Plant Genetic Diversity Studies: Insights from DNA Marker Analyses
Abstract
1. Introduction
2. DNA Markers
3. Application of Markers for Genetic Diversity and Population Structure Studies
3.1. Utilisation of Single Molecular Marker Systems
3.1.1. Random Amplification of Polymorphic DNA (RAPD)
3.1.2. Restriction Fragment Length Polymorphism (RFLP)
3.1.3. Amplified Fragment Length Polymorphism (AFLP)
3.1.4. CAAT Box-Derived Polymorphism (CBDP)
3.1.5. Sequence-Related Amplified Polymorphism (SRAP)
3.1.6. Start Codon-Targeted Polymorphism (SCoT)
3.1.7. Cleaved Amplified Polymorphism Sequence (CAPS)
3.1.8. Inter-Primer Binding Site (iPBS)
3.1.9. Simple Sequence Repeats (SSRs)
3.1.10. Inter-Retrotransposon Amplified Polymorphism (IRAP)
3.1.11. Conserved DNA-Derived Polymorphism (CDDP)
3.1.12. Diversity Array Technique (DArT)
3.1.13. Internal Transcribed Spacer (ITS)
3.1.14. Directed Amplification of Minisatellite DNA (DAMD)
3.2. Utilization of Combined Molecular Markers
3.2.1. Cumulative Applications of Dominant Markers
| Molecular Markers | Applications | Plants Investigated | References | |
|---|---|---|---|---|
| AFLP | Amplified fragment length polymorphism: uses restriction enzymes and primers specific to genomic DNA to amplify DNA fragments of different sizes. | Detects genetic variation within and among populations, linkage mapping, discrimination of cultivars, and association analyses. | Tectona grandis; Brassica oleracea; Glehnia littoralis; Solanum tuberosum; Daucus carota | [173,174,175,176,177] |
| ISSR | Inter-simple sequence repeat: uses primers specific to inter-microsatellite regions to amplify DNA fragments of different sizes. | Evaluates genetic variation within and among populations, linkage mapping, and association analyses. | Lepidium sativum; Balanites aegyptiaca; Prunus armeniaca; Vigra unguiculata; Camellia yuhsienensis; Clitaria ternatea | [178,179,180,181,182,183] |
| RAPD | Random amplified polymorphic DNA: uses arbitrary primers to amplify DNA fragments of different sizes. | Detects genetic variation within and among populations and genetic similarity. | Carica papaya; Coffee canephora; Allium sativum; Dendrobium species; Nigella sativa | [184,185,186,187,188] |
| SSR | Simple sequence repeat: uses primers specific to microsatellite regions to amplify DNA fragments of different sizes. | Ascertains genetic variation within and among populations, linkage mapping, association analyses, and plant breeding. | Solanum tuberosum; Cajanus cajan; Vicia amoena; Allium sativum; Curcuma longa | [119,126,189,190,191] |
| RFLP | Restriction fragment length polymorphism: uses restriction enzymes to cut DNA at specific sites, and the resulting fragments are separated via gel electrophoresis. | Detects genetic variation within and among populations and DNA fingerprinting. | Oryza sativa; Fragaria x Ananassa; Brassica juncea | [192,193,194] |
| DArT | Diversity array technology: a high-throughput marker technology that uses a combination of restriction enzymes and a microarray platform. | Determines genetic variation within and among populations and marker-assisted selection. | Lesquerella species; Glycine max; Vigna unguiculata; Camellia sinensis | [195,196,197,198] |
| SCAR | Sequence-characterized amplified region: uses primers specific to a known DNA sequence to amplify a fragment of a specific size. | Detects specific genes or alleles in a population and marker-assisted selection. | Calanthe species; Poa pratensis; Dendrobium officinale; Musa species; Moringa oleifera | [199,200,201,202,203] |
| CAPS | Cleaved amplified polymorphic sequence: uses restriction enzymes and primers specific to a known DNA sequence to amplify a fragment of a specific size. | Detects specific genes or alleles in a population, identification of cultivars, and marker-assisted selection. | Glycyrrhiza species; Lathyrus sativum; Citrullus lanatus; Zingiber officinale; Capsicum annum | [100,204,205,206,207] |
| IRAP | Inter retrotransposon amplified polymorphism: uses primers specific to transposable elements to amplify DNA fragments of different sizes. | Evaluates genetic variation within and among populations. | Sorghum bicolor; Piper nigrum; Hordeum vulgare; Pinus sylvestris; Sakura species | [208,209,210,211,212] |
| CDDP | Conserved DNA-derived polymorphism: uses a single primer constructed with a conserved area of functional genes. | Ascertains genetic variation within and among populations. | Salix taishanensis; Pistacia vera; Musa species; Arachis hypogaea; Amomum tsao-kosaleh | [138,213,214,215,216] |
| DAMD | Directed amplification of minisatellite-region DNA: uses a single primer specific to inter-microsatellite regions. | Assesses genetic variation within and among populations. | Capsicum; Origanum syriacum; Salvia officinalis; Ficus sycomorus | [157,217,218,219] |
| SRAP | Sequence-related amplified polymorphism: uses arbitrary forward and reverse primer combinations targeting ORFs to amplify a coding region. | Detects genetic variation within and among populations, mapping and tagging genes, germplasm identification, and sex determination. | Cuminum cyminum; Pinus yunnanensis; Lavandula angustifolia; Aspergillus flavus; Zea mays | [220,221,222,223,224] |
| SCoT | Start codon-targeted polymorphism: uses a short-conserved region flanking the start codon, producing highly reproducible amplification of targeted DNA fragments of different sizes. | Detects genetic variation within and among populations, determines population structures, identifies cultivars, QTL mapping, and DNA fingerprinting. | Ardisia crenata; Avena nuda; Scutellaria baicalensis; Trigonella species; Triticum aestivum; Crataegus monogyna | [225,226,227,228,229,230] |
| ITS2 | Internal transcribed spacer 2: a segment of the internal transcribed spacer (ITS) region, utilized as an alternative for species differentiation, involves the spacer DNA located within the tandem repeats separating the small and large subunits of ribosomal RNA (rRNA). ITS primers are designed to amplify the gene sequence containing the fastest-evolving region of the rRNA gene, resulting in fragments of varying sizes for differentiating species. | Evaluates genetic variation within and among populations, intraspecific variation, species identification, authentication of plant variety, and detection of adulterants. | Dendrobium species; Physalis species; Astragalus species; | [231,232,233] |
| iPBS | Inter-primer binding site: uses the primer binding site for the reverse transcription enzyme of the LTR retrotransposon. No prior sequence information to amplify DNA fragments of different sizes, a preferred universal marker system. | Detects genetic differentiation at both the intra-specific and inter-specific levels, marker-assisted selection, and breeding. | Abelmoschus esculentus; Alfalfa; Phaseolus vulgaris; Triticum species; Brassica species; Castanea sativa | [234,235,236,237,238,239] |
| CBDP | CAAT-box derived polymorphism: Uses the CAAT box consensus sequence of the plant promoter upstream of the start codon to amplify DNA fragments of different sizes. | Detects genetic diversity among and within species/populations, cultivar identification, linkage map construction, and marker-assisted selection. | Triticum durum; Salvia species; Lens culinaris | [73,74,240] |
| STS | Sequence-tagged site: Short DNA sequences of known locations that are easily detectable using PCR and serve as landmarks in the physical map of the genome. | Variation analysis, gene expression, genome mapping, and gene silencing. | Cenchrus species; Triticum aestivum; Oryza sativa; Agropyron cristatum; Secale cereale; Thinopyrum intermedium | [241,242,243,244,245,246] |
3.2.2. Cumulative Application of Dominant and Co-Dominant Markers
4. Drawbacks and Recent Developments in DNA Marker Technology
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tikendra, L.; Koijam, A.S.; Nongdam, P. Molecular Markers based Genetic Fidelity Assessment of Micropropagated Dendrobium chrysotoxum Lindl. Meta Gene 2019, 20, 100562. [Google Scholar] [CrossRef]
- Hussain, H.; Nisar, M. Assessment of Plant Genetic Variations using Molecular Markers: A Review. J. Appl. Biol. Biotechnol. 2020, 8, 099–109. [Google Scholar] [CrossRef]
- Tuvesson, S.D.; Larsson, C.T.; Ordon, F. Use of Molecular Markers for Doubled Haploid Technology: From Academia to Plant Breeding Companies. In Double Haploid Technology; Segui-Simarro, J.M., Ed.; Humana: New York, NY, USA, 2021; Volume 2288, pp. 49–72. ISBN 9781071613351. [Google Scholar] [CrossRef]
- Amom, T.; Tikendra, L.; Potshangbam, A.M.; Bidyananda, N.; Devi, R.S.; Dey, A.; Sahoo, M.R.; Vendrame, W.A.; Jamir, I.; Nongdam, P. Conservation strategies for endemic Dendrocalamus manipureanus: A study on genetic diversity and population structure based on molecular and phytochemical markers. S. Afr. J. Bot. 2023, 152, 106–123. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Vilanova, S.; Fita, A.; Prohens, J.; Rodríguez-Burruezo, A. Genetic diversity, population structure, and relationships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS). Hort. Res. 2019, 6, 54. [Google Scholar] [CrossRef]
- Apana, N.; Amom, T.; Tikendra, L.; Angamba, P.; Dey, A.; Nongdam, P. Genetic diversity and population structure of Clerodendrum serratum (L.) Moon using CBDP, iPBS and SCoT markers. J. Appl. Res. Med. 2021, 25, 100349. [Google Scholar] [CrossRef]
- Gyani, P.C.; Bollinedi, H.; Gopala Krishnan, S.; Vinod, K.K.; Sachdeva, A.; Bhowmick, P.K.; Ellur, R.K.; Nagarajan, M.; Singh, A.K. Genetic Analysis and Molecular Mapping of the Quantitative Trait Loci Governing Low Phytic Acid Content in a Novel LPA Rice Mutant, PLM11. Plants 2020, 9, 1728. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Amom, T.; Nongdam, P. The use of molecular marker methods in plants: A review. Int. J. Curr. Res. Rev. 2017, 9, 1–7. [Google Scholar]
- Avise, J.C. Molecular Markers, Natural History and Evolution, 2nd ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2004; ISBN 9780412037719. [Google Scholar]
- Ramesh, P.; Mallikarjuna, G.; Sameena, S.; Kumar, A.; Gurulakshmi, K.; Reddy, B.V.; Reddy, P.C.O.; Sekhar, A.C. Advancements in molecular marker technologies and their applications in diversity studies. J. Biosci. 2020, 45, 1–15. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.N.; Latif, M.A. A Review of Microsatellite Markers and Their Applications in Rice Breeding Programs to Improve Blast Disease Resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, S.; Okubo, H.; Fujieda, K. Isozyme variation in Cucumber (Cucumis sativus L.). J. Jpn. Soc. Hortic. Sci. 1992, 61, 595–601. [Google Scholar] [CrossRef]
- Nongdam, P.; Nirmala, C. Genetic variability in four species of Cymbidium based on isozyme markers. Physiol. Mol. Biol. Plants 2007, 13, 65–68. [Google Scholar]
- Zarei, A.; Erfani-Moghadam, J. SCoT markers provide insight into the genetic diversity, population structure and phylogenetic relationships among three Pistacia species of Iran. Genet. Resour. Crop Evol. 2021, 68, 1625–1643. [Google Scholar] [CrossRef]
