Co-Selection of Heavy Metal and Antibiotic Resistance in Soil Bacteria from Agricultural Soils in New Zealand
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Samples
2.2. Plate Culture
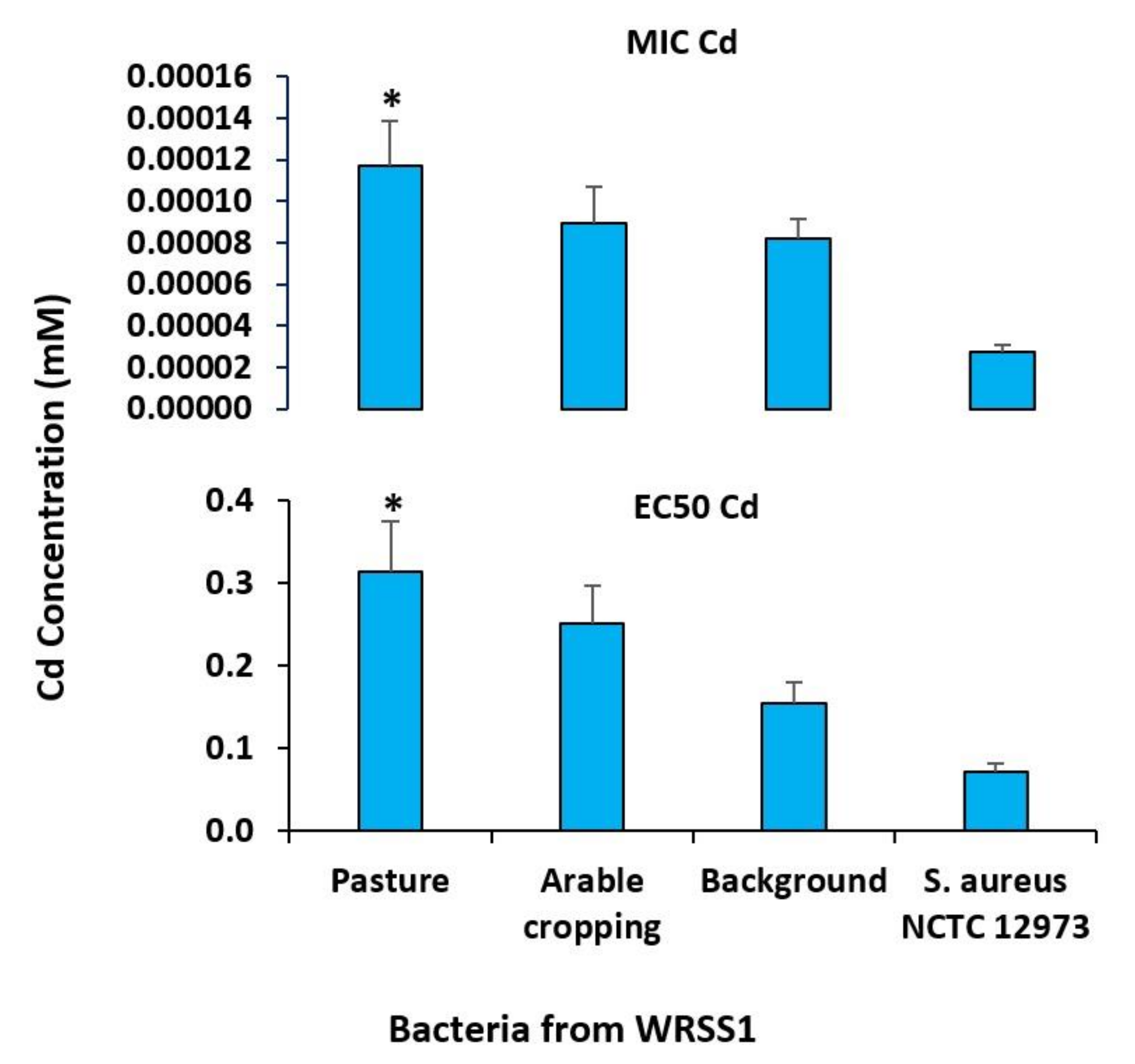
2.3. Minimum Inhibitory Concentration (MIC) and EC50 Determination
2.4. Pollution Induced Community Tolerance (PICT) Analysis
2.5. Terminal Restriction Fragment Length Polymorphism (TRFLP)
2.6. Genetic Mobility of Cd Resistance by Horizontal Gene Transfer
2.7. Statistical Analysis
3. Results
3.1. Physicochemical Properties of Soil
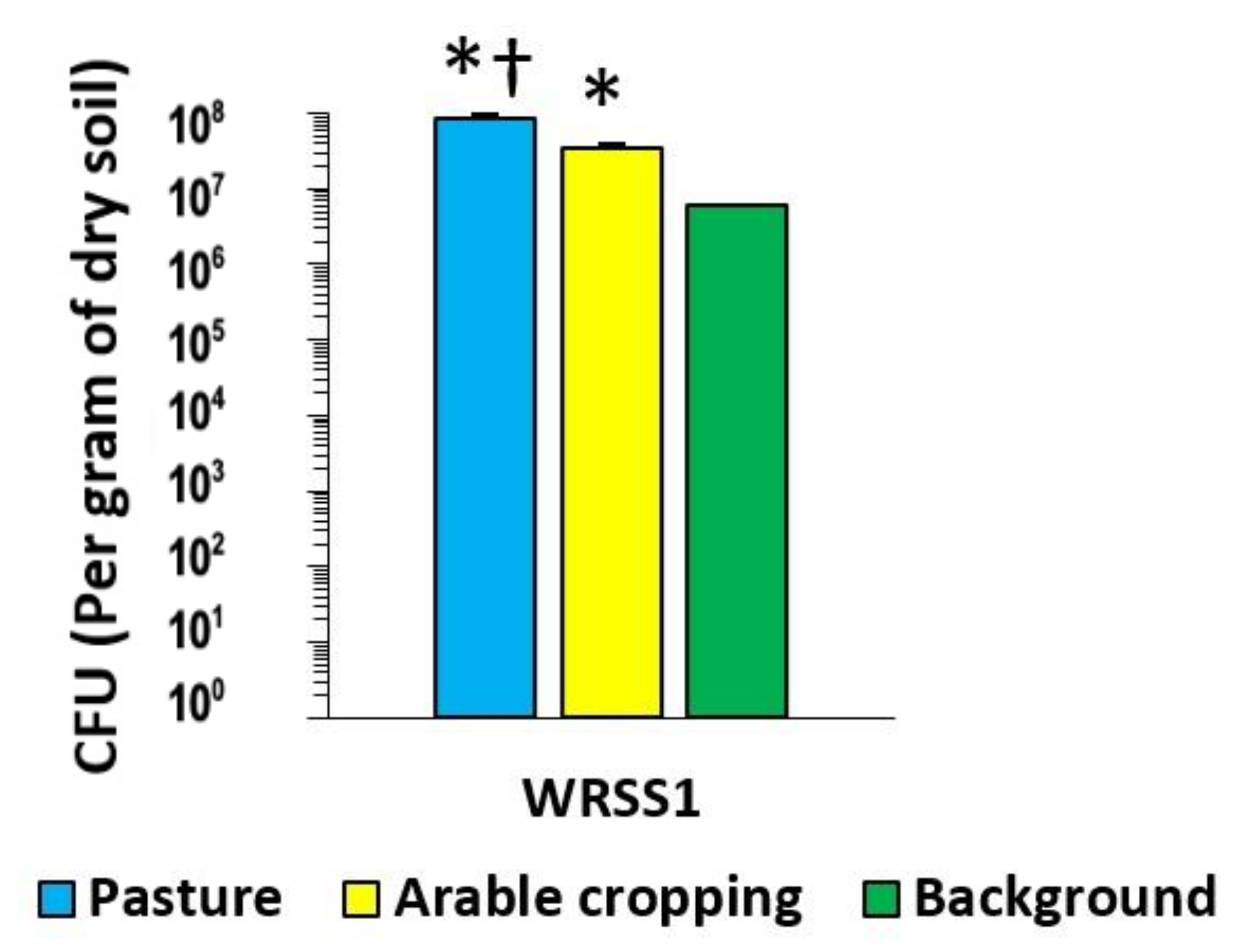
3.2. Bacteriological Characterisation of Soil Samples
