1. Introduction
Machine learning—a method where a computer discovers rules to execute a data processing task, given training examples—can generally be divided into two categories: Shallow learning and deep learning methods [
1]. Deep learning uses many successive layered representations of data (i.e., hundreds of convolutions/filters), while shallow learning typically uses one or two layered representations of the data [
1]. Deep learning has shown great promise for tackling many tasks such as image recognition, natural language processing, speech recognition, superhuman Go playing, and autonomous driving [
1,
2,
3].
In remote sensing, shallow learning for landcover classification is a well-established method. Countless studies have used random forest [
4], support vector machine (SVM) [
5], boosted regression trees [
6,
7], and many other algorithms to classify landcover from Earth observation and remote sensing data. These algorithms typically work on a pixel-level or object-level. Pixel-level algorithms extract the numerical value of the remote sensing inputs (i.e., vegetation index value, radar backscatter, relative elevation) and match that to a known landcover class (i.e., forest). With this numerical data, a shallow learning model can be built through methods such as kernel methods/decision boundaries [
8], decision trees [
9], and gradient boosting [
10]. These models can then be used to predict the landcover class of an unknown pixel with a unique numerical representation. In a somewhat different fashion, object-based shallow learning methods use features, such as texture, edge detection, and contextual information [
11,
12], of a given area to classify landcover. Once segmented (using k-Nearest-Neightbours, spectral clustering, etc.), these object-based algorithms often use similar shallow learning techniques to predict a given landcover class [
13]. Both pixel- and object-based methods are widely used and both have their pros and cons [
12].
The use of deep learning in remote sensing is a more recent development and it is gaining popularity [
14,
15,
16,
17,
18]. Of the many types of deep learning approaches that exist, we have selected a segmentation convolutional neural network (CNN) method. In the literature, there are two commonly used CNN models for landcover classification [
17]: Patch-based CNNs [
19] and fully convolutional networks (FCNs) [
20]. In the patch-based CNNs, a remote sensing scene is divided into several patches and a typical CNN model is applied to predict a single label for the center of each patch. On the other hand, in FCN models, the fully connected layers are replaced with convolutional layers. Therefore, FCNs can maintain a two-dimensional output image structure and this increases its training efficiency. Typically, CNN architectures can incorporate multiple stacked layers as inputs, which can be from different sources (i.e., optical and synthetic aperture radar (SAR) sensors) [
21]. These training patches then undergo a series of filters/convolutions/poolings to extract hierarchical features from the data, such as low-level edge and curved features, along with higher level textural or unique spectral features. Our specific segmentation CNN architecture used in this study is known as a U-Net style [
22].
In this study, we focus on the prediction of wetland class (bog, fen, marsh, and swamp), as defined by the Canadian Wetland Classification System [
23], and the prediction of upland and open water classes to fill out the classified map. Uses of spatially-explicit wetland inventories have become even more imperative lately due to their increased applications for carbon pricing, ecosystem services, and biodiversity conservation efforts [
24,
25,
26]. Wetlands store a much higher relative percentage of soil carbon when compared to other vegetated habitats such as forest or agriculture [
24]. They are vital for hydrological services, such as flood remediation and filtering of freshwater [
26]. Wetland carbon and hydrological processes are also both important in the context of climate change, as wetlands have the ability to store or release vast amounts of carbon [
27,
28] and may be able to mitigate changes to hydrology caused by variations in precipitation [
29]. These represent some of the reasons why the Government of Alberta seeks to update the current provincial wetland inventory, the Alberta Merged Wetland Inventory [
30], to yield a consistent baseline state of the environment product, which can be leveraged for monitoring and reporting purposes, and to aid policy-makers in the strategic management of wetlands. The update will follow the Canadian Wetland Classification System (CWCS) wetland classes, whilst simultaneously conforming to defined minimum mapping unit and classification accuracy criteria. The machine learning methods investigated in the current study demonstrate high relevance in the creation of such a product.
Spatial wetland inventories such as the Alberta Merged Wetland Inventory need to consider the class of wetland, because certain wetland classes are more sensitive to certain types of disturbance. For example, fens—peatlands with flowing water—are very sensitive to anthropogenic pressure, i.e., new roads and linear features acting like dams to moving water [
31]. Bogs—peatlands with stagnant water—are more sensitive to climate-driven changes in precipitation patterns because bogs are recharged through precipitation [
32]. Marshes and swamps are also sensitive to climate change due to their reliance on predictable seasonal flooding cycles [
33].