- Kumar, A.; Shahina, P.S.; Singh, R.S.; Singh, D.N. Prospect of molecular markers in precision plant breeding. In Recent Advances in Chemical Sciences and Biotechnology; New Delhi Publishers: New Delhi, India, 2020; pp. 131–142. [Google Scholar]
- Tikendra, L.; Rahaman, H.; Dey, A.; Sahoo, M.R.; Nongdam, P. Applicability of Molecular Markers in Ascertaining Genetic Diversity and Relationship between Five Edible Bamboos of North-East India. In Molecular Marker Techniques; : Kumar, N., Ed.; Springer: Singapore, 2023; pp. 141–160. [Google Scholar] [CrossRef]
- Amom, T.; Tikendra, L.; Apana, N.; Goutam, M.; Sonia, P.; Koijam, A.S.; Potshangbam, A.M.; Rahaman, H.; Nongdam, P. Efficiency of RAPD, ISSR, iPBS, SCoT and phytochemical markers in the genetic relationship study of five native and economical important bamboos of North-East India. Phytochemistry 2020, 174, 112330. [Google Scholar] [CrossRef]
- Tahir, N.; Lateef, D.; Rasul, K.; Rahim, D.; Mustafa, K.; Sleman, S.; Mirza, A.; Aziz, R. Assessment of genetic variation and population structure in Iraqi barley accessions using ISSR, CDDP, and SCoT markers. Czech J. Genet. Plant Breed. 2023, 59, 148–159. [Google Scholar] [CrossRef]
- Aly, A.A.; Eliwa, N.E.; Borik, Z.M.; Safwat, G. Physiological variation of irradiated red raddish plants and their phylogenic relationship using SCoT and CDDP marker. Not. Bot. Hortic. Agrobot. Cluj Napoca 2021, 49, 12396. [Google Scholar] [CrossRef]
- Abaza, N.O.; Yousief, S.S.; Moghaieb, R.E.A. The efficiency of SCoT, ISSR, and SRAP markers for detecting genetic polymorphism among Egyptian barley genotypes. J. Pharm. Negat. Results 2022, 13, 1851–1863. [Google Scholar] [CrossRef]
- Pudake, R.N.; Kumari, M. Assessment of Genetic Diversity in Indigenous Plants from Northeast India Using Molecular Marker Technology. In Bioprospecting of Indigenous Bioresources of North-East India; Purkayastha, J., Ed.; Springer: Singapore, 2016; pp. 181–192. [Google Scholar] [CrossRef]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef] [PubMed]
- Nilkanta, H.; Amom, T.; Tikendra, L.; Rahaman, H.; Nongdam, P. ISSR marker-based population genetic study of Melocanna baccifera (Roxb.) Kurz: A commercially important bamboo of Manipur, North-East India. Scientifica 2017, 2017, 3757238. [Google Scholar] [CrossRef] [PubMed]
- Amom, T.; Tikendra, L.; Rahaman, H.; Potshangbam, A.; Nongdam, P. Evaluation of genetic relationship between 15 bamboo species of North-East India based on ISSR marker analysis. Mol. Biol. Res. Commun. 2018, 7, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kordrostami, M.; Rahimi, M. Molecular markers in plants: Concepts and applications. Genet. Millenn. 2015, 13, 4024–4031. [Google Scholar]
- Chen, W.; Hou, L.; Zhang, Z.; Pang, X.; Li, Y. Genetic diversity, population structure, and linkage disequilibrium of a core collection of Ziziphusjujuba assessed with genome-wide SNPs developed by genotyping-by-sequencing and SSR markers. Front. Plant Sci. 2017, 8, 575. [Google Scholar]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Tikendra, L.; Potshangbam, A.M.; Dey, A.; Devi, T.R.; Sahoo, M.R.; Nongdam, P. RAPD, ISSR, and SCoT markers based genetic stability assessment of micropropagated Dendrobium fimbriatum Lindl. var. oculatum Hk. f.- an important endangered orchid. Physiol. Mol. Biol. Plants 2021, 27, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, M.; Ahmadi Afzadi, M.; Kordrostami, M. Genetic diversity in Sickleweed (Falcaria vulgaris) and using stepwise regression to identify marker associated with traits. Sci. Rep. 2023, 13, 12142. [Google Scholar] [CrossRef] [PubMed]
- Tikendra, L.; Amom, T.; Nongdam, P. Molecular genetic homogeneity assessment of micropropagated Dendrobium moschatum Sw.—A rare medicinal orchid, using RAPD and ISSR markers. Plant Gene 2019, 19, 100196. [Google Scholar] [CrossRef]
- Tikendra, L.; Potshangbam, A.M.; Amom, T.; Dey, A.; Nongdam, P. Understanding the genetic diversity and population structure of Dendrobium chrysotoxum Lindl. -An endangered medicinal orchid and implication for its conservation. S. Afr. J. Bot. 2021, 138, 364–376. [Google Scholar] [CrossRef]
- Chen, M.Y.; He, X.H.; Zhang, Y.L.; Lu, T.T.; He, W.Q.C.; Yang, J.H.; Huang, X.; Zhu, J.W.; Yu, H.X.; Luo, C. Genetic diversity and relationship analyses of mango (Mangifera indica L.) germplasm resources with ISSR, SRAP, CBDP and CEAP markers. Sci. Hortic. 2022, 301, 111146. [Google Scholar] [CrossRef]
- Pradhan, S.; Paudel, Y.P.; Qin, W.; Pant, B. Genetic fidelity assessment of wild and tissue cultured regenerants of a threatened orchid, Cymbidium aloifolium using molecular markers. Plant Gene 2023, 34, 100418. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.G.; de Souza, A.M.; de Almeida Vieira, F.; Estopa, R.A.; Reis, C.A.F.; de Carvalho, D. Using Random Amplified Polymorphic DNA to Assess Genetic Diversity and Structure of Natural Calophyllumbrasiliense (Clusiaceae). Populations in Riparian Forests. Int. J. For. Res. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Bibi, T.; Mustafa, H.S.B.; Hasan, E.U.; Rauf, S.; Mahmood, T.; Ali, Q. Analysis of genetic diversity in linseed using molecular markers. Life Sci. J. 2015, 12, 28–37. [Google Scholar]
- Dhakshanamoorthy, D.; Selvaraj, R.; Chidambaram, A. Utility of RAPD marker for genetic diversity analysis in gamma rays and ethyl methane sulphonate (EMS)-treated Jatropha curcas plants. C. R. Biol. 2015, 338, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Qiu, Y.; Kong, H. RAPD analysis for genetic diversity in Changium smyrnioides (Apiaceae), an endangered plant1. Bot. Bull. Acad. Sin. 2003, 44, 13–18. [Google Scholar]
- Zhang, C.; He, P.; He, J.; Zhang, Y.; Qiao, Y.; Zhang, M.; Shi, Z.; Hu, S. RAPD analysis for genetic diversity of medicinal plant Coptis omeiensis. Zhongguo Zhong Yao Za Zhi = Zhongguo Zhongyao Zazhi = China J. Chin. Mater. Medica 2010, 35, 138–141. [Google Scholar] [CrossRef]
- Ruiz-Chután, J.A.; Salava, J.; Janovská, D.; Žiarovská, J.; Kalousová, M.; Fernández, E. Assessment of genetic diversity in Sorghum bicolor using RAPD markers. Genetika 2019, 51, 789–803. [Google Scholar] [CrossRef]
- Mortazavi Moghadam, F.A.; Qaderi, A.; Sharifi-Sirchi, G.R. Evaluation of Genetic Diversity of 17 Populations (Lepidium sativum L.) Plant Collected from Different Regions of Iran by RAPD Marker. ACS Agric. Sci. Technol. 2021, 1, 684–690. [Google Scholar] [CrossRef]
- Ben-Ari, G.; Lavi, U. Marker-assisted selection in plant breeding. In Plant Biotechnology and Agriculture; Academic Press: Cambridge, MA, USA, 2012; pp. 163–184. [Google Scholar] [CrossRef]
- Miller, J.C.; Tanksley, S.D. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 1990, 80, 437–448. [Google Scholar] [CrossRef]
- Mondini, L.; Arshiya, N.; Pagnotta, M.A. Assessing Plant Genetic Diversity by Molecular Tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Mir, R.R.; Hiremath, P.J.; Riera-Lizarazu, O.; Varshney, R.K. Evolving molecular marker technologies in plants: From RFLPs to GBS. In Diagnostics in Plant Breeding; Springer: Dordrecht, The Netherlands, 2013; pp. 229–247. [Google Scholar]
- Cui, Y.X.; Xu, G.W.; Magill, C.W.; Schertz, K.F.; Hart, G.E. RFLP-based assay of Sorghum bicolor (L.) Moench genetic diversity. Theor. Appl. Genet. 1995, 90, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Maizura, I.; Rajanaidu, N.; Zakri, A.H.; Cheah, S.C. Assessment of Genetic Diversity in Oil Palm (Elaeis guineensis Jacq.) using Restriction Fragment Length Polymorphism (RFLP). Genet. Resour. Crop Evol. 2006, 53, 187–195. [Google Scholar] [CrossRef]
- Chang, C.; Bowman, J.L.; De John, A.W.; Lander, E.S.; Meyerowitz, E.M. Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 85, 6856–6860. [Google Scholar] [CrossRef] [PubMed]
- Benchimol, L.L.; de Souza, C.L., Jr.; Garcia, A.A.F.; Kono, P.M.S.; Mangolin, C.A.; Barbosa, A.M.M.; Coelho, A.S.G.; de Souza, A.P. Genetic diversity in tropical maize inbred lines: Heterotic group assignment and hybrid performance determined by RFLP markers. Plant Breed. 2000, 119, 491–496. [Google Scholar] [CrossRef]
- Zaharleva, M.; Santoni, S.; David, J. Use of RFLP markers to study genetic diversity and to build a core-collection of the wild wheat relative Ae. geniculata Roth (=Ae. ovata L.). Genet. Sel. Evol, 2001; 33, (Suppl. 1), S269–S288. [Google Scholar] [CrossRef]
- Bonierbale, M.W.; Plaisted, R.L.; Tanksley, S. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 1998, 120, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D.; Young, N.D.; Paterson, A.H.; Bonierbale, M.W. RFLP mapping in plant breeding: New tools for an old science. Biotechnology 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Lee, T.V.D.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Brugmans, B.; van der Hulst, R.G.; Visser, R.G.; Lindhout, P.; van Eck, H.J. A new and versatile method for the successful conversion of AFLP™ markers into simple single locus markers. Nucleic Acids Res. 2003, 31, e55. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, K.N.; Gnanesh, B.N.; Byre Gowda, M.; Venkatesha, S.C.; Gomashe, S.S.; Channamallikarjuna, V. AFLP analysis in pigeonpea (Cajanus cajan (L.) Millsp.) Revealed close relationship of cultivated genotypes with some of its wild relatives. Genet. Resour. Crop Evol. 2011, 58, 837–847. [Google Scholar] [CrossRef]
- Al-Nadabi, H.; Khan, M.; Al-Yahyai, R.A.; Al-Sadi, A.M. AFLP fingerprinting analysis of citrus cultivars and wild accessions from Oman suggests the presence of six distinct cultivars. Agriculture/Pol’nohospodárstvo 2018, 64, 173–182. [Google Scholar] [CrossRef]
- Huang, W.K.; Wan, F.H.; Guo, J.Y.; Gao, B.D.; Xie, B.Y.; Peng, D.L. AFLP analyses of genetic variation of Eupatorium adenophorum (Asteraceae) populations in China. Can. J. Plant Sci. 2009, 89, 119–126. [Google Scholar] [CrossRef]
- Murariu, D.; Plăcintă, D.D.; Simioniuc, V. Assessing genetic diversity in Romanian maize landraces, using molecular markers. Rom. Agric. Res. 2019, 36, 3–9. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Ghosh, S.; Mandi, S.S.; Kumaria, S.; Tandon, P. Genetic variability and association of AFLP markers with some important biochemical traits in Dendrobium thyrsiflorum, a threatened medicinal orchid. S. Afr. J. Bot. 2017, 109, 214–222. [Google Scholar] [CrossRef]
- El-Demerdash, E.S.S.; Elsherbeny, E.A.; Salama, Y.A.M.; Ahmed, M.Z. Genetic diversity analysis of some Egyptian Origanum and Thymus species using AFLP markers. J. Genet. Eng. Biotechnol. 2019, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.D.; Liu, W.H.; Sun, M.; Zhou, J.Q.; Liu, W.; Zhang, C.L.; Zhang, X.Q.; Peng, Y.; Huang, L.K.; Ma, X. Genetic diversity and structure of Elymus tangutorum accessions from western China as unraveled by AFLP markers. Hereditas 2019, 156, 8. [Google Scholar] [CrossRef] [PubMed]
- Assaeed, A.M.; Al-Faifi, S.A.; Migdadi, H.M.; El-Bana, M.I.; Al Qarawi, A.A.; Khan, M.A. Evaluation of genetic diversity of Panicumturgidum Forssk from Saudi Arabia. Saudi J. Biol. Sci. 2018, 25, 123–129. [Google Scholar] [CrossRef]
- Wang, Q.; Ruan, X.; Jiang, H.; Meng, Q.; Wang, L. Genetic diversity of different geographical populations of Rhodiolarosea based on AFLP markers. Zhongguo Zhong Yao Za Zhi = Zhongguo Zhongyao Zazhi = China J. Chin. Mater. Medica 2009, 34, 2279–2284. [Google Scholar]