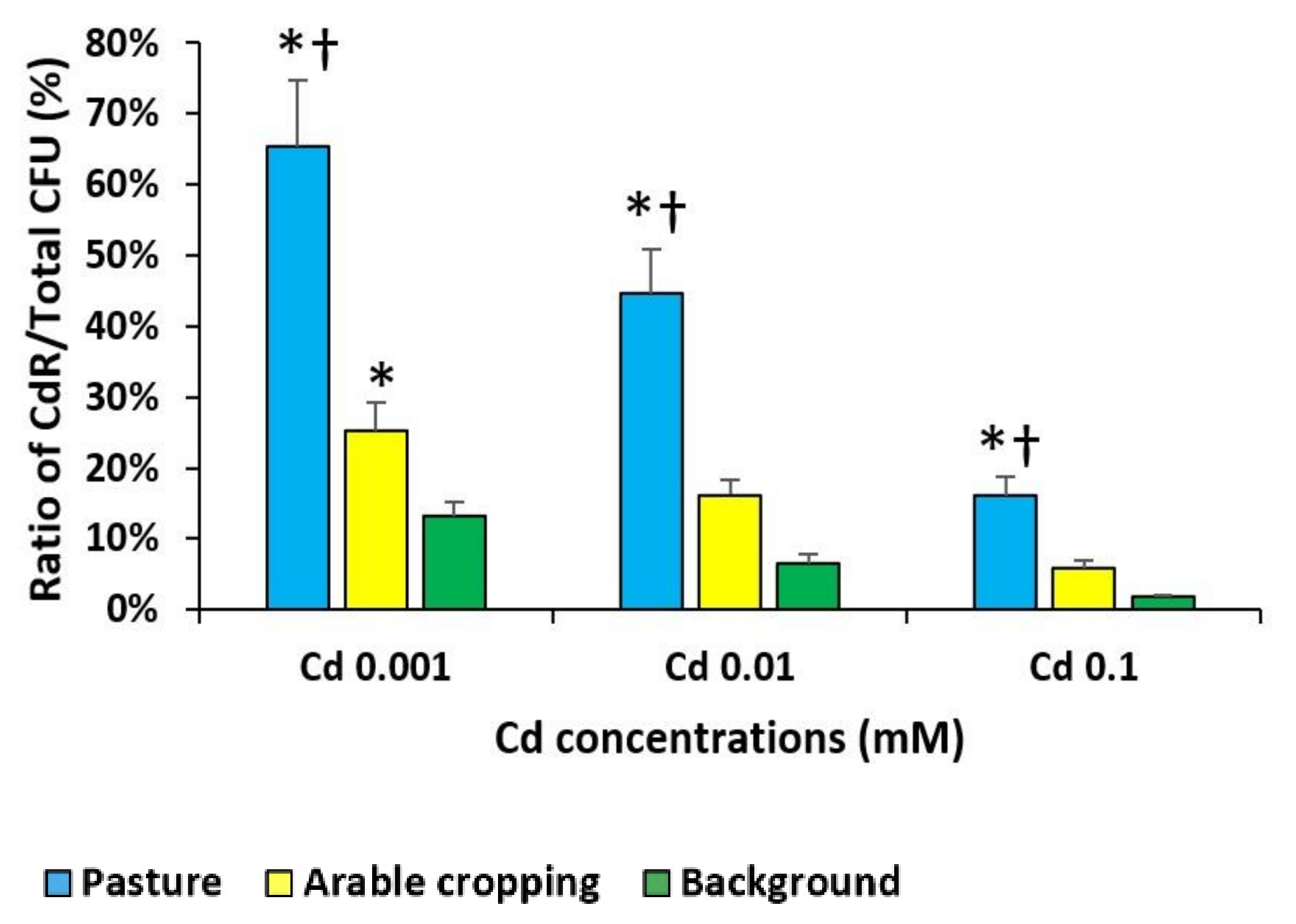
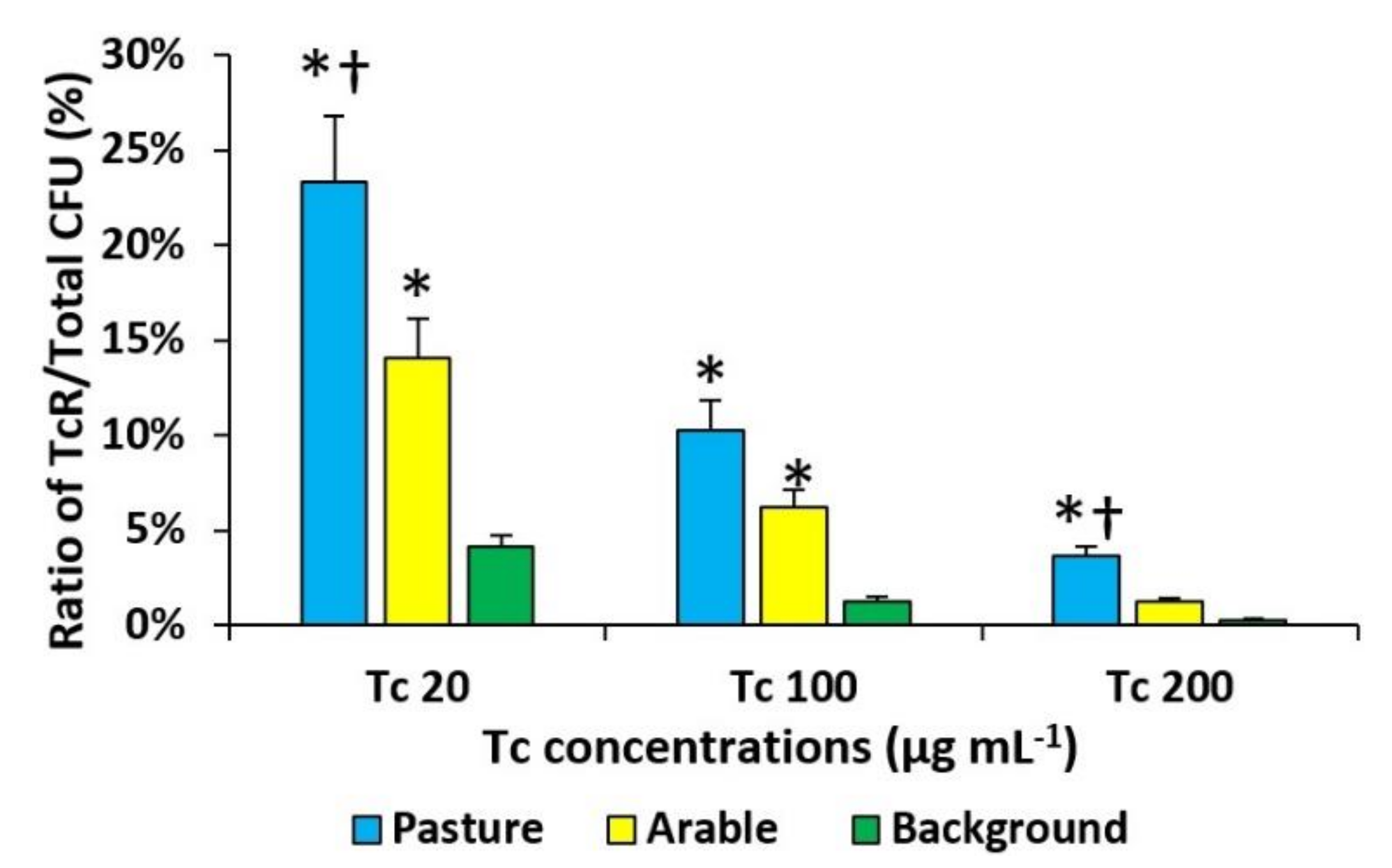
3.3. Pollution Induced Community Tolerance (PICT) Assay
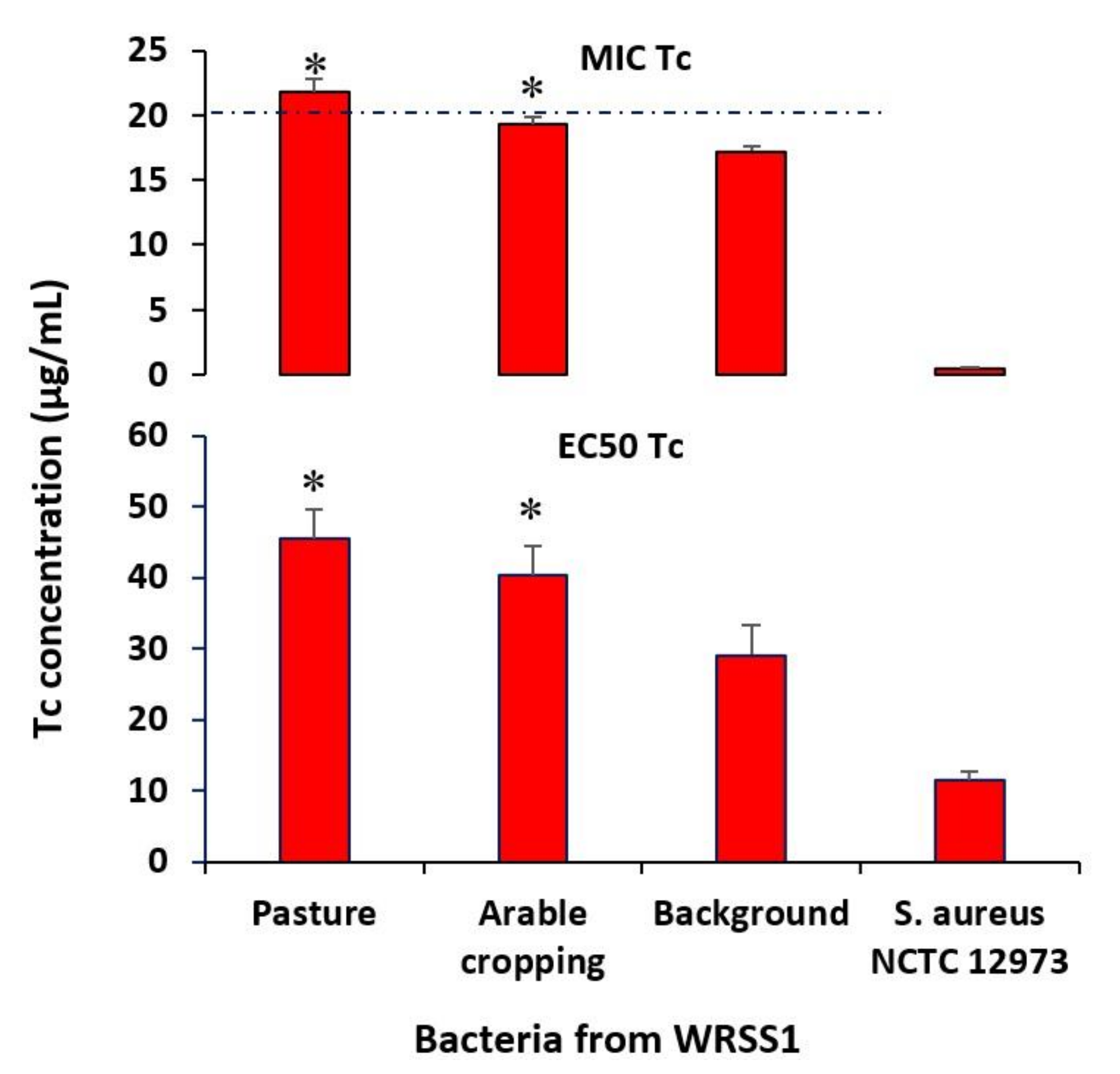
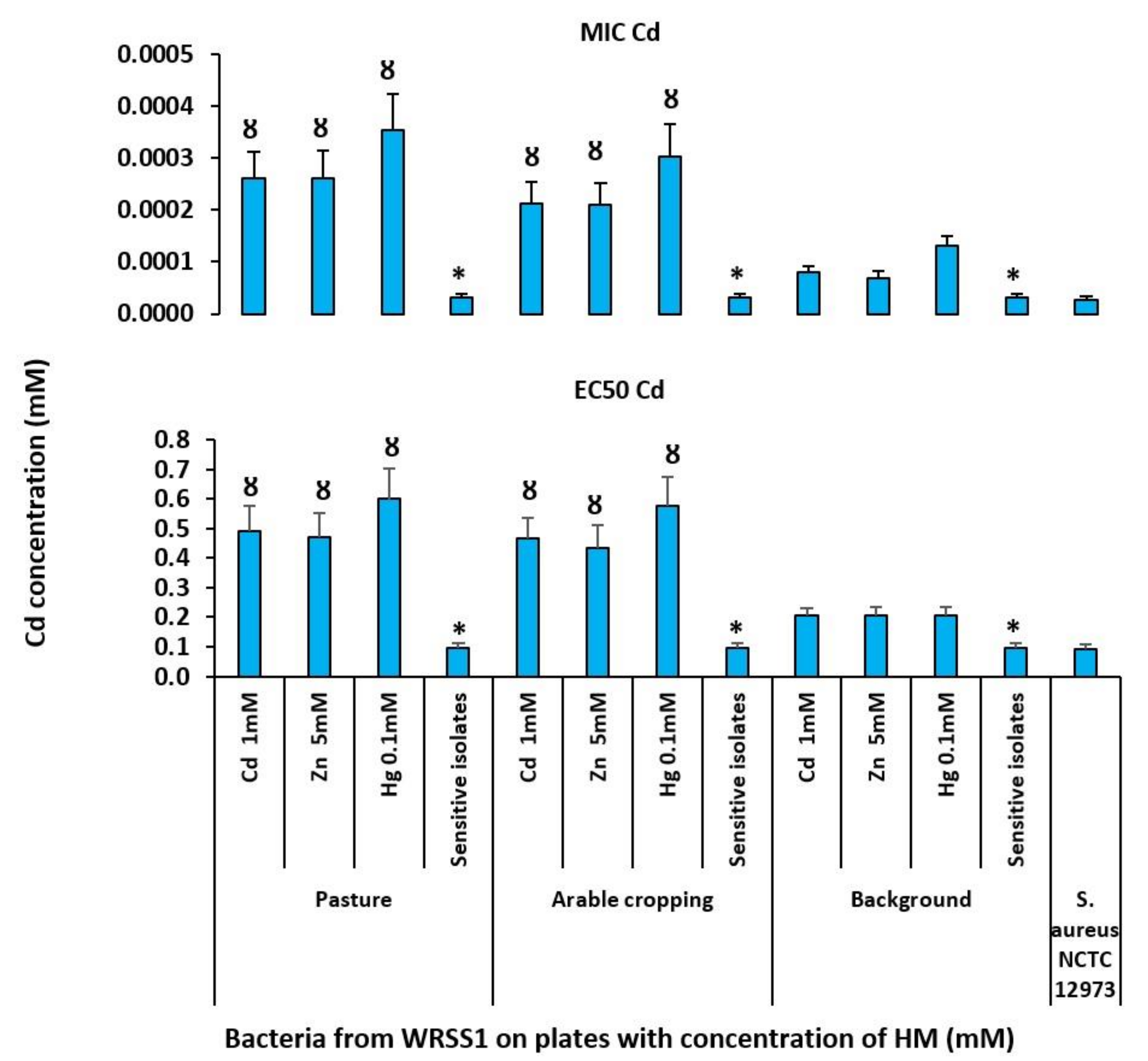
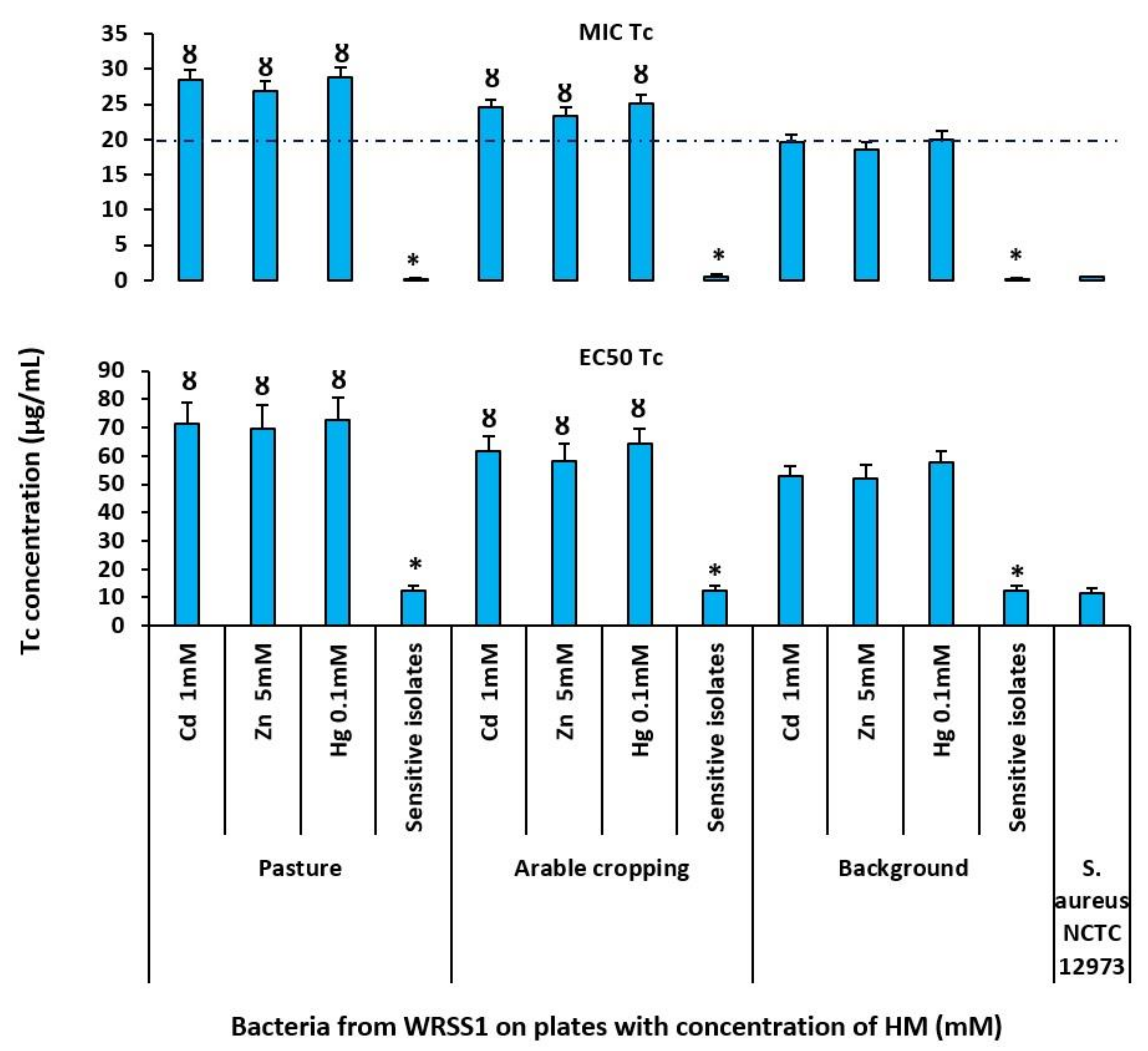
3.4. Characterisation of Resistant Isolates
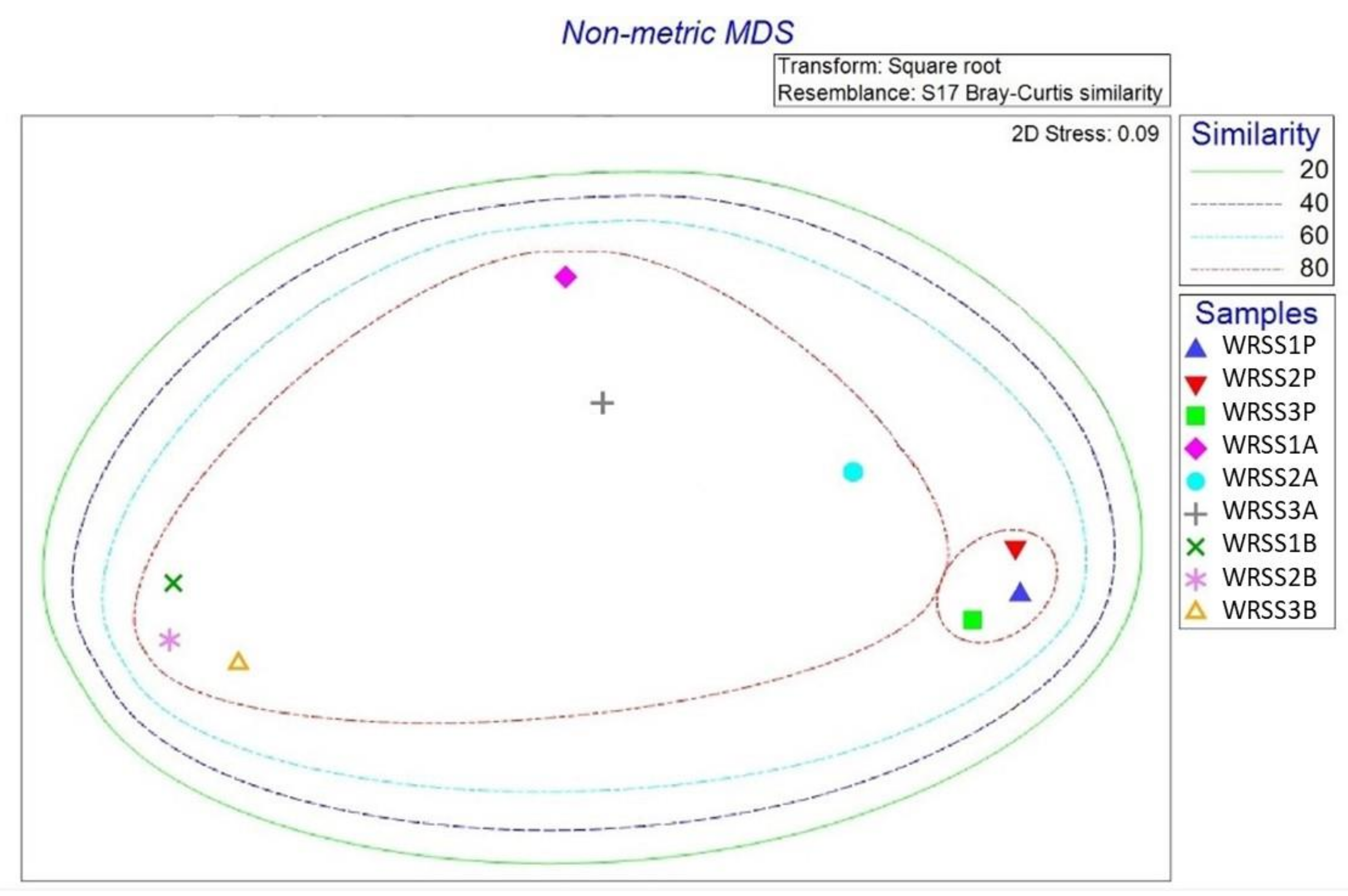
3.5. Bacterial Community Structure Investigation
3.6. Characterisation of Cd Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teitzel, G.M.; Parsek, M.R. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2003, 69, 2313–2320. [Google Scholar] [CrossRef] [PubMed]