Spatial wetland inventories at a country or provincial scale [
30,
31,
32,
33,
34] are not new, but having data that are reliable for land management and land planning decisions is a challenge. In Canada, mapping of wetlands via remote sensing is a well-studied topic [
35]. Initially, inventories were typically built through aerial image interpretation [
36]. While accurate, this methodology is usually very time consuming and costly. Given Canada’s commitment to, and involvement with, synthetic aperture radar (SAR) data, many studies have used SAR to map and monitor wetlands [
37,
38,
39,
40,
41,
42,
43,
44,
45] with varying degrees of success. It appears SAR data are most useful for monitoring the dynamics of wetlands. Other studies and projects have used moderate resolution optical data such as Landsat or Sentinel-2 to generate wetland inventories [
30,
46,
47]. Most modern approaches to large-scale wetland inventories utilize a fusion of data such as SAR and optical [
34,
39,
48] and, ideally, SAR, optical, plus topographic information [
6,
7,
49]. Theoretically the fusion of SAR, optical, and topographic information should give the most information on wetlands and wetland class because: (1) SAR is sensitive to the physical structure of vegetation and can detect the dynamic nature of wetlands with a rich time series stack; (2) optical data can capture variations in vegetation type and vegetation productivity, as it is sensitive to the molecular structure of vegetation; and (3) topographic information can provide data about hydrological patterns which drive wetland formation and function.
It is our understanding that all of the machine learning studies listed in the paragraph above used shallow learning methods, such as random forest, SVM, or boosted regression trees. It appears that distinguishing wetland class with remote sensing data and shallow machine learning is still a difficult task. This is likely due to the fact that one wetland class (i.e., fen) can have different vegetation types—forested, shrubby, graminoid [