- Zhao, B.; Yin, Z.F.; Xu, M.; Wang, Q.C. AFLP analysis of genetic variation in wild populations of five Rhododendron species in Qinling Mountain in China. Biochem. Syst. Ecol. 2012, 45, 198–205. [Google Scholar] [CrossRef]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Rana, M.K.; Singh, S.; Kumar, S.; Kumar, R.; Singh, R. CAAT box-derived polymorphism (CBDP): A novel promoter-targeted molecular marker for plants. J. Plant Biochem. Biotechnol. 2014, 23, 175–183. [Google Scholar] [CrossRef]
- Imani, A.; Ahmadi, J.; Heydari, M. Molecular variation and genetic relationships among Iranian and foreign pistachio cultivars using gene-targeted CAAT box-derived markers. Agric. Biotechnol. J. 2022, 14, 41–62. [Google Scholar]
- Tomar, P.; Malik, C.P. Genetic diversity assessment in Trachyspermum ammi L. Sprague using CDDP and CBDP markers. J. Plant Sci. Res. 2016, 32, 27. [Google Scholar]
- Ahmed, D.A.; Tahir, N.A.; Salih, S.H.; Talebi, R. Genome diversity and population structure analysis of Iranian landrace and improved barley (Hordeum vulgare L.) genotypes using arbitrary functional gene-based molecular markers. Genet. Resour. Crop Evol. 2021, 68, 1045–1060. [Google Scholar] [CrossRef]
- Etminan, A.; Pour-Aboughadareh, A.; Mohammadi, R.; Noori, A.; Ahmadi-Rad, A. Applicability of CAAT Box-derived Polymorphism (CBDP) Markers for Analysis of Genetic Diversity in Durum Wheat. Cereal Res. Commun. 2018, 46, 1–9. [Google Scholar] [CrossRef]
- Sarvmeili, J.; Saidi, A.; Farrokhi, N.; Pouresmael, M.; Talebi, R. Genetic diversity and population structure analysis of landrace and wild relatives of lentil germplasm using CBDP marker. Cytol. Genet. 2020, 54, 566–573. [Google Scholar] [CrossRef]
- Fabriki-Ourang, S.; Karimi, H. Assessment of genetic diversity and relationships among Salvia species using gene targeted CAAT box-derived polymorphism markers. J. Genet. 2019, 98, 75. [Google Scholar] [CrossRef] [PubMed]
- Eslamzadeh, M.; Omidi, M.; Rashidi, V.; Etminan, A. Evaluation of genetic diversity and population structure analysis in some Aegilops species using CBDP markers. IGS 2021, 16, 1–8. [Google Scholar]
- Zhou, Y.; Wang, X.; Zhang, X. Development and application of a SRAP marker for the identification of sex in Buchloe dactyloides. Euphytica 2011, 181, 261–266. [Google Scholar] [CrossRef]
- Robarts, D.W.; Wolfe, A.D. Sequence-related amplified polymorphism (SRAP) markers: A potential resource for studies in plant molecular biology1. Appl. Plant Sci. 2014, 2, 1400017. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; McVetty, P.B.E.; Quiros, C.F. SRAP molecular marker technology in plant science. In Plant Breeding from Laboratories to Fields; Andersen, S.B., Ed.; InTech: Rijeka, Croatia, 2013. [Google Scholar] [CrossRef][Green Version]
- Zhou, L.; Yarra, R.; Cao, H.; Zhao, Z. Sequence-Related Amplified Polymorphism (SRAP) Markers Based Genetic Diversity and Population Structure Analysis of Oil Palm (Elaeis guineensis Jacq.). Trop. Plant Biol. 2021, 14, 63–71. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, S.; Liu, J.; Zhao, Y.; Liu, J. Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE 2017, 12, e0177508. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Dong, Z.; Lei, Y.; Zhao, J.; Xiong, Y.; Yang, J.; Xiong, Y.; Gou, W.; Ma, X. Genetic diversity and molecular characterization of worldwide prairie grass (Bromus catharticus Vahl) accessions using SRAP markers. Agronomy 2021, 11, 2054. [Google Scholar] [CrossRef]
- Wang, D.; Shen, B.; Gong, H. Genetic diversity of Simao pine in China revealed by SRAP markers. PeerJ 2019, 7, e6529. [Google Scholar] [CrossRef]
- Agyenim-Boateng, K.G.; Lu, J.; Shi, Y.; Zhang, D.; Yin, X. SRAP analysis of the genetic diversity of wild castor (Ricinus communis L.) in South China. PLoS ONE 2019, 14, e0219667. [Google Scholar] [CrossRef] [PubMed]
- Suman, A.; Kimbeng, C.; Edmé, S.; Veremis, J. Sequence-related amplified polymorphism (SRAP) markers for assessing genetic relationships and diversity in sugarcane germplasm collections. Plant Genet. Resour. 2008, 6, 222–231. [Google Scholar] [CrossRef]
- Aseny, N.; Syamsuardi, S.; Nurainas, N. Molecular characterization of andalas tree dioecious plant [Morus macroura Miq.] using SRAP marker. IOP Conf. Ser. Earth Environ. Sci. 2021, 741, 012050. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Abdin, M.Z.; Arya, L.; Verma, M. Use of SCoT markers to assess the gene flow and population structure among two different populations of bottle gourd. Plant Gene 2017, 9, 80–86. [Google Scholar] [CrossRef]
- Luo, C.; He, X.H.; Chen, H.; Ou, S.J.; Gao, M.P. Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem. Syst. Ecol. 2010, 38, 1176–1184. [Google Scholar] [CrossRef]
- Mavlyutov, Y.M.; Shamustakimova, A.O.; Klimenko, I.A. Application of SCoT markers for accessing of genetic diversity of gramineous forage grass species. IOP Conf. Ser. Earth Environ. Sci. 2021, 901, 012038. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, W.; Wang, Y.; Zhao, X. Potential of start codon targeted (SCoT) markers to estimate genetic diversity and relationships among Chinese Elymus sibiricus accessions. Molecules 2015, 20, 5987–6001. [Google Scholar] [CrossRef] [PubMed]
- Satya, P.; Karan, M.; Jana, S.; Mitra, S.; Sharma, A.; Karmakar, P.G.; Ray, D.P. Start codon targeted (SCoT) polymorphism reveals genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fiber producing species. Meta Gene 2015, 3, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Rayan, W.A.; Osman, S.A. Phylogenetic relationships of some Egyptian soybean cultivars (Glycine max L.) using SCoT marker and protein pattern. Bull. Natl. Res. Cent. 2019, 43, 161. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, V.; Haq, S.U.; Jatav, P.K.; Kothari, S.L.; Kachhwaha, S. Assessment of genetic diversity in 29 rose germplasms using SCoT marker. J. King Saud Univ. Sci. 2019, 31, 780–788. [Google Scholar] [CrossRef]
- Sharma, V.; Thakur, M. Applicability of SCoT markers for detection of variations in Fusarium yellows resistant lines of ginger (Zingiber officinale Rosc.) induced through gamma irradiations. S. Afr. J. Bot. 2021, 140, 454–460. [Google Scholar] [CrossRef]
- Feng, S.; Zhu, Y.; Yu, C.; Jiao, K.; Jiang, M.; Lu, J.; Shen, C.; Ying, Q.; Wang, H. Development of species-specific SCAR markers, based on a SCoT analysis, to authenticate Physalis (Solanaceae) species. Front. Genet. 2018, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y.N. CAPS markers in plant biology. Russ. J. Genet. Appl. Res. 2016, 6, 279–287. [Google Scholar] [CrossRef]
- Konieczny, A.; Ausubel, F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993, 4, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y. Cleaved Amplified Polymorphic Sequences (CAPS) Markers in Plant Biology; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2014; 251p, ISBN 9781631175534. [Google Scholar]
- Liu, S.; Gao, P.; Zhu, Q.; Luan, F.; Davis, A.R.; Wang, X. Development of cleaved amplified polymorphic sequence markers and a CAPS-based genetic linkage map in watermelon (Citrullus lanatus [Thunb.] Matsum. And Nakai) constructed using whole-genome re-sequencing data. Breed. Sci. 2016, 66, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Jo, I.H.; Sung, J.; Hong, C.E.; Raveendar, S.; Bang, K.H.; Chung, J.W. Development of cleaved amplified polymorphic sequence (CAPS) and high-resolution melting (HRM) markers from the chloroplast genome of Glycyrrhiza species. 3 Biotech 2018, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Kushanov, F.N.; Pepper, A.E.; Yu, J.Z.; Buriev, Z.T.; Shermatov, S.E.; Saha, S.; Ulloa, M.; Jenkins, J.N.; Abdukarimov, A.; Abdurakhmonov, I.Y. Development, genetic mapping and QTL association of cotton PHYA, PHYB, and HY5-specific CAPS and dCAPS markers. BMC Genet. 2016, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Lee, H.Y.; Shim, D.; Choi, S.H.; Cho, H.; Hyun, T.K.; Jo, I.H.; Chung, J.W. Development of CAPS markers for evaluation of genetic diversity and population structure in the germplasm of button mushroom (Agaricus bisporus). J. Fungi 2021, 7, 375. [Google Scholar] [CrossRef] [PubMed]
- Erper, I.; Ozer, G.; Kalendar, R.; Avci, S.; Yildirim, E.; Alkan, M.; Turkkan, M. Genetic diversity and pathogenicity of Rhizoctonia spp. isolates associated with red cabbage in Samsun (Turkey). J. Fungus 2021, 7, 234. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Wang, Y.; Ma, T.; Kanzana, G.; Wu, F.; Zhang, J. Genome-wide identification and development of LTR retrotransposon-based molecular markers for the Melilotus Genus. Plants 2021, 10, 890. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Khapilina, O.; Turzhanova, A.; Danilova, A.; Tumenbayeva, A.; Shevtsov, V.; Kotukhov, Y.; Kalendar, R. Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult. BioTech 2021, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Başak, İ.; Özer, G.; Muradoğlu, F. Morphometric traits and iPBS based molecular characterizations of walnut (Juglans regia L.) genotypes. Genet. Resour. Crop Evol. 2022, 69, 2731–2743. [Google Scholar] [CrossRef]
- Baran, N.; Shimira, F.; Nadeem, M.A.; Altaf, M.T.; Andirman, M.; Baloch, F.S.; Temiz, M.G. Exploring the genetic diversity and population structure of upland cotton germplasm by iPBS-retrotransposons markers. Mol. Biol. Rep. 2023, 50, 4799–4811. [Google Scholar] [CrossRef]
- Shimira, F.; Boyaci, H.F.; Çilesiz, Y.; Nadeem, M.A.; Baloch, F.S.; Taşkin, H. Exploring the genetic diversity and population structure of scarlet eggplant germplasm from Rwanda through iPBS-retrotransposon markers. Mol. Biol. Rep. 2021, 48, 6323–6333. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Yılmaz, A.; Nadeem, M.A.; Habyarimana, E.; Subaşı, I.; Nawaz, M.A.; Chaudhary, H.J.; Shahid, M.Q.; Ercişli, S.; Zia, M.A.B.; et al. Mobile genomic element diversity in world collection of safflowers (Carthamus tinctorius L.) panel using iPBS-retrotransposon markers. PLoS ONE 2019, 14, e0211985. [Google Scholar] [CrossRef] [PubMed]
- Yıldız, M.; Koçak, M.; Baloch, F.S. Genetic bottlenecks in Turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment. Genet. Mol. Res. 2015, 14, 10588–10602. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.B.; Guo, D.L.; Guo, L.L.; Wei, D.F.; Hou, X.G. Genetic diversity analysis of tree peony germplasm using iPBS markers. Genet. Mol. Res. 2015, 14, 7556–7566. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, J.P.; Carvalho, A.; Martín, A.; Lima-Brito, J. Molecular characterization of Fagaceae species using inter-primer binding site (iPBS) markers. Mol. Biol. Rep. 2018, 45, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, N.K.; Singh, V.K.; Singh, D.P.; Singh, N.P. Tools for simple sequence repeat (SSR) markers. Agric. Update 2016, 11, 163–172. [Google Scholar] [CrossRef]