- Silver, S. Bacterial resistances to toxic metal ions—A review. Gene 1996, 179, 9–19. [Google Scholar] [CrossRef]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Sutton, N.B.; Maphosa, F.; Morillo, J.A.; Abu Al-Soud, W.; Langenhoff, A.A.; Grotenhuis, T.; Rijnaarts, H.H.; Smidt, H. Impact of long-term diesel contamination on soil microbial community structure. Appl. Environ. Microbiol. 2013, 79, 619–630. [Google Scholar] [CrossRef]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 2014, 5, e01918-14. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Wales, A.D.; Davies, R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef]
- Dickinson, A.W.; Power, A.; Hansen, M.G.; Brandt, K.K.; Piliposian, G.; Appleby, P.; O'Neill, P.A.; Jones, R.T.; Sierocinski, P.; Koskella, B.; et al. Heavy metal pollution and co-selection for antibiotic resistance: A microbial palaeontology approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef]
- Cen, T.; Zhang, X.; Xie, S.; Li, D. Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environ. Int. 2020, 138, 105544. [Google Scholar] [CrossRef]
- Spain, A.; Alm, E. Implications of microbial heavy metal tolerance in the environment. Rev. Undergrad. Res. 2003, 2, 1–6. [Google Scholar]
- Al Salah, D.M.M.; Laffite, A.; Sivalingam, P.; Pote, J. Occurrence of toxic metals and their selective pressure for antibiotic-resistant clinically relevant bacteria and antibiotic-resistant genes in river receiving systems under tropical conditions. Environ. Sci. Pollut. Res. Int. 2021. [Google Scholar] [CrossRef]
- Taylor, M.; Kim, N.; Smidt, G. Trace element contaminants and radioactivity from phosphate fertiliser. In Phosphorus in Agriculture: 100% Zero; Schnug, E., De Kok, L.J., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 231–266. [Google Scholar]
- Council, W.R. Cadmium and New Zealand Agriculture and Horticulture: A Strategy for Long-Term Risk Management; Ministry of Agriculture and Forestry: Wellington, New Zealand, 2011.
