Genomic Regions and Candidate Genes for Seed Iron and Seed Zinc Accumulation Identified in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Analysis for Seed Fe and Zn
2.3. DNA Isolation, SNP Genotyping, and Genetic Map Construction
2.4. Fe and Zn QTL Detection and Their Candidate Genes
2.5. Statistical Analysis
3. Results
3.1. ANOVA and Statistical Analysis
3.2. Fe and Zn QTL
3.3. Fe and Zn Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saleem, M.H.; Rizwan, M.; Jabri, H.A.; Alsafran, M. Functions and strategies for enhancing zinc availability in plants for sustainable agriculture. Front. Plant Sci. 2022, 13, 1033092. [Google Scholar] [CrossRef] [PubMed]
- Al-Mutairi, A.A.; Cavagnaro, T.R.; Khor, S.F.; Neumann, K.; Burton, R.A.; Watts-Williams, S. The effect of zinc fertilisation and arbuscular mycorrhizal fungi on grain quality and yield of contrasting barley cultivars. Funct. Plant Biol. 2020, 47, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Andresen, E.; Peiter, E.; Küpper, H. Trace metal metabolism in plants. J. Exp. Bot. 2018, 69, 909–954. [Google Scholar] [CrossRef] [PubMed]
- Mengel, K.; Kirkby, E.A. Principles of Plant Nutrition; International Potash Institute: Worblaufen, Switzerland, 1982. [Google Scholar]
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: London, UK, 1995. [Google Scholar]
- Bellaloui, N.; Smith, J.R.; Ray, J.D.; Gillen, A.M. Effect of maturity on seed composition in the early soybean production system as measured on near-isogenic soybean lines. Crop Sci. 2009, 49, 608–620. [Google Scholar] [CrossRef]
- Bellaloui, N.; Khandaker, L.; Akond, M.; Kantartzi, S.K.; Meksem, K.; Mengistu, M.; Lightfoot, D.A.; Kassem, M.A. Identification of QTL underlying seed micronutrients accumulation in ‘MD 96-5722’ by ‘Spencer’ recombinant inbred lines of soybean. Atlas J. Plant Biol. 2015, 1, 39–49. [Google Scholar] [CrossRef]
- Wang, H.; Jia, J.; Cai, Z.; Duan, M.; Jiang, Z.; Xia, Q.; Ma, Q.; Lian, T.; Nian, H. Identification of quantitative trait loci (QTLs) and candidate genes of seed Iron and zinc content in soybean [Glycine max (L.) Merr.]. BMC Genom. 2022, 146, 146. [Google Scholar] [CrossRef]
- King, K.E.; Peiffer, G.A.; Reddy, M.; Lauter, N.; Lin, S.F.; Cianzio, S.; Shoemaker, R.C. Mapping of iron and zinc quantitative trait loci in soybean for association to iron deficiency chlorosis resistance. J. Plant Nutr. 2013, 36, 2132–2153. [Google Scholar] [CrossRef]
- Grotz, N.; Guerinot, M. Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta 2006, 1763, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Kirkby, E.A.; Romheld, V. Micronutrients in Plant Physiology: Functions, Uptake, and Mobility; International Fertilizer Society: Cambridge, UK, 2004. [Google Scholar]
- Sousa, S.F.; Lopes, A.B.; Fernandes, P.A.; Ramos, M.J. The zinc proteome: A tale of stability and functionality. Dalton Trans. 2009, 38, 7946–7956. [Google Scholar] [CrossRef]
- Cakmak, I. Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 2000, 146, 185–205. [Google Scholar] [CrossRef]
- Prask, J.A.; Plocke, D.J.A. Role of zinc in the structural integrity of the cytoplasmic ribosomes of Euglena gracilis. Plant Physiol. 1971, 48, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.E. Zinc proteins: Enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. BioChem. 1992, 61, 897–946. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I.; Rosato, A. Metalloproteomes: A bio-informatic approach. Acc. Chem. Res. 2009, 42, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Sekimoto, H.; Hoshi, M.; Nomura, T.; Yokota, T. Zinc deficiency affects the levels of endogenous gibberellins in Zea mays L. Plant Cell Physiol. 1997, 38, 1087–1090. [Google Scholar] [CrossRef]
- Maret, W.; Li, Y. Coordination dynamics of zinc in proteins. Chem. Rev. 2009, 109, 4682–4707. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I.; Cavallaro, G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS ONE 2011, 6, e26325. [Google Scholar] [CrossRef]
- Lin, S.; Cianzio, S.; Shoemaker, R.C. Mapping genetic loci for iron deficiency chlorosis in soybean. Mol. Breed. 1997, 3, 219–229. [Google Scholar] [CrossRef]
