0.18 | max | maximum height on a CHM raster (1 m freeze distance) | [84] |
| No Att. | Corr. | RMSE | MAE | |
|---|---|---|---|---|
| 5 level, basic reference scheme (1–2–3–4–5) | 8 | 0.88 | 0.55 | 0.38 |
| 7 level, refined reference scheme (1–1.5–2–2.5–3–4–5) | 9 | 0.89 | 0.48 | 0.34 |
| Classified as --> | Level 1–3 | Level 4, 5 | Total | User’s Accuracy |
|---|---|---|---|---|
| Level 1–3 | 761 | 13 | 774 | 98.3 |
| Level 4, 5 | 19 | 102 | 121 | 84.3 |
| Total | 780 | 115 | 895 | |
| Producer’s Accuracy | 97.6 | 88.7 | 96.4 |
| No Crowns | WV2 Only 1 | WV2 and LiDAR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No Att. | Corr. 2 | RMSE | MAE | No Att. | Corr. 2 | RMSE | MAE | ||
| 7 level reference scheme (1–1.5–2–2.5–3–4–5) | 895 | 9 | 0.89 | 0.48 | 0.34 | 11 | 0.92 | 0.43 | 0.31 |
| First symptoms (1–1.5–2–2.5–3) | 774 | 9 | 0.72 | 0.37 | 0.28 | 6 | 0.76 | 0.34 | 0.26 |
| Crown Statistics | First Symptoms | 7-Level Symptoms | Description | Source | |||
|---|---|---|---|---|---|---|---|
| Imp. % | Corr. | Imp. % | Corr. | ||||
| NDVI75 | mean | 31.8 | −0.67 | 33.3 | −0.83 | normalized difference vegetation index (b7 −b5)/(b7 + b5) 2 | [99,100] |
| R_max_P50 | 21.5 | 0.61 | ratio maximum height and 50th percentile | [83] | |||
| intensity | mean | 26.9 | −0.60 | 16.8 | −0.71 | intensity values | [83] |
| MNF_b1 | st. dev. | 10.1 | 0.38 | 14.1 | 0.68 | 1st band of an MNF transformation | [102,105] |
| Pan_k7mn | st. dev. | 7.8 | −0.22 | mean of a 7 × 7 kernel on the PAN band | [104] | ||
| Cg1Cor7 | mean | 13.3 | −0.35 | 4.5 | −0.43 | correlation on a 7 × 7 kernel CHM 1 | [104] |
| CHM | max. | 10.0 | 0.14 | 3.8 | −0.18 | maximum crown height from a CHM | [83] |
| b7_k3Rg | mean | 3.5 | −0.05 | range of a 3 × 3 kernel on band 7 | [104] | ||
| Bright | mean | 2.4 | −0.14 | brightness layer (b2 + b3 + b5 + b7)/4 2 | [91] | ||
| Attribute Type | Min. Crown Diameter | No. Attributes | Overall Accuracy | Level 4 and 5 (Dying and Dead Trees) | |
|---|---|---|---|---|---|
| User’s Acc. | Prod. Acc. | ||||
| WV2 | 3 m | 5 | 94.9 | 73.6 | 90.2 |
| WV2 and LiDAR | 3 m | 7 | 96.7 | 87.1 | 90.4 |
| WV2 | 4 m | 5 | 96.4 | 84.3 | 88.7 |
| WV2 & LiDAR | 4 m | 6 | 97.2 | 86.0 | 92.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meiforth, J.J.; Buddenbaum, H.; Hill, J.; Shepherd, J.D.; Dymond, J.R. Stress Detection in New Zealand Kauri Canopies with WorldView-2 Satellite and LiDAR Data. Remote Sens. 2020, 12, 1906. https://doi.org/10.3390/rs12121906
Meiforth JJ, Buddenbaum H, Hill J, Shepherd JD, Dymond JR. Stress Detection in New Zealand Kauri Canopies with WorldView-2 Satellite and LiDAR Data. Remote Sensing. 2020; 12(12):1906. https://doi.org/10.3390/rs12121906
Chicago/Turabian StyleMeiforth, Jane J., Henning Buddenbaum, Joachim Hill, James D. Shepherd, and John R. Dymond. 2020. "Stress Detection in New Zealand Kauri Canopies with WorldView-2 Satellite and LiDAR Data" Remote Sensing 12, no. 12: 1906. https://doi.org/10.3390/rs12121906
APA StyleMeiforth, J. J., Buddenbaum, H., Hill, J., Shepherd, J. D., & Dymond, J. R. (2020). Stress Detection in New Zealand Kauri Canopies with WorldView-2 Satellite and LiDAR Data. Remote Sensing, 12(12), 1906. https://doi.org/10.3390/rs12121906