- Cavanagh, J. Working Towards New Zealand Risk-Based Soil Guideline Values for the Management of Cadmium Accumulation on Productive Land; Ministry for Primary Industries: Wellington, New Zealand, 2012.
- Vermeulen, V. Use of Zinc in Agriculture: An Assessment of Data for Evidence of Accumulation in Waikato Soils Surface Water and Sediments. Master Thesis, Massey University, Wellington, New Zealand, 2015. [Google Scholar]
- Alloway, B.J. Zinc in Soils and Crop Nutrition; International fertilizer Industry Association and International Zinc Association: Brussels, Belgium, 2004. [Google Scholar]
- McDowell, R.W.; Taylor, M.D.; Stevenson, B.A. Natural background and anthropogenic contributions of cadmium to New Zealand soils. Agric. Ecosyst. Environ. 2013, 165, 80–87. [Google Scholar] [CrossRef]
- Taylor, M.; Gibb, R.; Willoughby, J.; Hewitt, A.; Arnold, G. Soil Maps of Cadmium in New Zealand; Manaaki Whenua-Landcare Research: Wellington, New Zealand, 2007. [Google Scholar]
- Alloway, B.J. Copper and Zinc in soils: Too little or too much. In Proceedings of the New Zealand Trace Elements Group Conference, Hamilton, New Zealand, 13–15 February 2008; p. 10. [Google Scholar]
- Kim, N.; Robinson, B. Cadmium: A clandestine threat to food safety. Food Qual. Saf. 2015, 22, 49–51. [Google Scholar]
- Zhang, J.; Liu, J.; Lu, T.; Shen, P.; Zhong, H.; Tong, J.; Wei, Y. Fate of antibiotic resistance genes during anaerobic digestion of sewage sludge: Role of solids retention times in different configurations. Bioresour. Technol. 2019, 274, 488–495. [Google Scholar] [CrossRef]
- Ma, X.; Guo, N.; Ren, S.; Wang, S.; Wang, Y. Response of antibiotic resistance to the co-existence of chloramphenicol and copper during bio-electrochemical treatment of antibiotic-containing wastewater. Environ. Int.. 2019, 126, 127–133. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Derakshani, M.; Lukow, T.; Liesack, W. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 2001, 67, 623–631. [Google Scholar] [CrossRef]
- Dubé, J.S.; Boudreault, J.P.; Bost, R.; Sona, M.; Duhaime, F.; Éthier, Y. Representativeness of laboratory sampling procedures for the analysis of trace metals in soil. Environ. Sci. Pollut. Res. Int. 2015, 22, 11862–11876. [Google Scholar] [CrossRef]
- Kim, N.D.; Taylor, M.D.; Drewry, J.J. Anthropogenic fluorine accumulation in the Waikato and Bay of Plenty regions of New Zealand: Comparison of field data with projections. Environ. Earth Sci. 2016, 75, 147. [Google Scholar] [CrossRef]
- Kim, N.D. Cadmium Accumulation in Waikato Soils.; Waikato Regional Council: Hamilton, New Zealand, 2008. [Google Scholar]
- Zealand, S.N. Nitrogen and Phosphorus in Fertilisers; Statistics New Zealand: Wellington, New Zealand, 2019. [Google Scholar]
- Hewitt, A.E. New Zealand Soil Classification, 3rd ed.; Manaaki Whenua Press: Lincoln, New Zealand, 2010; p. 136. [Google Scholar]
- IUSS Working Group WRB. World Reference Base for Soil Resources; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- Mohamed, R.M.; Abo-Amer, A.E. Isolation and characterization of heavy-metal resistant microbes from roadside soil and phylloplane. J. Basic Microbiol. 2012, 52, 53–65. [Google Scholar] [CrossRef]
- Craig Maclean, R. Adaptive radiation in microbial microcosms. J. Evolut. Biol. 2005, 18, 1376–1386. [Google Scholar] [CrossRef]
- Nishioka, T.; Elsharkawy, M.M.; Suga, H.; Kageyama, K.; Hyakumachi, M.; Shimizu, M. Development of Culture Medium for the Isolation of Flavobacterium and Chryseobacterium from Rhizosphere Soil. Microbes Environ. 2016, 31, 104–110. [Google Scholar] [CrossRef]
- EUCAST. MIC and Zone Diameter Distributions and Ecoffs. Available online: https://www.eucast.org/mic_distributions_and_ecoffs/ (accessed on 9 November 2021).