- Kassem, M.A. Seed amino acids, macronutrients, micronutrients, sugars, and other compounds. In Soybean Seed Composition: Protein, Oil, Fatty Acids, Amino Acids, Sugars, Mineral Nutrients, Tocopherols, and Isoflavones; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Ramamurthy, R.K.; Jedlicka, J.; Graef, G.L.; Waters, B.M. Identification of new QTLs for seed mineral, cysteine, and methionine concentrations in soybean [Glycine max (L.) Merr.]. Mol. Breed. 2014, 34, 431–445. [Google Scholar] [CrossRef]
- Diers, B.W.; Cianzio, S.R.; Shoemaker, R.C. Possible identification of quantitative trait loci affecting iron efficiency in soybean. J. Plant Nutr. 1992, 10, 2127–2136. [Google Scholar] [CrossRef]
- Kumar, S.; Hash, C.T.; Thirunavukkarasu, N.; Singh, G.; Rajaram, V.; Rathore, A.; Senapathy, S.; Mahendrakar, M.D.; Yadav, R.S.; Srivastava, R.K. Mapping quantitative trait loci controlling high iron and zinc content in self and open pollinated grains of pearl millet [Pennisetum glaucum (L.) R. Br.]. Front. Plant Sci. 2016, 7, 1636. [Google Scholar] [CrossRef]
- Rai, K.N.; Govindaraj, M.; Rao, A.S. Genetic enhancement of grain iron and zinc content in pearl millet. Qual. Assur. Saf. Crops Foods 2012, 4, 119–125. [Google Scholar] [CrossRef]
- Qin, J.; Shi, A.; Mou, B.; Grusak, M.A.; Weng, Y.; Ravelombola, W.; Bhattarai, G.; Dong, L.; Yang, W. Genetic diversity and association mapping of mineral element concentrations in spinach leaves. BMC Genom. 2017, 18, 941. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, E.E.; Epps, J.M. Registration of ‘Forrest’ soybeans. Crop Sci. 1973, 13, 287. [Google Scholar] [CrossRef]
- Bernard, R.L.; Cremeens, C.R. Registration of Williams 82 soybean. Crop Sci. 1988, 28, 1027–1028. [Google Scholar] [CrossRef]
- Shultz, J.L.; Kurunam, D.; Shopinski, K.; Iqbal, M.J.; Kazi, S.; Zobrist, K.; Bashir, R.; Yaegashi, S.; Lavu, N.; Afzal, A.J.; et al. The soybean genome database (SoyGD): A browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res. 2006, 34, D758–D765. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Vuong, T.D.; Leroy, J.A.; Shannon, J.G.; Sleper, D.A.; Nguyen, H.T. Selection of a core set of RILs from Forrest x Williams to develop a framework map in soybean. Theor. Appl. Genet. 2011, 122, 1179–1187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knizia, D.; Yuan, J.; Bellaloui, N.; Vuong, T.; Usovsky, M.; Usovsky, M.; Song, Q.; Betts, F.; Register, T.; Williams, E.; et al. The soybean high density ‘Forrest’ by ‘Williams 82’ SNP-based genetic linkage map identifies QTL and candidate genes for seed isoflavone content. Plants 2021, 10, 2029. [Google Scholar] [CrossRef] [PubMed]
- Bellaloui, N.; Saha, S.; Tonos, J.L.; Scheffler, J.A.; Jenkins, J.N.; McCarty, J.C.; Stelly, D.M. Effects of interspecific chromosome substitution in upland cotton on cottonseed micronutrients. Plants 2020, 9, 1081. [Google Scholar] [CrossRef] [PubMed]
- Bellaloui, N.; Smith, J.R.; Gillen, A.M.; Ray, J.D. Effects of maturity, genotypic background, and temperature on seed mineral composition in near-isogenic soybean lines in the early soybean production system. Crop Sci. 2011, 51, 1161–1171. [Google Scholar] [CrossRef]
- Bellaloui, N.; Turley, R.B. Effects of fuzzless cottonseed phenotype on cottonseed nutrient composition in near isogenic cotton (Gossypium hirsutum L.) mutant lines under well-watered and water stress conditions. Front. Plant Sci. 2013, 4, 516. [Google Scholar] [CrossRef]
- Vuong, T.D.; Sleper, D.A.; Shannon, J.G.; Nguyen, H.T. Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor. Appl. Genet. 2010, 121, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Yan, L.; Quigley, C.; Fickus, E.; Wei, H.; Chen, L.; Dong, F.; Araya, S.; Liu, J.; Hyten, D.; et al. Soybean BARCSoySNP6K: An assay for soybean genetics and breeding research. Plant J. 2020, 104, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. Joinmap 4.0 Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Plant Research International: Wageningen, The Netherlands, 2006. [Google Scholar]
- Brown, A.V.; Conners, S.I.; Huang, W.; Wilkey, A.P.; Grant, D.; Weeks, N.T.; Cannon, S.B.; Graham, M.A.; Nelson, R.T. A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2020, 49, D1496–D1501. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef] [PubMed]
- Pilet-Nayel, M.L.; Muehlbauer, F.J.; McGee, R.J.; Kraft, J.M.; Baranger, A.; Coyne, C.J. Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 2002, 106, 28–39. [Google Scholar] [CrossRef] [PubMed]
- R Software. Available online: https://www.r-project.org (accessed on 15 June 2023).