35]. Additionally, wetlands of different classes can have identical vegetation and vegetation structure. These types are then only distinguished through their below-ground hydrological patterns. Finally, wetland classes do not have a defined boundary, since they gradually transition into another class or upland habitat [
50]. This makes spatially explicit inventories inherently inaccurate because a hard boundary must be identified. These issues with wetland and wetland class mapping are best summed up in
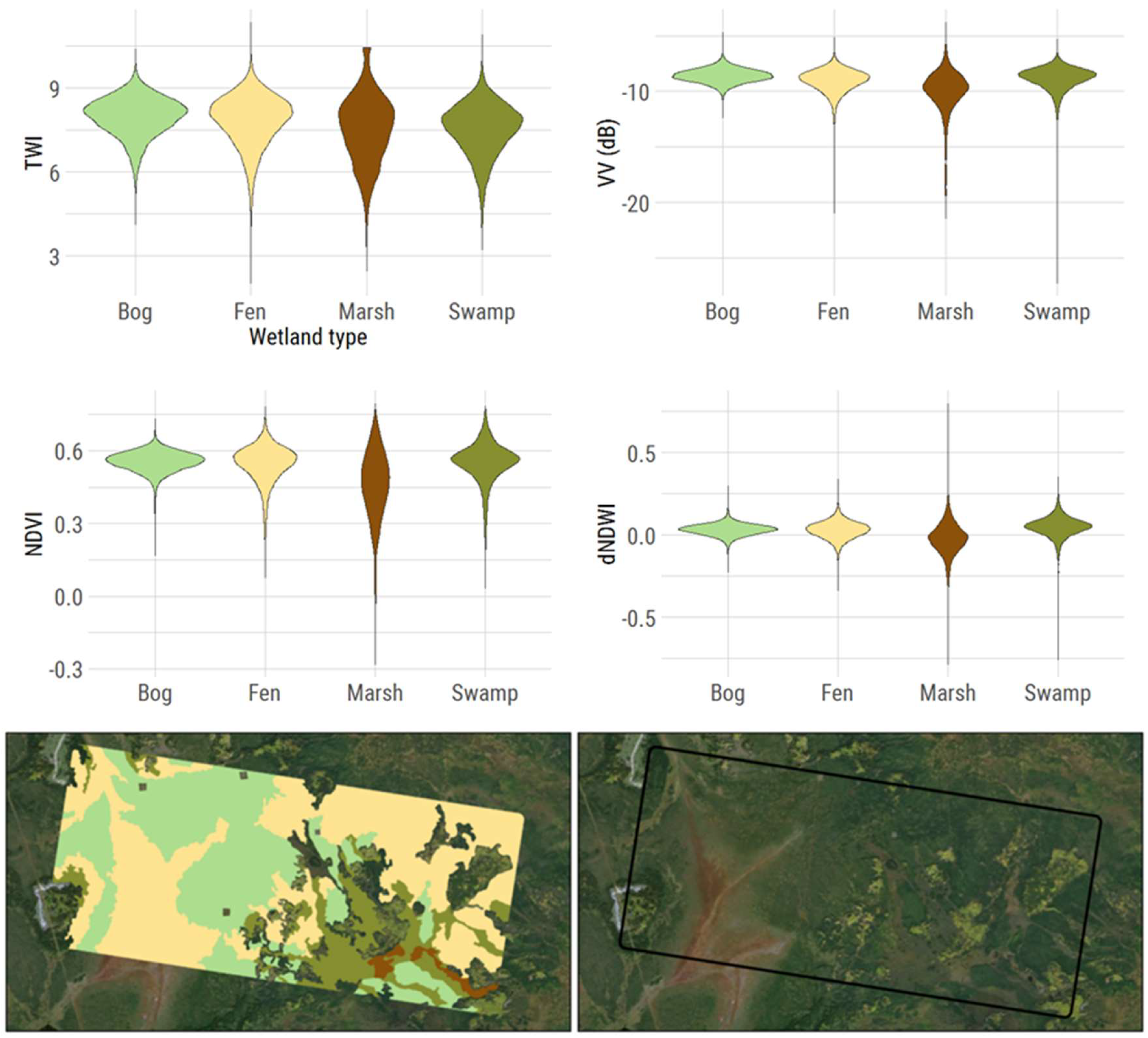
Figure 1. Given three different data sources (SAR, optical, and topographic) with static and multi-temporal remote sensing measures, the four wetland classes do not show any noticeable differences on a pixel level (top panel of
Figure 1). The violin plots show the distribution of numerical values for all four wetland classes (essentially a vertical histogram by class). Marshes may show a wider distribution of values, but fens, bogs, and swamps are almost identical. In the bottom panel of
Figure 1, even the visual identification of these wetland classes with high resolution imagery is difficult. Fens, in the bottom right of the image, can be seen visually (flow lines and lighter tan color), but then fens appear very different in the top right corner (dark green color and appear to be treed).
With the known difficulty of wetland classification with shallow learning (
Figure 1), we believe wetland class mapping is the perfect candidate for deep learning and CNNs. In practice, CNNs trained on a patch-level learn low- and high-level features from the remote sensing data. For example, waterline edges which delineate marshes and open water may only need simple edge detection convolution filters, while fens and bogs may be differentiated by subtle variations in texture or color (i.e., visible flow lines in fens). Within the last couple of years, a number of studies have attempted to use deep learning for wetland mapping in Canada over small areas and have achieved promising results when compared to alternative shallow learning methods [
20,
51,
52].
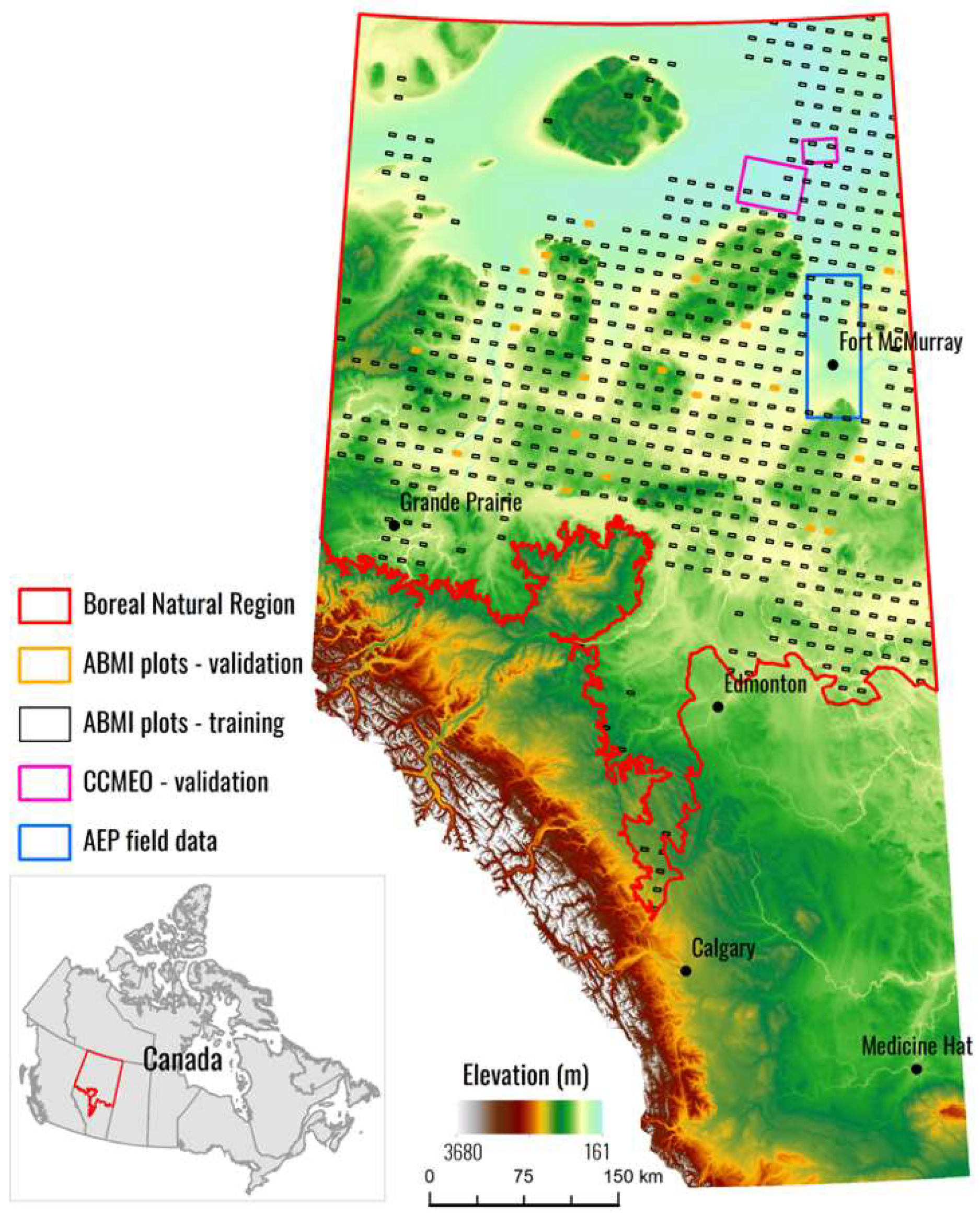