- Müsken, M.; Di Fiore, S.; Römling, U.; Häussler, S. A 96-well-plate–based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat. Protoc. 2010, 5, 1460. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Papiernik, S.K.; Gan, J.; Yates, S.R.; Yang, C.H.; Crowley, D.E. Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 2001, 67, 3245–3257. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, X.; Sun, G.; Zhang, Y.; Su, J.; Ye, J. Heavy Metal Induced Antibiotic Resistance in Bacterium LSJC7. Int. J. Mol. Sci. 2015, 16, 23390–23404. [Google Scholar] [CrossRef]
- Rousk, J.; Rousk, K. Responses of microbial tolerance to heavy metals along a century-old metal ore pollution gradient in a subarctic birch forest. Environ. Pollut. 2018, 240, 297–305. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Dymock, D.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef]
- Hauben, L.; Vauterin, L.; Swings, J.; Moore, E.R. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Bacteriol. 1997, 47, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Horswell, J.; Prosser, J.A.; Siggins, A.; van Schaik, A.; Ying, L.; Ross, C.; McGill, A.; Northcott, G. Assessing the impacts of chemical cocktails on the soil ecosystem. Soil Biol. Biochem. 2014, 75, 64–72. [Google Scholar] [CrossRef]
- Gielen, G.J.; Clinton, P.W.; Van den Heuvel, M.R.; Kimberley, M.O.; Greenfield, L.G. Influence of sewage and pharmaceuticals on soil microbial function. Environ. Toxicol. Chem. 2011, 30, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Marsh, T.L.; Saxman, P.; Cole, J.; Tiedje, J. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 2000, 66, 3616–3620. [Google Scholar] [CrossRef]
- Oger, C.; Berthe, T.; Quillet, L.; Barray, S.; Chiffoleau, J.F.; Petit, F. Estimation of the abundance of the cadmium resistance gene cadA in microbial communities in polluted estuary water. Res. Microbiol. 2001, 152, 671–678. [Google Scholar] [CrossRef]
- Ausubel, F.M. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology; Wiley: New York, NY, USA, 2002. [Google Scholar]
- Brophy, J.A.N.; Triassi, A.J.; Adams, B.L.; Renberg, R.L.; Stratis-Cullum, D.N.; Grossman, A.D.; Voigt, C.A. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol. 2018, 3, 1043–1053. [Google Scholar] [CrossRef]
- Aviv, G.; Rahav, G.; Gal-Mor, O. Horizontal Transfer of the Salmonella enterica Serovar Infantis Resistance and Virulence Plasmid pESI to the Gut Microbiota of Warm-Blooded Hosts. mBio 2016, 7, e01395-16. [Google Scholar] [CrossRef]
- Hung, C.W.; Martinez-Marquez, J.Y.; Javed, F.T.; Duncan, M.C. A simple and inexpensive quantitative technique for determining chemical sensitivity in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 11919. [Google Scholar] [CrossRef]
- Rys, G. A Cadmium Management Strategy for New Zealand. In Proceedings of the Fertiliser and Lime Conference, Palmerston North, New Zealand, 11–13 November 2011; p. 14. [Google Scholar]
- Association, N.Z.V. Antibiotic Judicious Use Guidelines for the New Zealand Veterinary Profession—Dairy; New Zealand Veterinary Association: Wellington, New Zealand, 2018. [Google Scholar]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Hermans, S.M.; Buckley, H.L.; Case, B.S.; Curran-Cournane, F.; Taylor, M.; Lear, G. Bacteria as Emerging Indicators of Soil Condition. Appl. Environ. Microbiol. 2017, 83, e02826-16. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Guo, D.; Fan, Z.; Lu, S.; Ma, Y.; Nie, X.; Tong, F.; Peng, X. Changes in rhizosphere bacterial communities during remediation of heavy metal-accumulating plants around the Xikuangshan mine in southern China. Sci. Rep. 2019, 9, 1947. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, S.; He, M. Bacterial community profile of contaminated soils in a typical antimony mining site. Environ. Sci. Pollut. Res. Int. 2018, 25, 141–152. [Google Scholar] [CrossRef]
- Yu, Z.; Gunn, L.; Wall, P.; Fanning, S. Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiol. 2017, 64, 23–32. [Google Scholar] [CrossRef]
- Taylor, M.D.; Kim, N. Dealumination as a mechanism for increased acid recoverable aluminium in Waikato mineral soils. Austral. J. Soil Res. 2009, 47, 828–838. [Google Scholar] [CrossRef]
- Li, L.G.; Xia, Y.; Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Zhao, Y.; Cocerva, T.; Cox, S.; Tardif, S.; Su, J.Q.; Zhu, Y.G.; Brandt, K.K. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 2019, 656, 512–520. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, W.; Schwarz, S.; Wang, C.; Liu, W.; Chen, F.; Luan, T.; Liu, S. Characterization of a blaIMP-4-carrying plasmid from Enterobacter cloacae of swine origin. J. Antimicrob. Chemother. 2019, 74, 1799–1806. [Google Scholar] [CrossRef]
- Liu, B.; Li, Y.; Gao, S.; Chen, X. Copper exposure to soil under single and repeated application: Selection for the microbial community tolerance and effects on the dissipation of antibiotics. J. Hazard. Mater. 2017, 325, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Henriques, I.; Tacão, M.; Leite, L.; Fidalgo, C.; Araújo, S.; Oliveira, C.; Alves, A. Co-selection of antibiotic and metal(loid) resistance in gram-negative epiphytic bacteria from contaminated salt marshes. Mar. Pollut. Bull. 2016, 109, 427–434. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Sandaa, R.A.; Enger, O.; Torsvik, V. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 1999, 65, 3293–3297. [Google Scholar] [CrossRef] [PubMed]
- Reith, F.; Zammit, C.M.; Pohrib, R.; Gregg, A.L.; Wakelin, S.A. Geogenic Factors as Drivers of Microbial Community Diversity in Soils Overlying Polymetallic Deposits. Appl. Environ. Microbiol. 2015, 81, 7822–7832. [Google Scholar] [CrossRef]
- Brodie, E.; Edwards, S.; Clipson, N. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 2002, 44, 260–270. [Google Scholar] [CrossRef]