- Feng, S.; He, R.; Lu, J.; Jiang, M.; Shen, X.; Jiang, Y.; Wang, Z.; Wang, H. Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front. Genet. 2016, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Tong, H.; Chen, Y.; Wang, J.; Chen, Y.; Sun, G.; He, J.; Wu, Y. Development of pineapple microsatellite markers and germplasm genetic diversity analysis. BioMed Res. Int. 2013, 2013, 317912. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhong, Q.; Tian, J.; Wang, L.; Zhao, M.; Li, L.; Sun, X. Characterization and development of EST-SSR markers to study the genetic diversity and populations’ analysis of Jerusalem artichoke (Helianthus tuberosus L.). Genes Genom. 2018, 40, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Kimaro, D.; Melis, R.; Sibiya, J.; Shimelis, H.; Shayanowako, A. Analysis of genetic diversity and population structure of pigeonpea [Cajanus cajan (L.) Millsp] accessions using SSR markers. Plants 2020, 9, 1643. [Google Scholar] [CrossRef] [PubMed]
- Romdhane, M.B.; Riahi, L.; Selmi, A.; Zoghlami, N. Patterns of genetic structure and evidence of gene flow among Tunisian Citrus species based on informative nSSR markers. C. R. Biol. 2016, 339, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Babaei, N.; Abdullah, N.A.P.; Saleh, G.; Abdullah, T.L. Isolation and characterization of microsatellite markers and analysis of genetic variability in Curculigo latifolia Dry and. Mol. Biol. Rep. 2012, 39, 9869–9877. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, Z.; Ma, X.; Wei, M.; Zhao, T.; Zhan, R.; Chen, W. SSR marker development and intraspecific genetic divergence exploration of Chrysanthemum indicum based on transcriptome analysis. BMC Genom. 2018, 19, 291. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Liu, J.; Yu, J.; Wan, H. High-throughput SSR marker development and the analysis of genetic diversity in Capsicum frutescens. Horticulturae 2021, 7, 187. [Google Scholar] [CrossRef]
- Rahman, M.M.; Quddus, M.R.; Ali, M.O.; Liu, R.; Li, M.; Yan, X.; Li, G.; Ji, Y.; Hossain, M.M.; Wang, C.; et al. Genetic diversity of Lathyrus sp collected from different geographical regions. Mol. Biol. Rep. 2022, 49, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Oliya, B.K.; Kim, M.Y.; Lee, S.H. Development of genic-SSR markers and genetic diversity of Indian lettuce (Lactuca indica L.) in South Korea. Genes Genom. 2018, 40, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Kumar, A.; Sharma, S.; Singh, B.; Sood, S.; Dipta, B.; Singh, R.; Siddappa, S.; Thakur, A.K.; Dalamu, D.; et al. Analysis of Genetic Diversity, Population Structure, and Association Mapping for Late Blight Resistance in Potato (Solanum tuberosum L.) Accessions Using SSR Markers. Agronomy 2023, 13, 294. [Google Scholar] [CrossRef]
- Ahmad, A.; Wang, J.D.; Pan, Y.B.; Sharif, R.; Gao, S.J. Development and use of simple sequence repeats (SSRs) markers for sugarcane breeding and genetic studies. Agronomy 2018, 8, 260. [Google Scholar] [CrossRef]
- Kalendar, R.; Grob, T.; Regina, M.; Suoniemi, A.; Schulman, A. IRAP and REMAP: Two new retrotransposon-based DNA fingerprinting techniques. Theor. Appl. Genet. 1999, 98, 704–711. [Google Scholar] [CrossRef]
- Alzohairy, A.M.; Gyulai, G.; Ramadan, M.F.; Edris, S.; Sabir, J.S.; Jansen, R.K.; Eissa, H.F.; Bahieldin, A. Retrotransposon-based molecular markers for assessment of genomic diversity. Funct. Plant Biol. 2014, 41, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Abedinpour, H.; Ranjbar, G.A.; Jelodar, N.B.; Golein, B. Evaluation of genetic diversity in Citrus genotypes by IRAP molecular marker. Int. J. Farm. Allied Sci. 2014, 3, 230–234. [Google Scholar]
- Widyawan, M.H.; Wulandary, S. Genetic diversity analysis of yardlong bean genotypes (Vigna unguiculata subsp. sesquipedalis) based on IRAP marker. Biodiversitas 2020, 21, d210333. [Google Scholar] [CrossRef]
- Boronnikova, S.V.; Kalendar, R.N. Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants. Russ. J. Genet. 2010, 46, 36–42. [Google Scholar] [CrossRef]
- Du, X.; Zhang, Q.; Luo, Z. Development of retrotransposon primers and their utilization for germplasm identification in Diospyros spp. (Ebenaceae). Tree Genet. Genomes 2009, 5, 235–245. [Google Scholar] [CrossRef]
- Flavell, A.J.; Knox, M.R.; Pearce, S.R.; Ellis, T.N. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 1998, 16, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Mackill, D.J. Conserved DNA-derived polymorphism (CDDP): A simple and novel method for generating DNA markers in plants. Plant Mol. Biol. Rep. 2009, 27, 558–562. [Google Scholar] [CrossRef]
- Bilčíková, J.; Farkasová, S.; Žiarovská, J. Genetic variability of commercially important apple varieties (Malus x domestica Borkh.) assessed by CDDP markers. Acta Fytotech. Zootech. 2021, 24, 21–26. [Google Scholar]
- Haffar, S.; Baraket, G.; Usai, G.; Aounallah, A.; Ben Mustapha, S.; Ben Abdelkrim, A.; Salhi Hannachi, A. Conserved DNA-derived polymorphism as a useful molecular marker to explore genetic diversity and relationships of wild and cultivated Tunisian figs (Ficus carica L.). Trees 2022, 36, 723–735. [Google Scholar] [CrossRef]
- Liu, H.; Zang, F.; Wu, Q.; Ma, Y.; Zheng, Y.; Zang, D. Genetic diversity and population structure of the endangered plant Salix taishanensis based on CDDP markers. Glob. Ecol. Conserv. 2020, 24, e01242. [Google Scholar] [CrossRef]
- Novoselović, D.; Bentley, A.R.; Šimek, R.; Dvojković, K.; Sorrells, M.E.; Gosman, N.; Horsenell, R.; Drezner, G.; Šatović, Z. Characterizing Croatian wheat germplasm diversity and structure in a European context by DArT markers. Front. Plant Sci. 2016, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Alexander, H.; Wittenberg, J.; Van, T.; Lee, D.; Cayla, C.; Kilian, A.; Visser, R.G.F.; Schouten, H.J. Validation of the high-throughput marker technology DArT using the model plant Arabidopsis thaliana. Mol. Genet. Genom. 2005, 274, 30. [Google Scholar] [CrossRef]
- Gawroński, P.; Pawełkowicz, M.; Tofil, K.; Uszyński, G.; Sharifova, S.; Ahluwalia, S.; Tyrka, M.; Wędzomy, M.; Kilian, A.; Bolibok-Brągozewska, H. DArT markers effectively target gene space in the rye genome. Front. Plant Sci. 2016, 7, 1600. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sevilla, J.F.; Horvath, A.; Botella, M.A.; Gaston, A.; Folta, K.; Kilian, A.; Denoyes, B.; Amaya, I. Diversity arrays technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria x ananassa). PLoS ONE 2015, 10, e0144960. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Gao, L.; Mann, H.; Traini, A.; Chiusano, M.L.; Kilian, A.; Aversano, R.; Carputo, D.; Bradeen, J.M. A DArT marker-based linkage map for wild potato Solanum bulbocastanum facilitates structural comparisons between Solanum A and B genomes. BMC Genet. 2014, 15, 123. [Google Scholar] [CrossRef] [PubMed]
- Schouten, H.J.; Van De Weg, W.E.; Carling, J.; Khan, S.A.; McKay, S.J.; van Kaauwen, M.P.; Wittenberg, A.H.J.; Koehorst-van Putten, H.J.J.; Noordijk, Y.; Gao, Z.; et al. Diversity arrays technology (DArT) markers in apple for genetic linkage maps. Mol. Breed. 2012, 29, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Deres, D.; Feyissa, T. Concepts and applications of diversity array technology (DArT) markers for crop improvement. J. Crop Improv. 2023, 37, 913–933. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Q.; Qiu, S.; Dai, J.; Gao, X. DNA barcoding: An efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin. Med. 2022, 17, 112. [Google Scholar] [CrossRef]
- Banchi, E.; Ametrano, C.G.; Greco, S.; Stanković, D.; Muggia, L.; Pallavicini, A. PLANiTS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, 2020, baz155. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.F.; Han, Q.B.; Zhao, Z.L.; Wang, Z.T.; Xu, L.S.; Xu, H.X. Sequence analysis based on ITS1 region of nuclear ribosomal DNA of Amomum villosum and ten species of Alpinia. J. Food Drug Anal. 2009, 17, 1. [Google Scholar] [CrossRef]
- Selvaraj, D.; Shanmughanandhan, D.; Sarma, R.K.; Joseph, J.C.; Srinivasan, R.V.; Ramalingam, S. DNA barcode ITS effectively distinguishes the medicinal plant Boerhavia diffusa from its adulterants. Genomics Proteomics Bioinform. 2012, 10, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Manokar, J.; Balasubramani, S.P.; Venkatasubramanian, P. Nuclear ribosomal DNA–ITS region based molecular marker to distinguish Gmelina arborea Roxb. Ex Sm. from its substitutes and adulterants. J. Ayurveda Integr. Med. 2018, 9, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Hynniewta, M.; Malik, S.K.; Rao, S.R. Genetic diversity and phylogenetic analysis of Citrus (L.) from north-east India as revealed by meiosis, and molecular analysis of internal transcribed spacer region of rDNA. Meta Gene 2014, 2, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.J.; Wang, X.; Wang, J.R.; Su, N.; Zhang, L.; Ma, Y.P.; Chang, Z.Y.; Zhao, L.; Potter, D. Efficient identification of Pulsatilla (Ranunculaceae) using DNA barcodes and micro-morphological characters. Front. Plant Sci. 2019, 10, 1196. [Google Scholar] [CrossRef] [PubMed]
- Heath, D.D.; Lwama, G.K.; Devlin, R.H. PCR primed with VNTR core sequences yields species specific patterns and hypervariable probes. Nucleic Acids Res. 1993, 21, 5782–5785. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.B.; Li, J.W.; Wang, L.J.; Liu, L.J.; Si, S.W. Utilization of a set of high-polymorphism DAMD markers for genetic analysis of a cucumber germplasm collection. Acta Physiol. Plant 2011, 33, 227–231. [Google Scholar] [CrossRef]
- Ince, A.G.; Karaca, M.; Onus, A.N. Development and utilization of diagnostic DAMD-PCR markers for Capsicum accessions. Genet Resour. Crop Evol. 2009, 56, 211–221. [Google Scholar] [CrossRef]
- Pınar, H.; Bulut, M.; Altunoz, D.; Uzun, A.; Seday, U.; Yılmaz, K.U. Determination of genetic diversity and relationships within citrus and related genera using DAMD markers. Bangladesh J. Bot. 2017, 46, 163–170. [Google Scholar]
- Pinar, H.; Uzun, A.; Unlu, M.; Yaman, M. Genetic diversity in Turkish banana (Musa cavendishii) genotypes with DAMD markers. Fresenius Environ. Bull. 2019, 28, 459–463. [Google Scholar]
- İnce, A.G.; Karaca, M. Td-DAMD-PCR assays for fingerprinting of commercial carnations. Turk. J. Biol. 2015, 39, 290–298. [Google Scholar] [CrossRef]
- Jain, J.R.; Timsina, B.; Satyan, K.B.; Manohar, S.H. A comparative assessment of morphological and molecular diversity among Sechium edule (Jacq.) Sw. accessions in India. 3 Biotech 2017, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Saleh, B. Genetic diversity of Salvia tomentosa Miller (Lamiaceae) species using touch-down directed amplification of minisatellite DNA (Td-DAMD) molecular markers. Acta Biol. Szeged. 2019, 63, 135–141. [Google Scholar] [CrossRef]
- Goswami, B.; Gadi, B.R.; Rao, S.R. Morphological and molecular markers-based assessment of genetic diversity of a valuable endemic plant Lasiurus sindicus Henr. in the arid region of Rajasthan, India. Arid Land Res. Manag. 2022, 36, 298–313. [Google Scholar] [CrossRef]
- Hromadová, Z.; Gálová, Z.; Mikolášová, L.; Balážová, Ž.; Vivodík, M.; Chňapek, M. Efficiency of RAPD and SCoT markers in the Genetic Diversity Assessment of the Common Bean. Plants 2023, 12, 2763. [Google Scholar] [CrossRef] [PubMed]
- Mansoory, A.; Khademi, O.; Naji, A.M.; Rohollahi, I.; Sepahvand, E. Evaluation of genetic diversity in three Diospyros species, collected from different regions in Iran, using ISSR and SCoT molecular markers. Int. J. Fruit Sci. 2022, 22, 235–248. [Google Scholar] [CrossRef]