With the current status of machine learning and the history of Canadian wetland mapping in mind, we propose a simple goal for this study: To compare deep learning (CNN) classifications with shallow learning (XGBoost) classifications for wetland class mapping over a large region of Alberta, Canada (397,958 km2) using the most up-to-date, open source, fusion of data sources from Sentinel-1 (SAR), Sentinel-2 (optical), and the Advanced Land Observing Satellite (ALOS) digital elevation model (DEM) (topographic). To reach a strong conclusion, we plan to validate our results against three validation data sets: Two generated from photo interpretation and one field validation data set. These results will guide future large-scale spatial wetland inventory efforts in Canada. If deep learning techniques are found to generate better products, new wetland inventories projects should adopt deep learning workflows. If shallow learning methods still produce comparable results, they should continue to be used in wetland inventory workflow; however, deep learning architectures should still remain an active area of research with regard to methods for wetland mapping.
3. Results
The wetland classification results from the CNN and XGB models were compared against the photo-interpreted validation data sets (see
Table 3). The CNN model showed an accuracy of 81.3%, Kappa statistic of 0.57, and mean F1-score of 0.56, while the XGB model showed an accuracy of 75.6%, 0.49 Kappa statistic, and mean F1-score of 0.52 when compared to the ABMI plot data. The CNN model showed an accuracy of 80.3%, Kappa statistic of 0.52, and mean F1-score of 0.59, while the XGB model showed an accuracy of 72.1%, 0.41 Kappa statistic, and mean F1-score of 0.52 when compared to the CCMEO data. In terms of overall accuracy, the CNN model was 5.7% more accurate than the XGB model when compared to the ABMI data, and 8.2% more accurate when compared to the CCMEO data (
Table 3). Overall accuracy with uplands excluded (i.e., just wetland classes) was 60.0% for the CNN model and 45.6% for the XGB model. Full results of the accuracy assessment can be in
Appendix A.