- Lu, X.M.; Lu, P.Z. Distribution of antibiotic resistance genes in soil amended using Azolla imbricata and its driving mechanisms. Sci. Total Environ. 2019, 692, 422–431. [Google Scholar] [CrossRef]
- Udo, E.E.; Jacob, L.E.; Mathew, B. A cadmium resistance plasmid, pXU5, in Staphylococcus aureus, strain ATCC25923. FEMS Microbiol. Lett. 2000, 189, 79–80. [Google Scholar] [CrossRef][Green Version]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef]
- Fu, L.M.; Fu-Liu, C.S. Is Mycobacterium tuberculosis a closer relative to Gram-positive or Gram-negative bacterial pathogens? Tuberculosis 2002, 82, 85–90. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Li, X.; Yan, Z.; Gu, D.; Li, D.; Tao, Y.; Zhang, D.; Su, L.; Ao, Y. Characterization of cadmium-resistant rhizobacteria and their promotion effects on Brassica napus growth and cadmium uptake. J. Basic Microbiol. 2019, 59, 579–590. [Google Scholar] [CrossRef]
- Karelová, E.; Harichová, J.; Stojnev, T.; Pangallo, D.; Ferianc, P. The isolation of heavy-metal resistant culturable bacteria and resistance determinants from a heavy-metal-contaminated site. Biologia 2011, 66, 18–26. [Google Scholar] [CrossRef]
- Kaci, A.; Petit, F.; Lesueur, P.; Boust, D.; Vrel, A.; Berthe, T. Distinct diversity of the czcA gene in two sedimentary horizons from a contaminated estuarine core. Environ. Sci. Pollut. Res. Int. 2014, 21, 10787–10802. [Google Scholar] [CrossRef] [PubMed]

| Samples | Acronym | Date Sampled | Indigenous Forest Site | Arable Site | Pasture Site |
|---|---|---|---|---|---|
| 1st sample set | WRSS1 | February 2014 | EW-73 | EW-85 | EW-69 |
| 2nd sample set | WRSS2 | August 2014 | EW-73 | EW-86 | EW-135 |
| 3rd sample set | WRSS3 | June 2015 | EW-73 | EW-85 | EW-69 |
| Site No. | EW-73 (Indigenous Forest) | EW-85 (Arable) | EW-86 (Arable) | EW-69 (Pasture) | EW-135 (Pasture) |
|---|---|---|---|---|---|
| pH | 5.60 | 6.07 | 5.74 | 5.01 | 5.76 |
| Total C (%) | 8.00 | 3.79 | 3.40 | 10.10 | 8.63 |
| Total N (%) | 0.48 | 0.30 | 0.30 | 0.94 | 0.84 |
| C:N | 16.7 | 12.5 | 11.4 | 10.8 | 10.3 |
| Olsen P * | 6.00 | 110 | 89.0 | 54.0 | 73.0 |
| Cd * | 0.09 | 0.54 | 0.49 | 0.82 | 1.11 |
| Hg * | 0.19 | 0.31 | 0.42 | 0.20 | 0.21 |
| Zn * | 27.00 | 39.00 | 40.00 | 65.00 | 62.00 |
| Fe * | 28,000 | 42,000 | 53,000 | 59,000 | 39,000 |
| P * | 290 | 1850 | 1540 | 2300 | 2500 |
| Soil Samples | Number of CdR Isolates | Cd Resistance Genes | |
|---|---|---|---|
| cadA | czcA | ||
| WRSS1 pasture | 15 | 1 (6.6%) | 4 (28.5%) |
| WRSS2 pasture | 15 | 0 | 3 (20%) |
| WRSS3 pasture | 15 | 0 | 3 (20%) |
| WRSS1 arable | 15 | 0 | 3 (20%) |
| WRSS2 arable | 15 | 0 | 2 (13.3%) |
| WRSS3 arable | 15 | 0 | 2 (13.3%) |
| WRSS1 background | 15 | 1 (6.6%) | 1 (6.6%) |
| WRSS2 background | 15 | 0 | 0 |
| WRSS3 background | 15 | 0 | 1 (6.6%) |
| Soil Samples | Number of Transconjugants Carrying Cd Resistance Genes | |
|---|---|---|
| cadA | czcA | |
| WRSS1 pasture | 1 | 2 |
| WRSS2 pasture | 0 | 1 |
| WRSS3 pasture | 0 | 2 |
| WRSS1 arable | 0 | 2 |
| WRSS2 arable | 0 | 1 |
| WRSS3 arable | 0 | 0 |
| WRSS1 background | 1 | 0 |
| WRSS2 background | 0 | 0 |
| WRSS3 background | 0 | 0 |
| Bacterial Isolate ID & Source | Cd Resistance Gene | Description | Accession Number | Percent Identity |
|---|---|---|---|---|
| MUW002 WRSS1, pasture | cadA | Rhodococcus erythropolis partial 16S rRNA gene, strain SBUG 107. | FR745420.1 | 99.10% |
| MUW003 WRSS1, pasture | czcA | Pseudomonas azotoformans strain P45A chromosome, complete genome. | CP041236.1 | 99.80% |
| MUW004 WRSS1, pasture | czcA | Chryseobacterium rhizosphaerae strain WTB5 16S ribosomal RNA gene, partial sequence. | MK240433.1 | 98.90% |
| MUW005 WRSS2, pasture | czcA | Stenotrophomonas maltophilia strain Tj 16S ribosomal RNA gene, partial sequence. | MF280131.1 | 99.40% |
| MUW006 WRSS3, pasture | czcA | Bacterium strain BS1294 16S ribosomal RNA gene, partial sequence. | MK824482.1 | 97.73% |
| MUW007 WRSS3, pasture | czcA | Chryseobacterium lactis partial 16S rRNA gene, strain R-52618. | LN995695.1 | 99.20% |
| MUW008 WRSS1, arable | czcA | Bacterium strain BS1294 16S ribosomal RNA gene, partial sequence. | MK824482.1 | 99.32% |
| MUW009 WRSS1, arable | czcA | Cupriavidus sp. strain JS3054 16S ribosomal RNA gene, partial sequence. | MH588163.1 | 99.40% |
| MUW010 WRSS2, arable | czcA | Achromobacter xylosoxidans strain E2 16S ribosomal RNA gene, partial sequence. | MK849863.1 | 99.20% |
| MUW011 WRSS1, background | cadA | Microbacterium sp. strain PHIL_400ppmZn_ML16 16S ribosomal RNA gene, partial sequence. | MK652511.1 | 99.59% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heydari, A.; Kim, N.D.; Horswell, J.; Gielen, G.; Siggins, A.; Taylor, M.; Bromhead, C.; Palmer, B.R. Co-Selection of Heavy Metal and Antibiotic Resistance in Soil Bacteria from Agricultural Soils in New Zealand. Sustainability 2022, 14, 1790. https://doi.org/10.3390/su14031790
Heydari A, Kim ND, Horswell J, Gielen G, Siggins A, Taylor M, Bromhead C, Palmer BR. Co-Selection of Heavy Metal and Antibiotic Resistance in Soil Bacteria from Agricultural Soils in New Zealand. Sustainability. 2022; 14(3):1790. https://doi.org/10.3390/su14031790
Chicago/Turabian StyleHeydari, Ali, Nick D. Kim, Jacqui Horswell, Gerty Gielen, Alma Siggins, Matthew Taylor, Collette Bromhead, and Barry R. Palmer. 2022. "Co-Selection of Heavy Metal and Antibiotic Resistance in Soil Bacteria from Agricultural Soils in New Zealand" Sustainability 14, no. 3: 1790. https://doi.org/10.3390/su14031790
APA StyleHeydari, A., Kim, N. D., Horswell, J., Gielen, G., Siggins, A., Taylor, M., Bromhead, C., & Palmer, B. R. (2022). Co-Selection of Heavy Metal and Antibiotic Resistance in Soil Bacteria from Agricultural Soils in New Zealand. Sustainability, 14(3), 1790. https://doi.org/10.3390/su14031790