- Alzahrani, O.R.; Alshehri, M.A.; Alasmari, A.; Ibrahim, S.D.; Oyouni, A.A.; Siddiqui, Z.H. Evaluation of genetic diversity among Saudi Arabian and Egyptian cultivars of alfalfa (Medicago sativa L.) using ISSR and SCoT markers. J. Taibah Univ. Sci. 2023, 17, 2194187. [Google Scholar] [CrossRef]
- Guan, C.; Chachar, S.; Zhang, P.; Hu, C.; Wang, R.; Yang, Y. Inter-and intra-specific genetic diversity in Diospyros using SCoT and IRAP markers. Hortic. Plant J. 2020, 6, 71–80. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Kumaria, S.; Tandon, P. Applicability of ISSR and DAMD markers for phyto-molecular characterization and association with some important biochemical traits of Dendrobium nobile, an endangered medicinal orchid. Phytochem. 2015, 117, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Khodaee, L.; Azizinezhad, R.; Etminan, A.R.; Khosroshahi, M. Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J. Genet. Eng. Biotechnol. 2021, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arya, L.; Narayanan, R.K.; Verma, M.; Singh, A.K.; Gupta, V. Genetic diversity and population structure analyses of Morinda tomentosa Heyne, with neutral and gene-based markers. Genet. Resour. Crop Evol. 2014, 61, 1469–1479. [Google Scholar] [CrossRef]
- El-Mansy, A.B.; Abd El-Moneim, D.; ALshamrani, S.M.; Safhi, F.A.; Abdein, M.A.; Ibrahim, A.A. Genetic diversity analysis of tomato (Solanum lycopersicum L.) with morphological, cytological, and molecular markers under heat stress. Horticulturae 2021, 7, 65. [Google Scholar] [CrossRef]
- Tiwari, G.; Singh, R.; Singh, N.; Choudhury, D.R.; Paliwal, R.; Kumar, A.; Gupta, V. Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in Kalmegh [Andrographis paniculata (Burm. f.) Nees]. Ind. Crops Prod. 2016, 86, 1–11. [Google Scholar] [CrossRef]
- Vaishnaw, V.; Mohammad, N.; Wali, S.A.; Kumar, R.; Tripathi, S.B.; Negi, M.S.; Ansari, S.A. AFLP markers for analysis of genetic diversity and structure of teak (Tectona grandis) in India. Can. J. For. Res. 2015, 45, 297–306. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Germaine, K.; Bourke, P.; Malone, R. AFLP analysis of genetic diversity and phylogenetic relationships of Brassica oleracea in Ireland. C. R. Biol. 2016, 339, 163–170. [Google Scholar] [CrossRef]
- Li, B.; Wang, A.; Zhang, P.; Li, W. Genetic diversity and population structure of endangered Glehnia littoralis (Apiaceae) in China based on AFLP analysis. Biotechnol. Biotechnol. Equip. 2019, 33, 331–337. [Google Scholar] [CrossRef]
- del Rio, A.; Bamberg, J. An AFLP Marker Core Subset for the Cultivated Potato Species Solanum phureja ( Solanum tuberosum L. subsp. andigenum). Am. J. Potato Res. 2021, 98, 374–383. [Google Scholar] [CrossRef]
- Domblides, A.; Domblides, E. Rapid Genetic Assessment of Carrot Varieties Based on AFLP Analysis. Horticulturae 2023, 9, 298. [Google Scholar] [CrossRef]
- Kumar, V.; Yadav, H.K. Assessment of genetic diversity in Lepidium sativum L. using inter simple sequence repeat (ISSR) marker. Physiol. Mol. Biol. Plants 2019, 25, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Mint Abdelaziz, S.; Medraoui, L.; Alami, M.; Pakhrou, O.; Makkaoui, M.; Ould Mohamed Salem Boukhary, A.; Filali-Maltouf, A. Inter simple sequence repeat markers to assess genetic diversity of the desert date (Balanites aegyptiaca Del.) for Sahelian ecosystem restoration. Sci. Rep. 2020, 10, 14948. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, Z.N.; Sharma, V.; Shah, R.A.; Sharma, N.; Summuna, B.; Al-Misned, F.A.; El-Serehy, H.A.; Mir, J.I. Genetic diversity analysis and population structure in apricot (Prunus armeniaca L.) grown under the north-western Himalayas using ISSR markers. Saudi J. Biol. Sci. 2021, 28, 5986–5992. [Google Scholar] [CrossRef]
- Saxena, A.; Rukam, T.S. Assessment of genetic diversity in cowpea (Vigna unguiculata L. Walp.) through ISSR marker. Res. J. Biotechnol. 2020, 15, 66–71. [Google Scholar]
- Zou, Y.; Ge, X.; Yan, C.; Zhong, Q.; Chen, D.; Chen, Z.; Yuan, Y.; Guo, H.; Zhou, Y.; Wang, J.; et al. Assessment of genetic diversity of Camellia yuhsienensis based on leaf structure and inter simple sequence repeat (ISSR) markers. Genet. Resour. Crop Evol. 2024, 19, 1–4. [Google Scholar] [CrossRef]
- Nurhasanah; Hindersah, R.; Suganda, T.; Concibido, V.; Sundari; Karuniawan, A. The First Report on the Application of ISSR Markers in Genetic Variance Detection among Butterfly Pea (Clitoria ternatea L.) Accession in North Maluku Province, Indonesia. Horticulturae 2023, 9, 1059. [Google Scholar] [CrossRef]
- Kumar, D.; Yadav, P.; Priyanka, Y.A.; Dwivedi, U.N.; Yadav, K. Genetic diversity analysis among papaya (Carica papaya L.) varieties using RAPD markers. Int. J. Tradit. Med. App. 2019, 1, 22–27. [Google Scholar] [CrossRef]
- Ramadiana, S.; Hapsoro, D.; Evizal, R.; Setiawan, K.; Karyanto, A.; Yusnita, Y. Genetic diversity among 24 clones of Robusta coffee in Lampung based on RAPD markers. Biodiversitas 2021, 22, d220614. [Google Scholar] [CrossRef]
- Sheuly, K.N.; Hoque, M.E.; Syfullah, K.; Bashar, M.A.; Rahman, M.H.; Siddique, A.B. Genetic Diversity Analysis of Garlic (Allium sativum L.) Genotypes Using Rapd Markers. Plant Cell Biotechnol. Mol. Biol. 2022, 13, 61–68. [Google Scholar] [CrossRef]
- Hartati, S.; Samanhudi, S.; Cahyono, O. Genetic similarity among Dendrobium species from Indonesia using RAPD markers. Biodiversitas 2023, 24, d240945. [Google Scholar] [CrossRef]
- Aydın, A. Determining the genetic diversity of some black cumin genotypes collected in different regions of Türkiye using RAPD markers. Int. J. Agric. Environ. Food Sci. 2024, 8, 294–300. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, S.; Gao, Q.; Liu, F.; Wang, J.; Wang, X. Genetic diversity and population structure analysis in a large collection of Vicia amoena in China with newly developed SSR markers. BMC Plant Biol. 2021, 21, 1–12. [Google Scholar]
- Li, X.; Qiao, L.; Chen, B.; Zheng, Y.; Zhi, C.; Zhang, S.; Pan, Y.; Cheng, Z. SSR markers development and their application in genetic diversity evaluation of garlic (Allium sativum) germplasm. Plant Divers. 2022, 44, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, A.; Munda, S.; Paw, M.; Begum, T.; Siddiqui, M.H.; Gaafar, A.-R.Z.; Kesawat, M.S.; Lal, M. Molecular genetic divergence analysis amongst high curcumin lines of Golden Crop (Curcuma longa L.) using SSR marker and use in trait-specific breeding. Sci. Rep. 2023, 13, 19690. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Ebana, K.; Fukuoka, S.; Nagamine, T.; Kawase, M. Development of an RFLP-based rice diversity research set of germplasm. Breed. Sci. 2005, 55, 431–440. [Google Scholar] [CrossRef]
- Kunihisa, M.; Fukino, N.; Matsumoto, S. Development of PCR-RFLP marker on strawberry and the identification of Cultivars and their progeny. Acta Hortic. 2006, 708, 517–522. [Google Scholar] [CrossRef]
- Mir, J.I.; Shahidul, I.; Rajdeep, K. Evaluation of genetic diversity in Brassica juncea (L.) using protein profiling and molecular marker (RFLP). Int. J. Plant Breed. Genet. 2015, 9, 77–85. [Google Scholar]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop Lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef] [PubMed]
- Lukanda, M.M.; Dramadri, I.O.; Adjei, E.A.; Edema, R.; Ssemakula, M.O.; Tukamuhabwa, P.; Tusiime, G. Genetic Diversity and Population Structure of Ugandan Soybean (Glycine max L.) Germplasm Based on DArTseq. Plant Mol. Biol. Rep. 2023, 41, 417–426. [Google Scholar] [CrossRef]
- Gbedevi, K.M.; Boukar, O.; Ishikawa, H.; Abe, A.; Ongom, P.O.; Unachukwu, N.; Rabbi, I.; Fatokun, C. Genetic diversity and population structure of cowpea [Vigna unguiculata (L.) Walp.] germplasm collected from Togo based on DArT markers. Genes 2021, 12, 1451. [Google Scholar] [CrossRef] [PubMed]
- Malebe, M.; Mphangwe, N.; Myburg, A.; Apostolides, Z. Assessment of genome-wide DArT-seq markers for tea Camellia sinensis (L.) O. Kuntze germplasm analysis. Tree Genet. Genomes 2019, 15, 48. [Google Scholar] [CrossRef]
- Nutthapornnitchakul, S.; Peyachoknagul, S.; Sangin, P.; Kongbungkerd, A.; Punjansing, T.; Nakkuntod, M. Genetic relationship of orchids in the Calanthe group based on sequence-related amplified polymorphism markers and development of sequence-characterized amplified regions markers for some genus/species identification. Agric. Nat. Resour. 2019, 53, 340–347. [Google Scholar]
- Mingyue, T.U.; Yali, H.E.; Xiaoli, L.I.; Ying, Z.O.U.; Xiaojun, Y.U.A.N. Development of SCAR markers related to heat tolerance in Kentucky bluegrass. Not. Bot. Horti Agrobot. Cluj-Napoca 2020, 48, 509–522. [Google Scholar]
- Zheng, K.; Cai, Y.; Chen, W.; Gao, Y.; Jin, J.; Wang, H.; Feng, S.; Lu, J. Development, identification, and application of a germplasm specific SCAR Marker for Dendrobium officinale Kimura et Migo. Front. Plant Sci. 2021, 12, 669458. [Google Scholar] [CrossRef] [PubMed]
- Qv, M.; Feng, G.; Chen, S.; Chen, H.; Chen, C.; Wang, F.; Lv, S.; Dai, L.; Liu, H.; Huang, B.; et al. The development and utilization of two SCAR markers linked to the resistance of banana (Musa spp. AAA) to Fusarium oxysporum f. sp. cubense race 4. Euphytica 2024, 220, 69. [Google Scholar] [CrossRef]
- Ravi, D.; Siril, E.A.; Nair, B.R. SCAR Marker Development for the Identification of Elite Germplasm of Moringa Oleifera Lam.-A Never Die Plant. Plant Mol. Biol. Rep. 2021, 39, 850–861. [Google Scholar] [CrossRef]
- Shiferaw, E.; Porceddu, E.; Pé, E.; Ponnaiah, M. Application of CAPS markers for diversity assessment in grass pea (L.). Biodivers. Res. Conserv. 2017, 48, 11–18. [Google Scholar] [CrossRef]
- Osae, B.A.; Amanullah, S.; Liu, H.; Liu, S.; Saroj, A.; Zhang, C.; Liu, T.; Gao, P.; Luan, F. CAPS marker-base genetic linkage mapping and QTL analysis for watermelon ovary, fruit and seed-related traits. Euphytica 2022, 218, 39. [Google Scholar] [CrossRef]
- Kang, J.-N.; Lee, G.-H.; Yu, J.; Choi, M.-H.; Lee, S. Development of Cleaved Amplified Polymorphic Sequence Markers for Classifying Ginger (Zingiber officinale) Cultivars Using Reference Sequencing. Plant Breed. Biotechnol. 2023, 11, 130–140. [Google Scholar] [CrossRef]
- Bongiorno, G.; Di Noia, A.; Ciancaleoni, S.; Marconi, G.; Cassibba, V.; Albertini, E. Development and Application of a Cleaved Amplified Polymorphic Sequence Marker (Phyto) Linked to the Pc5.1 Locus Conferring Resistance to Phytophthora capsici in Pepper (Capsicum annuum L.). Plants 2023, 12, 2757. [Google Scholar] [CrossRef]
- Martiwi, I.N.A.; Nugroho, L.H.; Daryono, B.S.; Susandarini, R. Genotypic variability and relationships of Sorghum bicolor accessions from Java Island, Indonesia based on IRAP markers. Biodiversitas 2020, 21, d211220. [Google Scholar] [CrossRef]
- Dongare, M.D.; Alex, S.; Soni, K.B.; Sindura, K.P.; Nair, D.S.; Stephen, R.; Jose, E. Cross-species transferability of IRAP retrotransposon markers and polymorphism in black pepper (Piper nigrum L.). Genet. Resour. Crop Evol. 2023, 70, 2593–2605. [Google Scholar] [CrossRef]
- Zayed, E.M.; Ghonaim, M.M.; Attya, A.M.; Morsi, N.A.; Hussein, K.A. IRAP-PCR technique for determining the biodiversity between Egyptian barley cultivars. Egyptian J. Bot. 2022, 62, 359–370. [Google Scholar] [CrossRef]