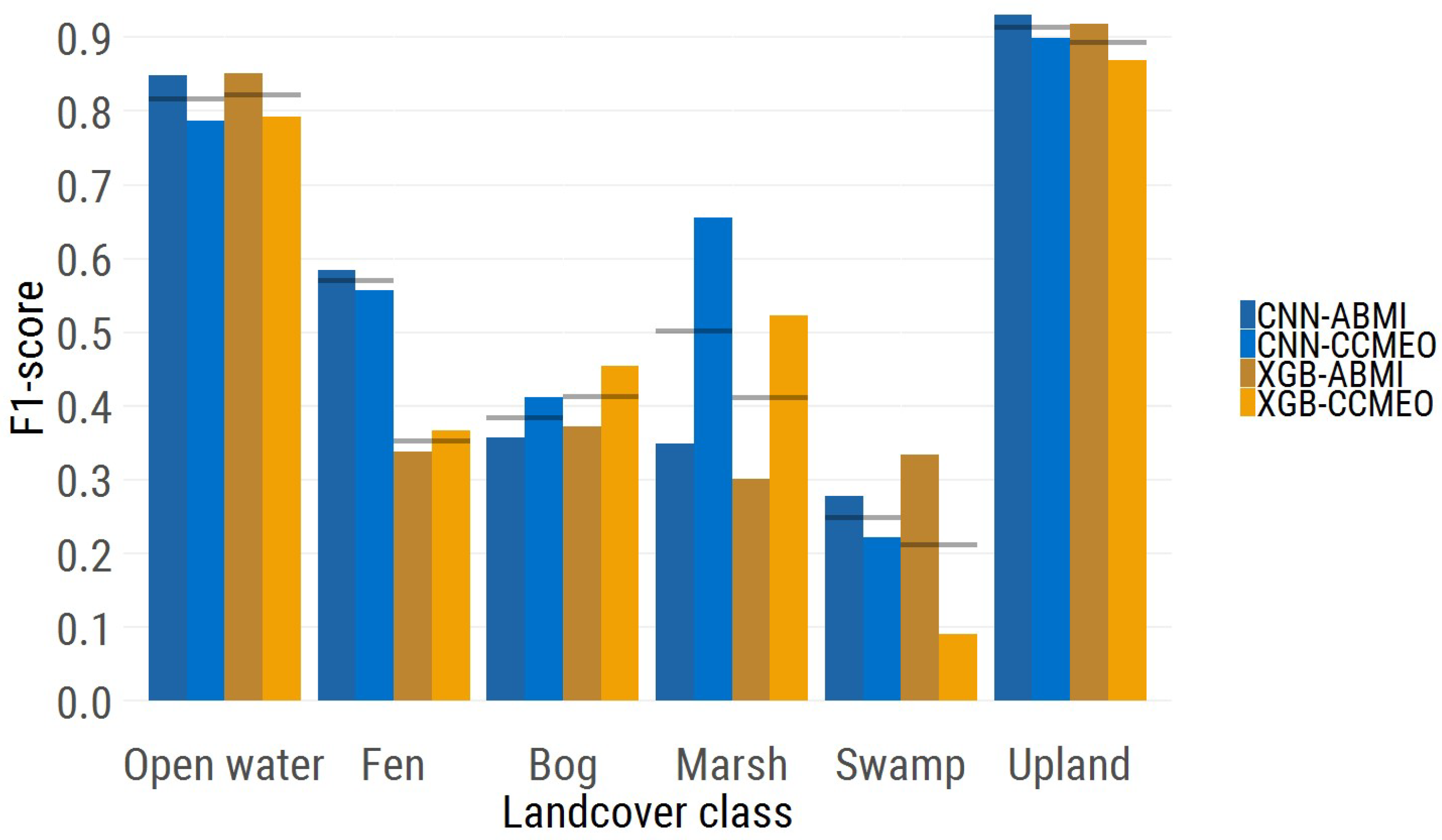
The per-class F1-scores for the ABMI and CCMEO data are seen in
Figure 3 (blue showing the F1-score for the CNN model and orange the F1-score for the XGB model). The open water class shows almost equal F1-scores between the two models. The fen class F1-score is much higher in the CNN model (0.57) than the XGB model (0.35), while the bog score proved to be slightly higher in the XGB model. The marsh and swamp class both had a higher F1-score in the CNN model, although the F1-score were pretty poor for the swamp class (0.25 and 0.21 scores). Finally, the most numerous class, upland, showed a slightly higher F1-score in the CNN model.
When compared to the field validation data (AEP data), both products show a 50% overall accuracy on 22 points. In the CNN data, fen is correctly identified in six sites, but fen is also incorrectly predicted in five out of the seven bog sites (
Table 4). In the XGB data, fen is predicted correctly in two of the six sites. XGB bog is more accurate than the CNN model, being correct in three of the seven sites. Open water appears to be similarly predicted in both products, with five out of nine sites being accurately predicted. Open water was incorrectly predicted as swamp or marsh in both models.
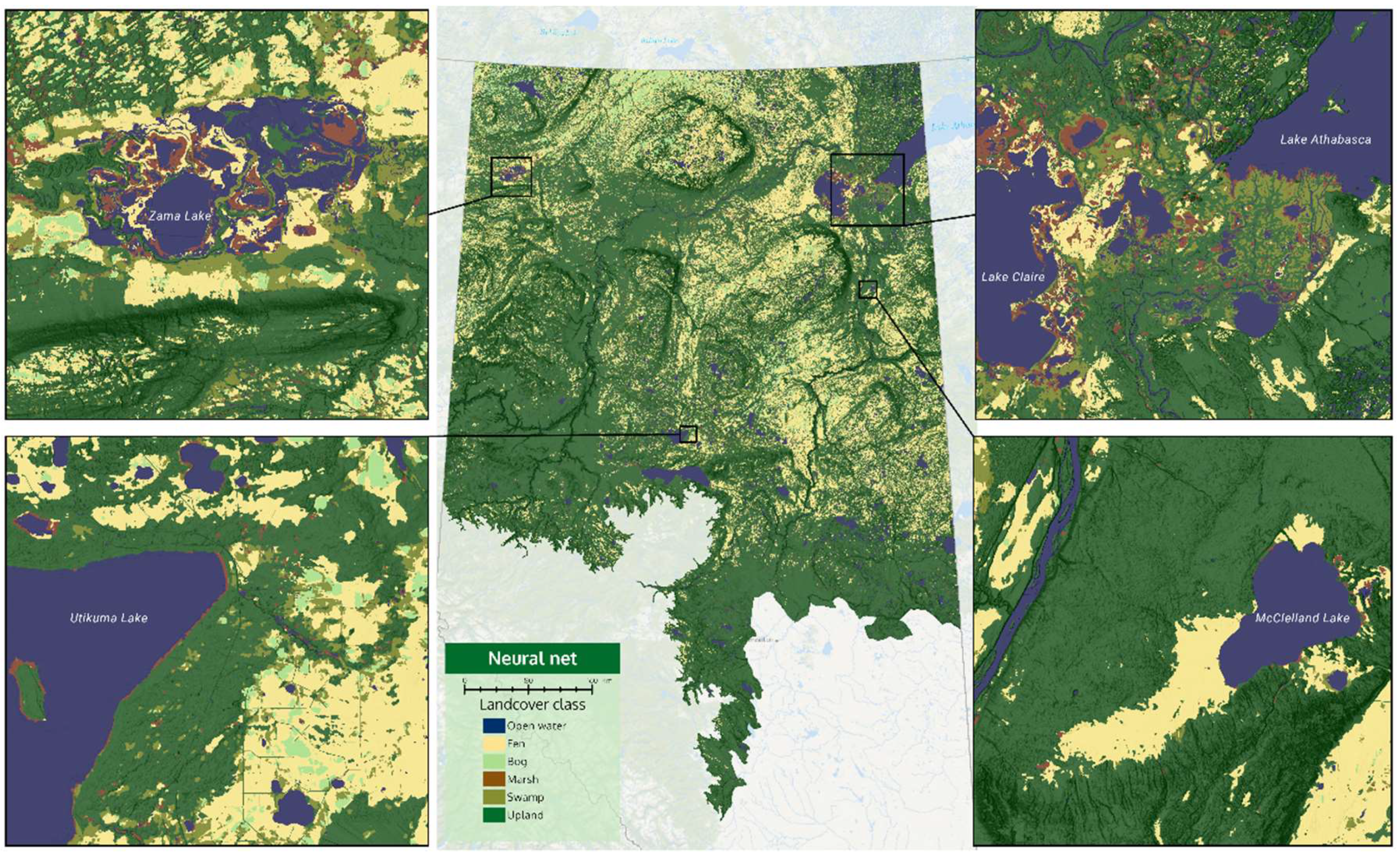
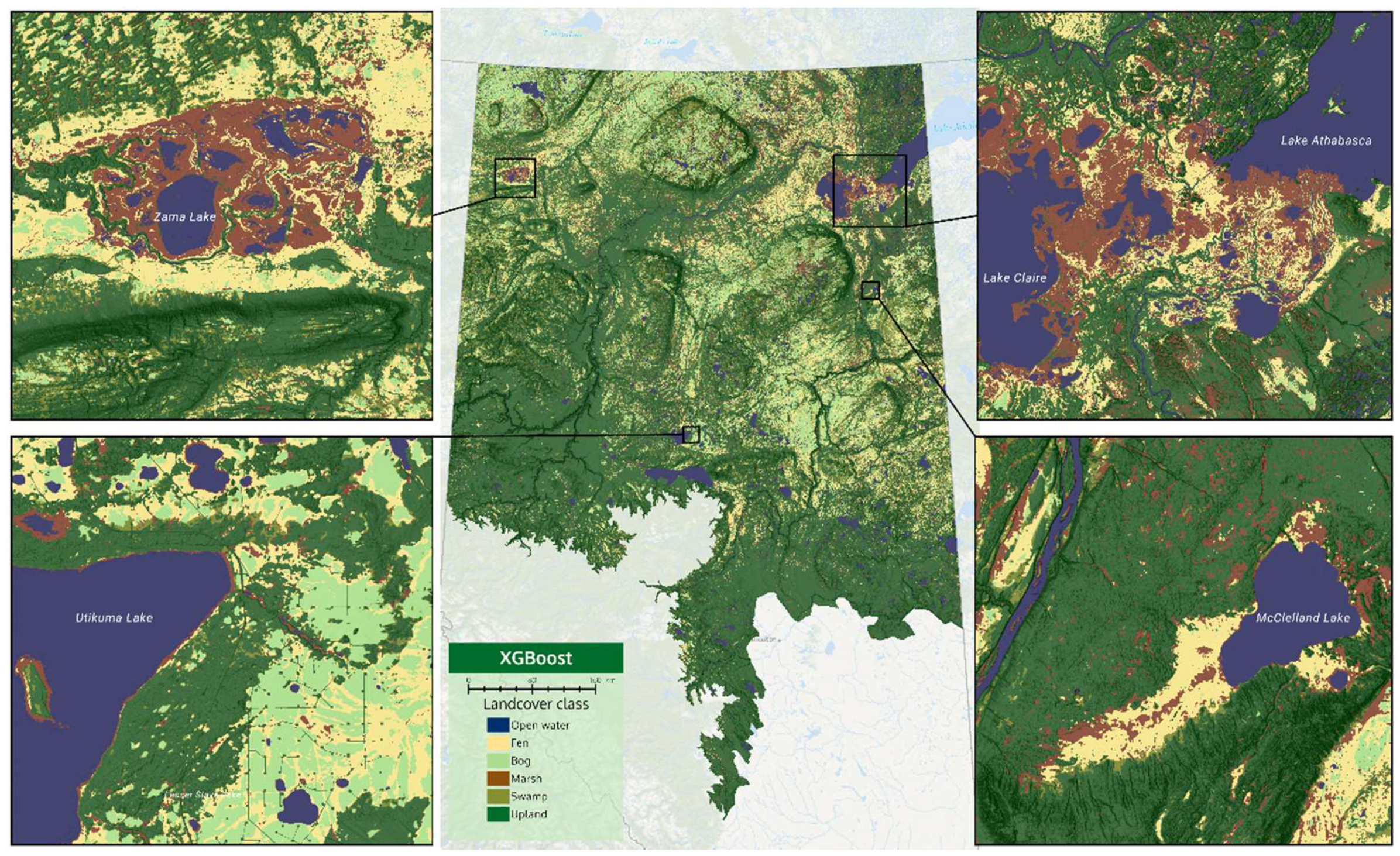
Visual results of the two products can be seen in
Figure 4 and
Figure 5. The four insets zoom into important wetland habitats in Alberta, which are described in the figure captions. An interactive side-by-side comparison of the two products can also be seen on a web map via this link (
https://abmigc.users.earthengine.app/view/cnn-xgb). Additionally, the CNN product can be downloaded via this link (
https://bit.ly/2X3Ao6N). We encourage readers to assess the visual aspects of these two products.
With reference to
Figure 4 and
Figure 5, and the web map, it is evident that CNN predictions produce smoother boundaries between wetland classes. The XGB model appears to produce speckled marsh at some locations (see McClelland fen—bottom-right inset of
Figure 5). One of the major differences is the amount of bog versus fen predicted in the north-west portion of the province. The XGB model predicts massive areas of bog with small ribbons of fen, while the CNN model predicts about equal parts of fen and bog in these areas. Overall, the CNN model predicts: 4.8% open water, 19.0% fen, 3.0% bog, 1.0% marsh, 4.0% swamp, and 68.2% upland. The XGB model predicts: 4.4% open water, 10.8% fen, 9.3% bog, 5.0% marsh, 10.3% swamp, and 60.2% upland.
4. Discussion
This study produced two large-scale wetland inventory products using a fusion of open-access satellite data, and machine learning methods. The two machine learning approaches that were compared—convolutional neural networks and XGBoost—demonstrate a decent ability to predict wetland classes and upland habitat across a large region. Some wetland classes, such as bog and swamp, proved to be much harder to map. This is made clear in the relative F1-scores of the wetland classes (
Figure 3). In the comparisons to the photo-interpretation validation data sets (
Table 3), it is clear that the CNN model outperforms the XGB model in terms of overall accuracy, Kappa statistic, and per-class F1-score. As expected, the ABMI validation data proved to be slightly more accurate than the CCMEO validation data by 1–3%. This is still surprisingly close, given that the CCMEO data were completely independent from the model training process. The ABMI data were also an independent validation data set, but it is a subset of the data used to train the model. The CCMEO validation demonstrated a larger gap between the two models, with the CNN outperforming the XGB model by 8.2%. The ABMI data still showed a large, 5.7%, difference. The gap between the models is even more apparent when removing uplands, as the CNN model was 60.0% accurate, while the XGB model was 45.6% accurate. In terms of model development, both models took similar amounts of time to optimize, train, and predict (
Table 2).