- Voronova, A.; Ruņģis, D. Development and Characterisation of Irap Markers From Expressed Retrotransposon-like sequences in Pinus sylvestris L. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 2014, 67, 485–492. [Google Scholar]
- Stepanov, I.; Balapanov, I.; Drygina, A. Search of effective IRAP markers for sakura genotyping. BIO Web Conf. 2020, 25, 03006. [Google Scholar] [CrossRef]
- Aouadi, M.; Guenni, K.; Abdallah, D.; Louati, M.; Chatti, K.; Baraket, G.; Salhi Hannachi, A. Conserved DNA-derived polymorphism, new markers for genetic diversity analysis of Tunisian Pistacia vera L. Physiol. Mol. Biol. Plants 2019, 25, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Igwe, D.O.; Ihearahu, O.C.; Osano, A.A.; Acquaah, G.; Ude, G.N. Genetic diversity and population assessment of Musa L. (Musaceae) employing CDDP markers. Plant Mol. Biol. Rep. 2021, 39, 801–820. [Google Scholar] [CrossRef]
- Klongová, L.; Kyseľ, M.; Fialková, V.; Fernández-Cusimamani, E.; Kovacik, A.; Ziarovska, J. Utilization of CDDP Markers in analysis of genetic variability of Arachis hypogaea L. J. Microbiol. Biotechnol. Food Sci. 2023, 13, E9884. [Google Scholar] [CrossRef]
- Ma, M.; Yan, Z.; Lu, B. Assessment of genetic diversity of the medicinal and aromatic crop, Amomum Tsao-Ko, using paap and CDDP markers. Agriculture 2022, 12, 1536. [Google Scholar] [CrossRef]
- Saleh, B. Genetic diversity of Origanum syriacum L. (Lamiaceae) species through Touch-Up Direct Amplification of Minisatellite-region DNA (TU-DAMD) marker. Not. Sci. Biol. 2022, 14, 11174. [Google Scholar] [CrossRef]
- Saleh, B. Genetic Diversity of Salvia officinalis L. (Lamiaceae) and its Related Species using TU-DAMD Analysis. Open Agric. J. 2023, 17, e187433152305080. [Google Scholar] [CrossRef]
- Saleh, B. Molecular characterization using Directed Amplification of Minisatellite-region DNA (DAMD) Marker in Ficus sycomorus L.(Moraceae). Open Agric. J. 2019, 13, 74–81. [Google Scholar] [CrossRef]
- Bhatt, J.; Kumar, S.; Patel, S.; Solanki, R. Sequence-related amplified polymorphism (SRAP) markers based genetic diversity analysis of cumin genotypes. Ann. Agrarian Sci. 2017, 15, 434–438. [Google Scholar] [CrossRef]
- Wang, X.; Gao, C.; Li, K. Strategy for constructing Pinus yunnanensis germplasm bank for timber based on the SRAP molecular marker. Plant Sci. J. 2019, 37, 211–220. [Google Scholar]
- Zagorcheva, T.; Stanev, S.; Rusanov, K.; Atanassov, I. SRAP markers for genetic diversity assessment of lavender (Lavandula angustifolia mill.) varieties and breeding lines. Biotechnol. Biotechnol. Equip. 2020, 34, 303–308. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, L.; Wang, M.; Tang, Y.; Zhou, H.; Sun, Q.; Yu, Q.; Zhang, J. Evaluation of SRAP markers efficiency in genetic diversity of Aspergillus flavus from peanut-cropped soils in China. OCS 2022, 7, 135–141. [Google Scholar] [CrossRef]
- Fareghi, S.; Mirlohi, A.F.; Saeidi, G.; Khamisabadi, H. Evaluation of SRAP marker efficiency in identifying the relationship between genetic diversities of corn inbred lines with seed quantity and quality in derived hybrids. CMB 2019, 65, 6–14. [Google Scholar] [CrossRef]
- Suparman, S.; Pornpongrungrueng, P.; Chantaranothai, P. Molecular studies of coralberry (Ardisia crenata Sims; Primulaceae) from Thailand based on SCoT markers. Biodiversitas 2023, 24, d240611. [Google Scholar] [CrossRef]
- Chňapek, M.; Mikolášova, L.; Vivodík, M.; Gálová, Z.; Hromadová, Z.; Ražná, K.; Balážová, Ž. Genetic diversity of oat genotypes using SCoT markers. Biol. Life Sci. Forum 2021, 11, 29. [Google Scholar] [CrossRef]
- Gawroński, J.; Dyduch-Siemińska, M. Potential of In Vitro Culture of Scutellaria baicalensis in the Formation of Genetic Variation Confirmed by ScoT Markers. Genes 2022, 13, 2114. [Google Scholar] [CrossRef] [PubMed]
- Mirzahosein-Tabrizi, M.; Ghanavati, F.; Azizinezhad, R.; Etminan, A. Genetic diversity revealed by phytochemical and molec-ular analyses among and within eight Trigonella sp. J. Crop. Sci. Biotechnol. 2022, 26, 345–357. [Google Scholar] [CrossRef]
- Emam, M.A.; Abd El-Mageed, A.M.; Niedbała, G.; Sabrey, S.A.; Fouad, A.S.; Kapiel, T.; Piekutowska, M.; Mahmoud, S.A. Genetic characterization and agronomic evaluation of drought tolerance in an Egyptian wheat (Triticum aestivum L.) Cultivars. Agronomy 2022, 12, 1217. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, X.; Wang, J.; Xue, M.; Sun, C.; Dong, W. Evaluation of molecular and phenotypic diversity of Crataegus bretschneideri CK Schneid. and related species in China. Genet. Resour. Crop. Evol. 2023, 70, 221–234. [Google Scholar] [CrossRef]
- Singh, H.K.; Parveen, I.; Raghuvanshi, S.; Babbar, S.B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res. Notes 2012, 5, 42. [Google Scholar] [CrossRef]
- Feng, S.; Jiang, M.; Shi, Y.; Jiao, K.; Shen, C.; Lu, J.; Ying, Q.; Wang, H. Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 2016, 7, 1047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jiang, B. Species identification in complex groups of medicinal plants based on DNA barcoding: A case study on Astragalus spp. (Fabaceae) from southwest China. Conserv. Genet. Resour. 2020, 12, 469–478. [Google Scholar] [CrossRef]
- Kantar, F.; Yemşen, S.N.; Bülbül, C.; Yilmaz, N.; Mutlu, N. Phenotypic and iPBS-retrotransposon marker diversity in okra (Abelmoschus esculentus (L.) Moench) germplasm. Biotech Stud. 2021, 30, 7–15. [Google Scholar] [CrossRef]
- Eren, B.; Keskin, B.; Demirel, F.; Demirel, S.; Türkoğlu, A.; Yilmaz, A.; Haliloğlu, K. Assessment of genetic diversity and population structure in local alfalfa genotypes using iPBS molecular markers. Genet. Resour. Crop Evol. 2023, 70, 617–628. [Google Scholar] [CrossRef]
- Haliloğlu, K.; Türkoğlu, A.; Öztürk, H.I.; Özkan, G.; Elkoca, E.; Poczai, P. iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye. Genes 2022, 13, 1147. [Google Scholar] [CrossRef] [PubMed]
- Demirel, F.; Yıldırım, B.; Eren, B.; Demirel, S.; Türkoğlu, A.; Haliloğlu, K.; Nowosad, K.; Bujak, H.; Bocianowski, J. Revealing Genetic Diversity and Population Structure in Türkiye’s Wheat Germplasm Using iPBS-Retrotransposon Markers. Agronomy. 2024, 14, 300. [Google Scholar] [CrossRef]
- Sameeullah, M.; Kayaçetin, F.; Khavar, K.M.; Perkasa, A.Y.; Maesaroh, S.; Waheed, M.T.; Çiftçi, V. Decoding genetic diversity and population structure of Brassica species by inter primer binding site (iPBS) retrotransposon markers. Genet. Resour. Crop Evol. 2024. [Google Scholar] [CrossRef]
- Orhan, E.; Kara, D. Use of retrotransposon based iPBS markers for determination of genetic relationship among some Chestnut Cultivars (Castanea sativa Mill.) in Türkiye. Mol. Biol. Rep. 2023, 50, 8397–8405. [Google Scholar] [CrossRef] [PubMed]
- Etminan, A.; Pour-Aboughadareh, A.; Mehrabi, A.A.; Shooshtari, L.; Ahmadi-Rad, A.; Moradkhani, H. Molecular characterization of the wild relatives of wheat using CAAT-box derived polymorphism. Plant Biosyst. 2019, 153, 398–405. [Google Scholar] [CrossRef]
- Wang, L.; Li, G.; Peña, R.J.; Xia, X.; He, Z. Development of STS markers and establishment of multiplex PCR for Glu-A3 alleles in common wheat (Triticum aestivum L.). J. Cereal Sci. 2010, 51, 305–312. [Google Scholar] [CrossRef]
- Lee, S.; Heo, H.; Kwon, Y.; Lee, B. Development of gene-based STS markers in wheat. Korean J. Crop Sci. 2012, 57, 71–77. [Google Scholar] [CrossRef][Green Version]
- Dewi, A.K.; Rahayu, S.; Dwimahyani, I.; Reflinur, R. Analysis of yield and genetic diversity among Kewal local rice mutant lines based on STS markers. AIP Conf. Proc. 2021, 2381, 020012. [Google Scholar]
- Zhang, J.; Liu, W.; Lu, Y.; Liu, Q.; Yang, X.; Li, X.; Li, L. A resource of large-scale molecular markers for monitoring Agropyron cristatum chromatin introgression in wheat background based on transcriptome sequences. Sci. Rep. 2017, 7, 11942. [Google Scholar] [CrossRef]
- Wang, C.M.; Li, L.H.; Zhang, X.T.; Gao, Q.; Wang, R.F.; An, D.G. Development and application of EST-STS markers specific to chromosome 1RS of Secale cereale. Cereal Res. Commun. 2009, 37, 13–21. [Google Scholar] [CrossRef]
- Qiao, L.; Liu, S.; Li, J.; Li, S.; Yu, Z.; Liu, C.; Li, X.; Liu, J.; Ren, Y.; Zhang, P.; et al. Development of sequence-tagged site marker set for identification of J, JS, and St sub-genomes of Thinopyrum intermedium in wheat background. Front. Plant Sci. 2021, 12, 685216. [Google Scholar] [CrossRef]
- Srivastava, N.; Bajpai, A.; Chandra, R.; Rajan, S.; Muthukumar, M.; Srivastava, M.K. Comparison of PCR based marker systems for genetic analysis in different cultivars of mango. J. Environ. Biol. 2012, 33, 159. [Google Scholar]
- Talebi, R.; Nosrati, S.; Etminan, A.; Naji, A.M. Genetic diversity and population structure analysis of landrace and improved safflower (Cartamus tinctorious L.) germplasm using arbitrary functional gene-based molecular markers. Biotechnol. Biotechnol. Equip. 2018, 32, 1183–1194. [Google Scholar] [CrossRef]
- Aravanopoulos, F.A. Clonal identification based on quantitative, codominant, and dominant marker data: A comparative analysis of selected willow (Salix L.) clones. Int. J. For. Res. 2010, 2010, 906310. [Google Scholar] [CrossRef]
- Garcia, A.A.; Benchimol, L.L.; Barbosa, A.M.; Geraldi, I.O.; Souza Jr, C.L.; Souza, A.P.D. Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet. Mol. Biol. 2004, 27, 579–588. [Google Scholar] [CrossRef]
- Ho, W.S.; Wickneswari, R.; Mahani, M.C.; Shukor, M.N. Comparative genetic diversity studies of Shorea curtisii (Dipterocarpaceae): An assessment using SSR and DAMD markers. J. Trop. For. Sci. 2006, 22–35. [Google Scholar]
- Luo, Y.; Zhang, X.; Xu, J.; Zheng, Y.; Pu, S.; Duan, Z.; Li, Z.; Liu, G.; Chen, J.; Wang, Z. Phenotypic and molecular marker analysis uncovers the genetic diversity of the grass Stenotaphrum secundatum. BMC Genet. 2020, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, S.; Kumar, R.; Kumar, V.; Prasad, L.; Kumar, N.; Singh, D. Identification of blast resistance expression in rice genotypes using molecular markers (RAPD & SCAR). Afr. J. Biotechnol. 2010, 9, 3501–3509. [Google Scholar]
- Basu, A.; Ghosh, M.; Meyer, R.; Powell, W.; Basak, S.L.; Sen, S.K. Analysis of genetic diversity in cultivated jute determined by means of SSR markers and AFLP profiling. Crop Sci. 2004, 44, 678–685. [Google Scholar] [CrossRef]
- Ahmed, N.; Mir, J.I.; Mir, R.R.; Rather, N.A.; Rashid, R.; Wani, S.H.; Shafi, W.; Mir, H.; Sheikh, M.A. SSR and RAPD analysis of genetic diversity in walnut (Juglans regia L.) genotypes from Jammu and Kashmir, India. Physiol. Mol. Biol. Plants 2012, 18, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.M.; Farhat, S.; Mahajan, R.; Bhakhri, A.; Sharma, A. Unraveling the efficiency of RAPD and SSR markers in diversity analysis and population structure estimation in common bean. Saudi J. Biol. Sci. 2016, 23, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Mudigunda, S.; Mittal, P.K.; Arumugam, N. Comparative assessment of genetic diversity in Sesamum indicum L. using RAPD and SSR markers. 3 Biotech 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, W.F.; Rodrigues, J.F.; Koehler, S.; Gepts, P.; Veasey, E.A. Spatially structured genetic diversity of the Amerindian yam (Dioscorea trifida L.) assessed by SSR and ISSR markers in Southern Brazil. Genet. Resour. Crop Evol. 2013, 60, 2405–2420. [Google Scholar] [CrossRef]
- Hammami, R.; Jouve, N.; Soler, C.; Frieiro, E.; González, J.M. Genetic diversity of SSR and ISSR markers in wild populations of Brachypodium distachyon and its close relatives B. stacei and B.hybridum (Poaceae). Plant Syst. Evol. 2014, 300, 2029–2040. [Google Scholar] [CrossRef]
- Ramzan, M.; Sarwar, S.; Kauser, N.; Saba, R.; Hussain, I.; Shah, A.A.; Aslam, M.N.; Alkahtani, J.; Alwahibi, M.S. Assessment of Inter simple sequence repeat (ISSR) and simple sequence repeat (SSR) markers to reveal genetic diversity among Tamarix ecotypes. J. King Saud Univ. Sci. 2020, 32, 3437–3446. [Google Scholar] [CrossRef]
- Nazir, M.; Mahajan, R.; Hashim, M.J.; Iqbal, J.; Alyemeni, M.N.; Ganai, B.A.; Zargar, S.M. Deciphering allelic variability and population structure in buckwheat: An analogy between the efficiency of ISSR and SSR markers. Saudi J. Biol. Sci. 2021, 28, 6050–6056. [Google Scholar]
- Papaioannou, C.; Fassou, G.; Petropoulos, S.A.; Lamari, F.N.; Bebeli, P.J.; Papasotiropoulos, V. Evaluation of the Genetic Diversity of Greek Garlic (Allium sativum L.) Accessions Using DNA Markers and Association with Phenotypic and Chemical Variation. Agriculture 2023, 13, 1408. [Google Scholar] [CrossRef]
- Liu, S.; Feuerstein, U.; Luesink, W.; Schulze, S.; Asp, T.; Studer, B.; Becker, H.C.; Dehmer, K.J. DArT, SNP, and SSR analyses of genetic diversity in Lolium perenne L. using bulk sampling. BMC Genet. 2018, 19, 10. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, S.; Pulipati, S.; Ramasamy, K.; Jaganathan, D.; Venkatesan, S.D.; Vijay, G.; Kumari, K.; Raju, K.; Hariharan, G.N.; Venkataraman, G. Analysis of genetic diversity and population structure using SSR markers and validation of a Cleavage Amplified Polymorphic Sequences (CAPS) marker involving the sodium transporter OsHKT1;5 in saline tolerant rice (Oryza sativa L.) landraces. Gene 2019, 713, 143976. [Google Scholar] [CrossRef]
- Shahnazari, N.; Noormohammadi, Z.; Sheidai, M.; Koohdar, F. A new insight on genetic diversity of sweet oranges: CAPs-SSR and SSR markers. J. Genet. Eng. Biotechnol. 2022, 20, 105. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulou, A.; Ganopoulos, I.; Kalivas, A.; Nianiou-Obeidat, I.; Ralli, P.; Moysiadis, T.; Tsaftris, A.; Madesis, P. Comparative analysis of genetic diversity in Greek Genebank collection of summer squash (‘Cucurbita pepo’) landraces using start codon targeted (SCoT) polymorphism and ISSR markers. Aust. J. Crop Sci. 2015, 9, 14–21. [Google Scholar]
- Sharma, U.; Rai, M.K.; Shekhawat, N.S.; Kataria, V. Genetic homogeneity revealed in micropropagated Bauhinia racemosa Lam. using gene-targeted markers CBDP and SCoT. Physiol. Mol. Biol. Plants 2019, 25, 581–588. [Google Scholar] [CrossRef]
- Ghobadi, G.; Etminan, A.; Mehrabi, A.M.; Shooshtari, L. Molecular diversity analysis in hexaploid wheat (Triticum aestivum L.) and two Aegilops species (Aegilops crassa and Aegilops cylindrica) using CBDP and SCoT markers. J. Genet. Eng. Biotechnol. 2021, 19, 56. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al-Khayri, J.M.; Mahdy, E.M.B.; Taha, H.S.A.; Eldomiaty, A.S.; Abd-Elfattah, M.A.; Abdel Latef, A.A.H.; Rezk, A.A.; Shehata, W.F.; Almaghasla, M.I.; Shalaby, T.A.; et al. Genetic and Morphological Diversity Assessment of Five Kalanchoe Genotypes by SCoT, ISSR and RAPD-PCR Markers. Plants 2022, 11, 1722. [Google Scholar] [CrossRef] [PubMed]
- Osman, S.A.; Ali, H.B. Research Article Genetic Diversity of Five Lathyrus Species using RAPD, ISSR and SCoT Markers. Asian J. Plant Sci. 2020, 19, 152–165. [Google Scholar] [CrossRef]
- Sun, Q.B.; Li, L.F.; Li, Y.; Wu, G.J.; Ge, X.J. SSR and AFLP markers reveal low genetic diversity in the biofuel plant Jatropha curcas in China. Crop Sci. 2008, 48, 1865–1871. [Google Scholar] [CrossRef]
- Cao, J.; Zhou, Z.; Tu, J.; Cheng, S.; Yao, J.; Xu, F.; Wang, G.; Zhang, J.; Ye, J.; Liao, Y.; et al. Genetic diversity and population structure analysis of sand pear (Pyrus pyrifolia) ‘Nakai’varieties using SSR and AFLP markers. Not. Bot. Horti Agrobot. Cluj-Napoca 2019, 47, 970–979. [Google Scholar] [CrossRef]
- Saghir, K.; Abdelwahd, R.; Iraqi, D.; Lebkiri, N.; Gaboun, F.; El Goumi, Y.; Ibrahimi, M.; Abbas, Y.; Diria, G. Assessment of genetic diversity among wild rose in Morocco using ISSR and DAMD markers. J. Genet. Eng. Biotechnol. 2022, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Adu, B.G.; Akromah, R.; Amoah, S.; Nyadanu, D.; Yeboah, A.; Aboagye, L.M.; Amoah, R.A.; Owasu, E.G. High-density DArT-based Silico DArT, and SNP markers for genetic diversity and population structure studies in cassava (Manihot esculenta Crantz). PLoS ONE 2021, 16, e0255290. [Google Scholar] [CrossRef] [PubMed]
- Shaibu, A.S.; Ibrahim, H.; Miko, Z.L.; Mohammed, I.B.; Mohammed, S.G.; Yusuf, H.L.; Kamara, A.Y.; Omoigui, L.O.; Karikari, B. Assessment of the genetic structure and diversity of soybean (Glycine max L.) germplasm using diversity array technology and single nucleotide polymorphism markers. Plants 2021, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, J.; Su, D.; Yang, J.; Wang, E.; Wu, S.; Li, M.; Ma, L. Discrimination of tobacco cultivars using SCAR and RAPD markers. Czech J. Genet. Plant Breed. 2020, 56, 170–173. [Google Scholar] [CrossRef]
- Dou, J.; Lu, X.; Ali, A.; Zhao, S.; Zhang, L.; He, N.; Liu, W. Genetic mapping reveals a marker for yellow skin in watermelon (Citrullus lanatus L.). PLoS ONE 2018, 13, e0200617. [Google Scholar] [CrossRef] [PubMed]
- Katzir, N.; Tadmor, Y.; Tzuri, G.; Leshzeshen, E.; Mozes-Daube, N.; Danin-Poleg, Y.; Paris, H.S. Further ISSR and preliminary SSR analysis of relationships among accessions of Cucurbita pepo. In Proceedings of the VII Eucarpia Meeting on Cucurbit Genetics and Breeding, Ma’ale Ha Hamisha, Israel, 19–23 March 2000; Volume 510, pp. 433–440. [Google Scholar]
- Sagar, M.R.; Kumar, S.; Patidar, D.; Sakure, A.A. Morphological, physico-biochemical and marker-based diversity of desi cotton (Gossypium herbaceum L.) germplasm. J. King Saud Univ. Sci. 2022, 34, 102336. [Google Scholar] [CrossRef]
- Ghasemzadeh Baraki, S.; Nikzat Siahkolaee, S. Assessment of SCoT and DAMD molecular markers in genetic diversity and species delimitation of three moss species grown in Iran. Iran. J. Genet. Plant Breed. 2018, 7, 33–41. [Google Scholar]
- Alaaddin Ahmed, A.; Anwar Qadir, S.; Tahir, N.A.R. CDDP and ISSR markers-assisted diversity and structure analysis in Iraqi Mazu (Quercus infectoria Oliv.) accessions. All Life 2022, 15, 247–261. [Google Scholar] [CrossRef]
- Premjet, D.; Boonsrangsom, T.; Sujipuli, K.; Rattanasut, K.; Kongbungkerd, A.; Premjet, S. Morphological and Molecular Characterizations of Musa (ABB)’Mali-Ong’in Thailand. Biology 2022, 11, 1429. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.Y.; Tsai, Y.Z.; Lin, S.F. Development of STS and CAPS markers for variety identification and genetic diversity analysis of tea germplasm in Taiwan. Bot. Stud. 2014, 55, 12. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhai, L.; Long, H.; Chen, N.; Gao, C.; Ding, Z.; Jin, B. Genetic diversity of Bletilla striata assessed by SCoT and IRAP markers. Hereditas 2018, 155, 35. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.R.; Pandey, B.K. Potential of molecular markers in plant biotechnology. Plant Omics 2009, 2, 141–162. [Google Scholar]
- Jonah, P.M.; Bello, L.L.; Lucky, O.; Midau, A.; Moruppa, S.M. The importance of molecular markers in plant breeding programmes. Glob. J. Sci. Front. Res. 2011, 11, 5–12. [Google Scholar]
- Varshney, R.K.; Mahendar, T.; Aggarwal, R.K.; Börner, A. Genic Molecular Markers in Plants: Development and Applications. In Genomics-Assisted Crop Improvement; Varhney, R.K., Tuberosa, R., Eds.; Springer: Dordrecht, The Netherland, 2007; Volume 1, pp. 13–29. ISBN 9781402062957. [Google Scholar]
- Jin, J.; Lu, P.; Xu, Y.; Tao, J.; Li, Z.; Wang, S.; Yu, S.; Wang, C.; Xie, X.; Gao, J.; et al. PCMDB: A curated and comprehensive resource of plant cell markers. Nucleic Acids Res. 2022, 50, D1448–D1455. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, H.; Tao, J.; Ren, Y.; Xu, C.; Wu, K.; Zou, C.; Zhang, J.; Xu, Y. Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize. Mol. Breed. 2019, 39, 37. [Google Scholar] [CrossRef]
- Jagtap, A.B.; Vikal, Y.; Johal, G.S. Genome-wide development and validation of cost-effective KASP marker assays for genetic dissection of heat stress tolerance in maize. Int. J Mol. Sci. 2020, 21, 7386. [Google Scholar] [CrossRef] [PubMed]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef]
- Wakeley, J.; Nielsen, R.; Liu-Cordero, S.N.; Ardlie, K. The discovery of single-nucleotide polymorphisms—And inferences about human demographic history. Am. J. Hum. Genet. 2001, 69, 1332–1347. [Google Scholar] [CrossRef] [PubMed]
- Morgil, H.; Can Gercek, Y.; Tulum, I. Single nucleotide polymorphisms (SNPs) in plant genetics and breeding. In The Recent Topics in Genetic Polymorphisms; IntechOpen: London, UK, 2020; pp. 400–825. [Google Scholar] [CrossRef]
- Hinze, L.L.; Hulse-Kemp, A.M.; Wilson, I.W.; Zhu, Q.H.; Llewellyn, D.J.; Taylor, J.M.; Spriggs, A.; Fang, D.D.; Ulleoa, M.; Burke, J.J.; et al. Diversity analysis of cotton (Gossypium hirsutum L.) germplasm using the CottonSNP63K Array. BMC Plant Biol. 2017, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, G.; Shimelis, H.; Laing, M.; Lule, D.; Assefa, E.; Mathew, I. Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) landraces using SNP markers. S. Afr. J. Plant Soil 2020, 37, 220–226. [Google Scholar] [CrossRef]
- Oliveira, L.S.D.; Schuster, I.; Novaes, E.; Pereira, W.A. SNP genotyping for fast and consistent clustering of maize inbred lines into heterotic groups. Crop Breed. Appl. Biotechnol. 2021, 21, e367121110. [Google Scholar] [CrossRef]
- Niu, S.; Song, Q.; Koiwa, H.; Qiao, D.; Zhao, D.; Chen, Z.; Liu, X.; Wen, X. Genetic diversity, linkage disequilibrium, and population structure analysis of the tea plant (Camellia sinensis) from an origin center, Guizhou plateau, using genome-wide SNPs developed by genotyping-by-sequencing. BMC Plant Biol. 2019, 19, 328. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Zhao, K.; Wright, M.; Tung, C.W.; Ebana, K.; Thomson, M.; Reynolds, A.; Wang, D.; DeClerck, G.; Ali, M.L.; et al. Development of genome-wide SNP assays for rice. Breed. Sci. 2010, 60, 524–535. [Google Scholar] [CrossRef]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T.; et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wu, T.; Ye, J.; Sun, J.; Jiang, Y.; Yu, J.; Tang, J.; Chen, G.; Wang, C.; Wan, J. SNP-based analysis of genetic diversity reveals important alleles associated with seed size in rice. BMC Plant Biol. 2016, 16, 93. [Google Scholar]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef]