When comparing the products to field data, the results do not seem to be as promising. Both products had a 50% overall accuracy over 22 field sites. With reference to the field data, the CNN model clearly over-predicts fen, with 11 of the 13 wetland classes being predicted as fen, while the XGB model does not appear to have much ability to distinguish bog from fen (only 6 out of the 13 predicted correctly). We fully expect the overall accuracy of field data to go up if upland classes were included, but the main goal of this landcover classification was to map wetland classes. We believe less weight should be assigned to this accuracy assessment, given that is was just 22 points and it was just a small portion of the overall study area. Nevertheless, this does raise the question about how well landcover classifications actually match with what is seen on the ground. This may be something to test further when a larger field data set can be acquired. Right now, we cannot conclude if this is a real issue or a result of the small sample size. In the end, on the ground, wetland class is what actually matters for policy and planning, not what a photo-interpreter sees.
It appears that contextual information, texture, and convolutional filters help the CNN model better predict wetland class. The fen class is predicted much more accurately in the CNN model—0.57 versus 0.35 F1-score. This may be due to the parallel flow lines seen in many fen habitats (
Figure 1), which can potentially be captured by certain convolutional filters. Marshes are also predicted more accurately in the CNN model. Here the CNN model likely uses contextual information about marshes, given marshes often surround water bodies. Visually, the CNN model appears to produce more ecologically meaningful wetland class boundaries. Boreal wetland classes are generally large, complex habitats, which can have multiple different vegetation types within one class. A large fen can be treed on the edges, then transition into shrub and graminoid fens at the center. Overall, it appears that the natural complexities of wetlands are better captured with a CNN model than a traditional pixel-based shallow learning (XGB) method. It is possible that an object-based wetland classification may also capture these natural complexities, but that is a question for future studies, as it was not in the scope of this project. We would also like to point out that the reference CNN was not subjected to rigorous optimization, and it is likely that there is still room for improvement in this model. This does tell us, though, that a naïve implementation of a CNN does outperform traditional shallow learning approaches for large-scale wetland mapping. Future work should focus on the ideal inputs for CNN wetland classification (i.e., spectral bands only, spectral bands + SAR, or handcrafted spectral features + SAR + topography).
Other studies have attempted similar deep learning wetland classifications in Canadian Boreal ecosystems. Pouliot et al. [
51] tested a CNN wetland classification over a similar region in Alberta using Landsat data and reported 69% overall accuracy. Mahdianpari et al. [
52] achieved a 96% wetland class accuracy in Newfoundland with RapidEye data and an InceptionResNetV2 algorithm. Mohammadimanesh et al. [
20] reported a 93% wetland class accuracy again in Newfoundland using RADARSAT-2 data and a fully convolutional neural network. All of these demonstrated that deep learning results outperform other machine learning methods such as random forest. Non-neural network methods, such as the work in Amani et al. [
83], reported a 71% wetland class accuracy across all of Canada. Another study by Mahdianpari et al. [
34] achieved 88% wetland class accuracy using an object-based random forest algorithm across Newfoundland. In this study, the comparison between CNNs and shallow learning methods comes to the same conclusion as other recent studies; CNN/deep learning algorithms lead to better wetland classification results. The CNN product produced in this study does not achieve accuracies over 90% as seen in some smaller scale studies, likely because it is predicted across such a large area (397,958 km
2). When compared to other large-scale wetland predictions, it appears to be one of the more accurate products, and thus it may prove useful for provincial/national wetland inventories. It also comes with the benefit of being produced with completely open-access data from Sentinel-1, Sentinel-2, and ALOS, and thus it can be easily updated to capture the dynamics of a changing Boreal landscape.
Large-scale wetland/landcover mapping in Canada seems to be converging towards a common methodology. Most studies are now using a fusion of remote sensing data from SAR, optical, and DEM products [
6,
7,
34,
39,
49]. The easiest way to access provincial/national-scale data appears to be through Google Earth Engine; thus, many studies use Sentinel-1, Sentinel-2, Landsat, ALOS, or SRTM data [
6,
7,
34,
83]. Finally, many machine learning methods have been tested, but it appears that convolutional neural network frameworks produce better, more accurate wetland/landcover classifications [
51,
52]. This work contributes to these previous studies by confirming the value of CNNs. It also contributes to the greater goal of large-scale wetland mapping by demonstrating the ability to produce an accurate wetland inventory with a CNN and open-access satellite data.