- Fu, Y.B. Genetic diversity analysis of highly incomplete SNP genotype data with imputations: An empirical assessment. G3-Genes Genom Genet. 2014, 4, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Montero-Pau, J.; Blanca, J.; Esteras, C.; Martínez-Pérez, E.M.; Gómez, P.; Monforte, A.J.; Cañizares, J.; Picó, B. An SNP-based saturated genetic map and QTL analysis of fruit-related traits in Zucchini using Genotyping-by-sequencing. BMC Genom. 2017, 18, 94. [Google Scholar] [CrossRef] [PubMed]
- Imai, A.; Nonaka, K.; Kuniga, T.; Yoshioka, T.; Hayashi, T. Genome-wide association mapping of fruit-quality traits using genotyping-by-sequencing approach in citrus landraces, modern cultivars, and breeding lines in Japan. Tree Genet. Genomes 2018, 14, 24. [Google Scholar] [CrossRef]
- Tomar, V.; Dhillon, G.S.; Singh, D.; Singh, R.P.; Poland, J.; Joshi, A.K.; Tiwari, B.S.; Kumar, U. Elucidating SNP-based genetic diversity and population structure of advanced breeding lines of bread wheat (Triticum aestivum L.). PeerJ 2021, 9, e11593. [Google Scholar] [CrossRef] [PubMed]
- Díaz, B.G.; Zucchi, M.I.; Alves-Pereira, A.; de Almeida, C.P.; Moraes, A.C.L.; Vianna, S.A.; Azevedo-Filho, J.; Colombo, C.A. Genome-wide SNP analysis to assess the genetic population structure and diversity of Acrocomia species. PLoS ONE 2021, 16, e0241025. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z.; Li, J.; Liu, Y.; Song, M.; Lockhart, P.J.; Tian, Y.; Niu, M.; Wang, Q. RADseq-based population genomic analysis and environmental adaptation of rare and endangered recretohalophyte Reaumuria trigyna. Plant Genom. 2023, 17, e20303. [Google Scholar] [CrossRef]
- Serba, D.D.; Muleta, K.T.; St. Amand, P.; Bernardo, A.; Bai, G.; Perumal, R.; Bashir, E. Genetic diversity, population structure, and linkage disequilibrium of pearl millet. Plant Genome 2019, 12, 180091. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Slonecki, T.J.; Rutter, W.B.; Olukolu, B.A.; Yencho, G.C.; Jackson, D.M.; Wadl, P.A. Genetic diversity, population structure, and selection of breeder germplasm subsets from the USDA sweet potato (Ipomoea batatas) collection. Front. Plant Sci. 2023, 13, 1022555. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.P.; Sibiya, J.; Kutu, F. Genetic diversity and population structure of maize inbred lines using phenotypic traits and single nucleotide polymorphism (SNP) markers. Sci. Rep. 2023, 13, 17851. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, Y.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wong, Y.; Ji, J.; et al. Genetic diversity and population structure analysis of 161 broccoli cultivars based on SNP markers. Hortic. Plant J. 2021, 7, 423–433. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.W.; Huang, J.L.; Ya, J.D.; Zhe, M.Q.; Zeng, C.X.; Zhang, Z.R.; Zhang, S.B.; Li, D.Z.; Li, H.T.; et al. DNA barcoding of Cymbidium by genome skimming: Call for next-generation nuclear barcodes. Mol. Ecol. Resour. 2023, 23, 424–439. [Google Scholar] [CrossRef] [PubMed]
- ŞapcıSelamoğlu, H. DNA barcoding of two narrow endemic plants; Astragalus argaeus and Astragalus stenosemioides from Mount Erciyes, Turkey. Conserv. Genet. Resour. 2022, 14, 81–84. [Google Scholar] [CrossRef]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and applications of a high throughput genotyping tool for polyploid crops: Single nucleotide polymorphism (SNP) array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- LaFramboise, T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef]
- Heslot, N.; Rutkoski, J.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Impact of marker ascertainment bias on genomic selection accuracy and estimates of genetic diversity. PLoS ONE 2013, 8, e74612. [Google Scholar] [CrossRef] [PubMed]
- Albrechtsen, A.; Nielsen, F.C.; Nielsen, R. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 2010, 27, 2534–2547. [Google Scholar] [CrossRef] [PubMed]
- Chat, V.; Ferguson, R.; Morales, L.; Kirchhoff, T. Ultra-low-coverage whole-genome sequencing as an alternative to genotyping arrays in genome-wide association studies. Front. Genet. 2022, 12, 790445. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Chavarro, C.; Agarwal, G.; Bertioli, D.J.; Leal-Bertioli, S.C.M.; Pandey, M.K.; Vaughn, J.; Abernathy, B.; Barkley, N.A.; et al. Genome-wide SNP Genotyping Resolves Signatures of Selection and Tetrasomic Recombination in Peanut. Mol. Plant 2017, 10, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Cowan, R.S.; Chase, M.W.; Kress, W.J.; Savolainen, V. 300,000 species to identify: Problems, progress, and prospects in DNA barcoding of land plants. Taxon 2006, 55, 611–616. [Google Scholar] [CrossRef]
- Mursyidin, D.H.; Setiawan, A. Assessing diversity and phylogeny of Indonesian breadfruit (Artocarpus spp.) using internal transcribed spacer (ITS) region and leaf morphology. J. Genet. Eng. Biotechnol. 2023, 21, 15. [Google Scholar]
- Massicotte, R.; Whitelaw, E.; Angers, B. DNA methylation: A source of random variation in natural populations. Epigenetics 2011, 6, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Z.; Li, H.L.; Li, J.M.; Yu, F.H. Correlations between genetic, epigenetic and phenotypic variation of an introduced clonal herb. Heredity 2020, 124, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, K.J.; Jansen, J.J.; Van Dijk, P.J.; Biere, A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010, 185, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.S.; Choi, S.C.; Jun, T.H.; Kim, C. Genotyping-by-sequencing: A promising tool for plant genetics research and breeding. Hortic. Environ. Biotechnol. 2017, 58, 425–431. [Google Scholar] [CrossRef]
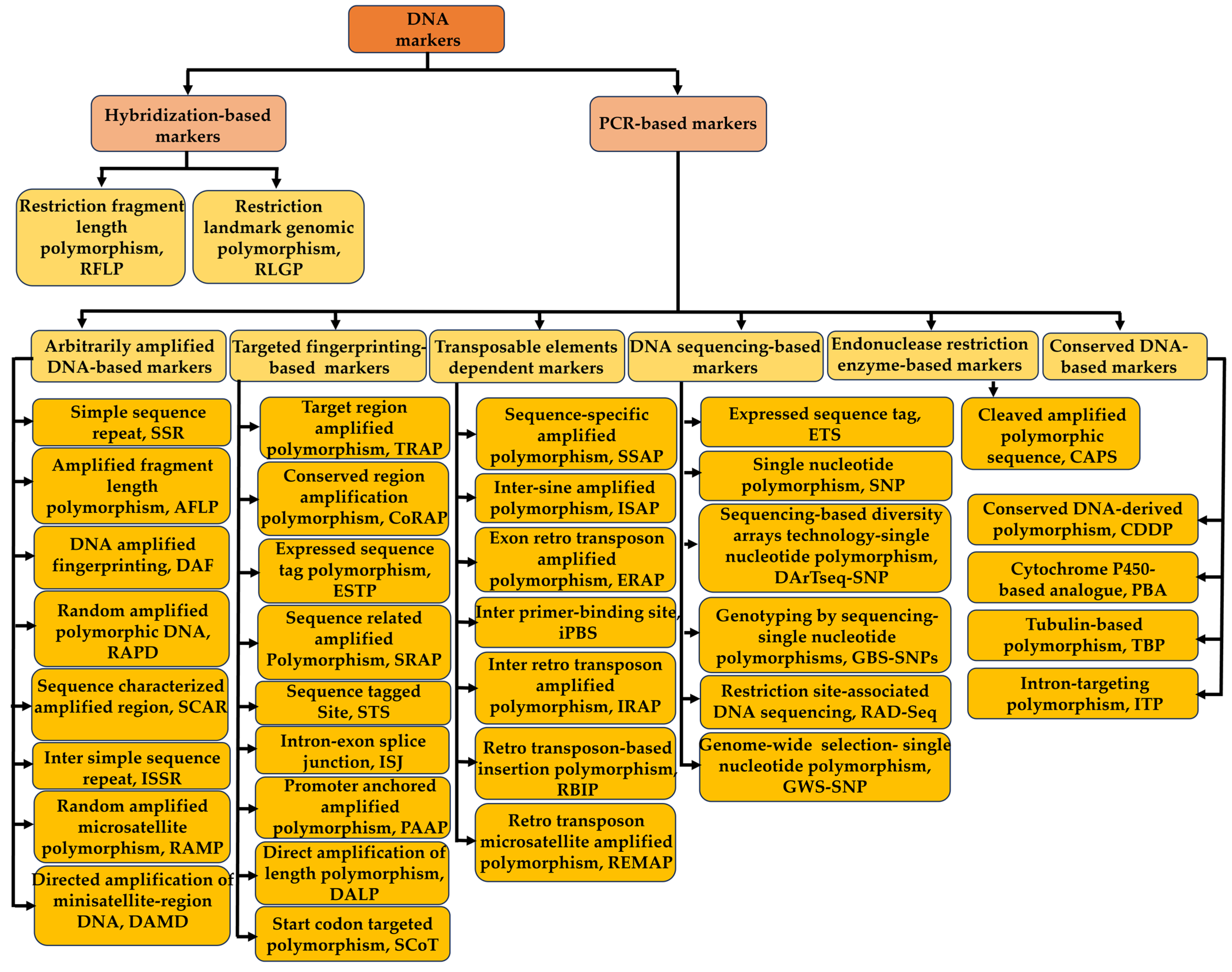
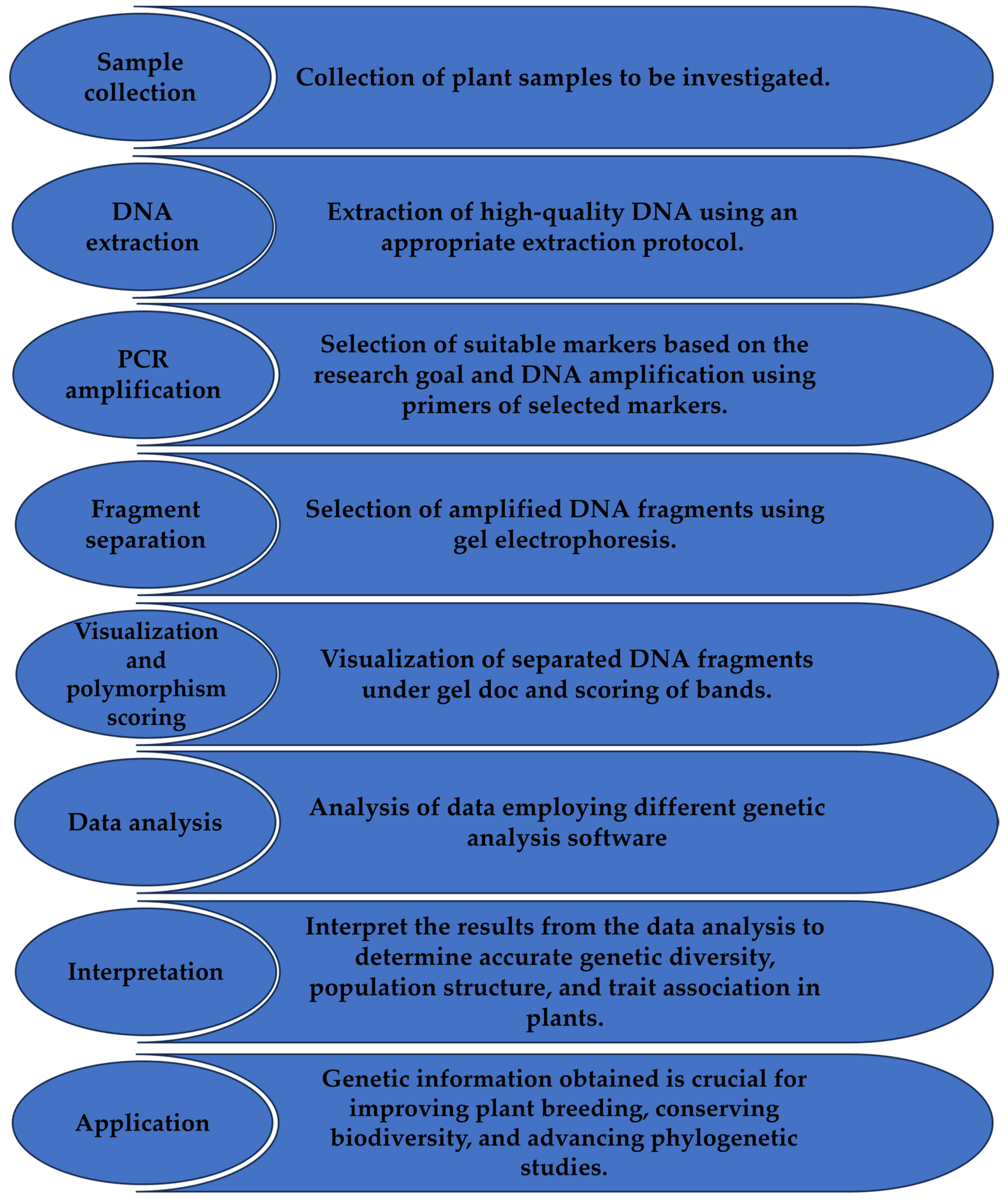
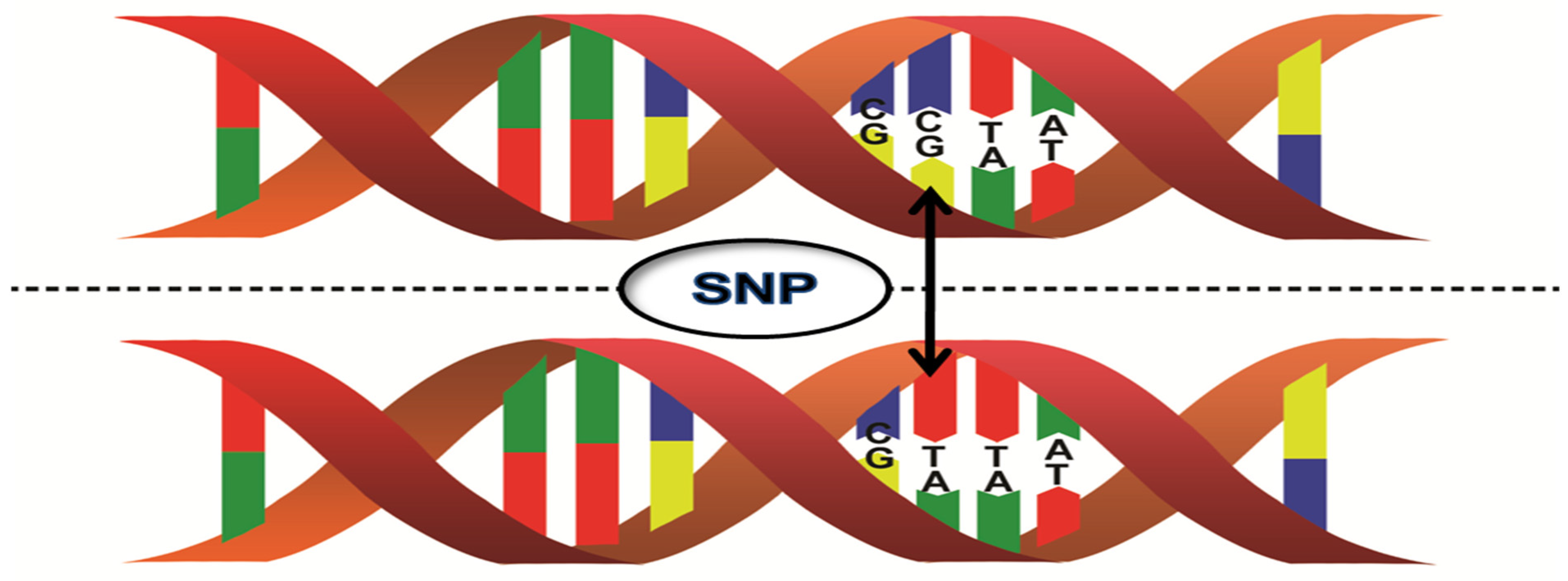

| Marker Combination | Marker Types | Number of Marker Systems | Plant Species | References |
|---|---|---|---|---|
| SCoT + ISSR | Dominant | Two | Dendrobium crysotoxum (SCoT = nine primers; ISSR = twenty primers; genetic diversity within population: ISSR = 86%; SCoT = 74% and between population: ISSR = 14%, SCoT = 26%) Diospyros species (ISSR = seven primers; SCoT = ten primers; average PIC: ISSR = 0.30, SCoT = 0.36; average marker index (MI): ISSR = 1.81; SCoT = 1.79) Cucurbita pepo (seven SCoT primers produced forty-nine polymorphic bands and six ISSR primers generated forty-two bands) | [33,165,266] |
| CBDP + SCoT | Dominant | Two | Bauhinia racemose (out of 25 CBDP primers, 21 produced 97 scorable bands, and for SCoT, 18 out of 36 primers produced 88 scorable bands) Triticum aestivum, Aegilops cylindrical, and A. crassa (CBDP = 15 primers; SCoT = 15 primers; PIC for SCoT: 0.31–0.39, CBDP: 0.28–0.36; cluster analysis: all samples were grouped based on their genomic constitution) | [267,268] |
| SCoT + ISSR + RAPD | Dominant | Three | Kalanchoe genotype (ScoT, ISSR, and RAPD = 10 primers each; polymorphism percentage: SCoT = 57%; ISSR = 15%, RAPD = 60.25%) Lathyrus species (SCoT = eight primers; ISSR = eight primers; RAPD = six primers; polymorphism: SCoT = 96%; ISSR = 96.81%; RAPD = 94.2%) | [269,270] |
| SSR + AFLP | Co-dominant and dominant | Two | Jatropha curcas (seven AFLP primer combinations produced seventy amplified polymorphic loci; thirty SSR primers were used, out of which seventeen were amplified in an appropriate size range) Pyrus pyrifolia (SSR; AFLP = 10 primers each; average PIC for SSR = 0.7585; polymorphism percentage for AFLP = 86.46%; genetic diversity: rich and highly representative) | [271,272] |
| ISSR + DAMD | Dominant | Two | Rosa species (ISSR = ten primers; DAMD = eight primers; genetic variation within population = 86%, between populations = 14%) | [273] |
| DArT + SNP | Dominant and co-dominant | Two | Manihot esculenta (DArT = 10,521 markers; SNP = 10,808 markers; average PIC for DArT = 0.36; SNP = 0.28) Glycine max (DArT = 16,116 markers; SNP = 19,505 markers; genetic variance: DArT = 98%; SNP = 97%) | [274,275] |
| DArT + SNP + SSR | Dominant and co-ominant | Three | Lolium perenne (DArT = 1384 markers; SNP = 182 markers; SSR = 48 markers; Genetic diversity: DArT = 0.26; SNP = 0.32; SSR = 0.45) | [263] |
| SCAR + RAPD | Co-dominant and dominant | Two | Nicotiana tabacum (two out of eight SCAR markers; seven out of two hundred RAPD markers efficiently discriminated a large number of Tobacco cultivars) | [276] |
| CAPS + SSR | Co-dominant | Two | Oryza sativa (a set of twenty-eight genome-wide SSR markers; eleven salt-responsive genic SSR markers; eight salt QT-linked SSR markers; CAPS markers: OsHKT1; 5v395) Citrus sinensis (a total of five markers; average genetic polymorphism = 98.46%; CAPs-SSR indicated more genetic variability) | [264,265] |
| CAPS + SSR + SNP | Co-dominant | Three | Citrullus lanatus (CAPS = fifteen markers; SSR = six markers; SNP = two markers; mapping confirmation of BSA-seq: yellow skin) | [277] |
| SSR + ISSR | Co-dominant and dominant | Two | A total of 28 accessions of Curcubita pepo were compared utilizing ISSR markers, detecting 90 polymorphic bands. Additionally, SSR markers were proposed to further elucidate infra-specific relationships within C. pepo. Gossypium herbaceum (SSR = thirteen markers; ISSR = five markers; average coefficient similarity = 0.32; low correlation and high variation) | [278,279] |
| SCoT + DAMD | Dominant and co-dominant | Two | Mosses (the inaugural genetic diversity study of three moss species incorporated the utilization of SCoT and DAMD markers to enhance the discriminatory power and precision within the species) | [280] |
| CDDP+ ISSR | Dominant | Two | Quercus infectoria (ISSR = twelve primers; CDDP = nine primers; population variance within: ISSR = 92.97%; CDDP = 94.17%) | [281] |
| ISSR + SRAP | Dominant | Two | Musa species (ISSR = eight primers; SRAP = seven primers; polymorphic bands: ISSR = 81.6%; SRAP = 87.7%) | [282] |
| STS + CAPS | Co-dominant | Two | Camelia sinensis (STS = two primers; CAPS = thirty-seven primers; high genetic diversity between the two varieties: C. sinensis var. sinensis and C. sinensis var. assamica) | [283] |
| SCoT + IRAP | Dominant | Three | Bletilla striata (SCoT = twenty primers; IRAP = eight primers; polymorphic bands: SCoT = 96.17%; IRAP = 94%) | [284] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bidyananda, N.; Jamir, I.; Nowakowska, K.; Varte, V.; Vendrame, W.A.; Devi, R.S.; Nongdam, P. Plant Genetic Diversity Studies: Insights from DNA Marker Analyses. Int. J. Plant Biol. 2024, 15, 607-640. https://doi.org/10.3390/ijpb15030046
Bidyananda N, Jamir I, Nowakowska K, Varte V, Vendrame WA, Devi RS, Nongdam P. Plant Genetic Diversity Studies: Insights from DNA Marker Analyses. International Journal of Plant Biology. 2024; 15(3):607-640. https://doi.org/10.3390/ijpb15030046
Chicago/Turabian StyleBidyananda, Nongthombam, Imlitoshi Jamir, Karolina Nowakowska, Vanlalrinchhani Varte, Wagner A. Vendrame, Rajkumari Sanayaima Devi, and Potshangbam Nongdam. 2024. "Plant Genetic Diversity Studies: Insights from DNA Marker Analyses" International Journal of Plant Biology 15, no. 3: 607-640. https://doi.org/10.3390/ijpb15030046
APA StyleBidyananda, N., Jamir, I., Nowakowska, K., Varte, V., Vendrame, W. A., Devi, R. S., & Nongdam, P. (2024). Plant Genetic Diversity Studies: Insights from DNA Marker Analyses. International Journal of Plant Biology, 15(3), 607-640. https://doi.org/10.3390/ijpb15030046