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, NCSU: Raleigh, NC, USA, 2012; Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 8 August 2023).
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTL. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- SoyBase Genome Browser. Available online: https://www.soybase.org/SequenceIntro.php (accessed on 9 April 2024).
- Statistical Analysis Systems (SAS); SAS Institute: Cary, NC, USA, 2002–2012.
- Ning, L.; Sun, P.; Wang, Q.; Ma, D.; Hu, Z.; Zhang, D.; Zhang, G.; Cheng, H.; Yu, D. Genetic architecture of biofortification traits in soybean (Glycine max L. Merr.) revealed through association analysis and linkage mapping. Euphytica 2015, 204, 353–369. [Google Scholar] [CrossRef]
- Dhanapal, A.P.; Ray, J.D.; Smith, J.R.; Purcell, L.C.; Fritschi, F.B. Identification of novel genomic loci associated with soybean shoot tissue macro and micronutrient concentrations. Plant Gen. 2018, 11, 170066. [Google Scholar] [CrossRef]
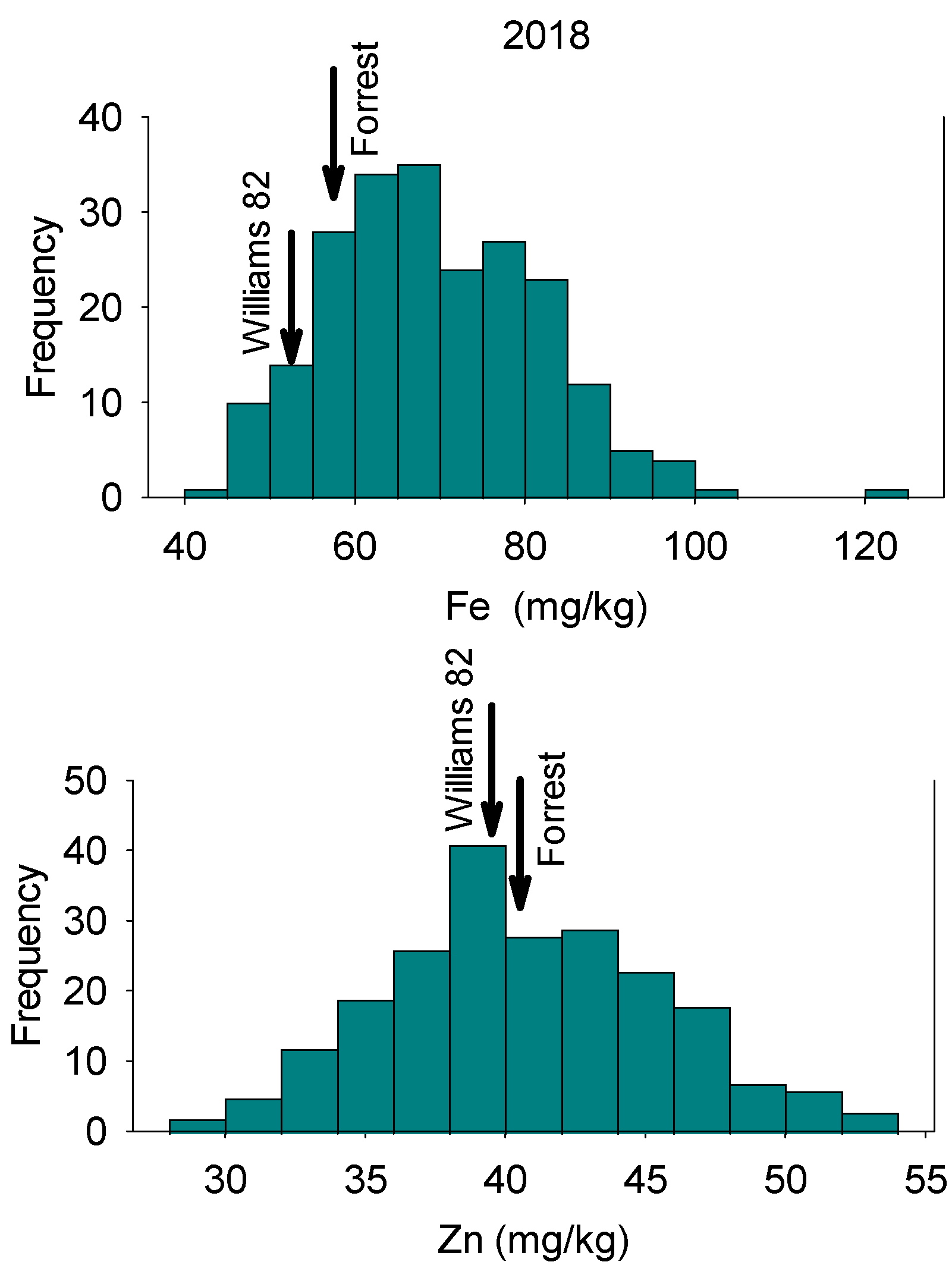
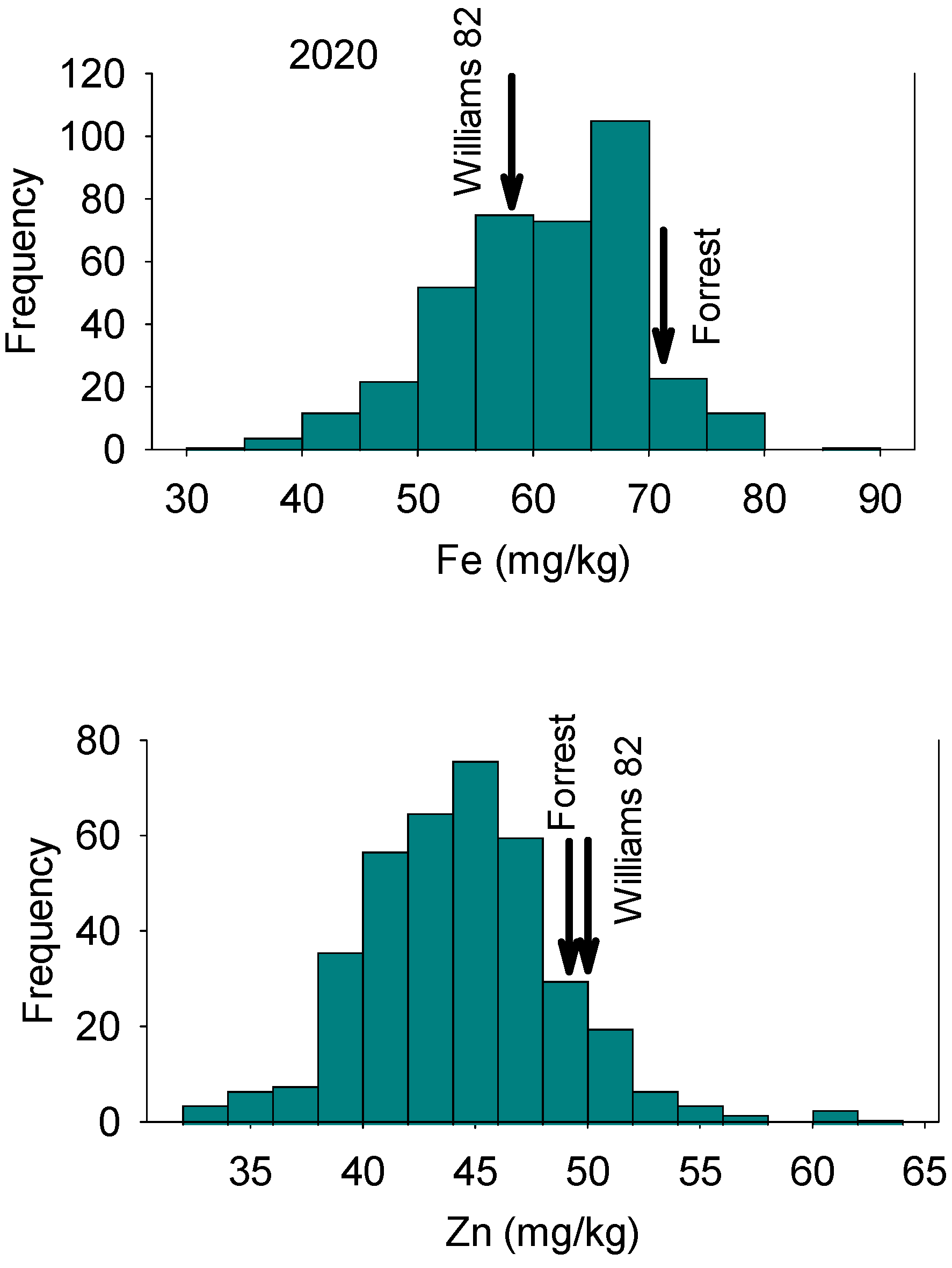

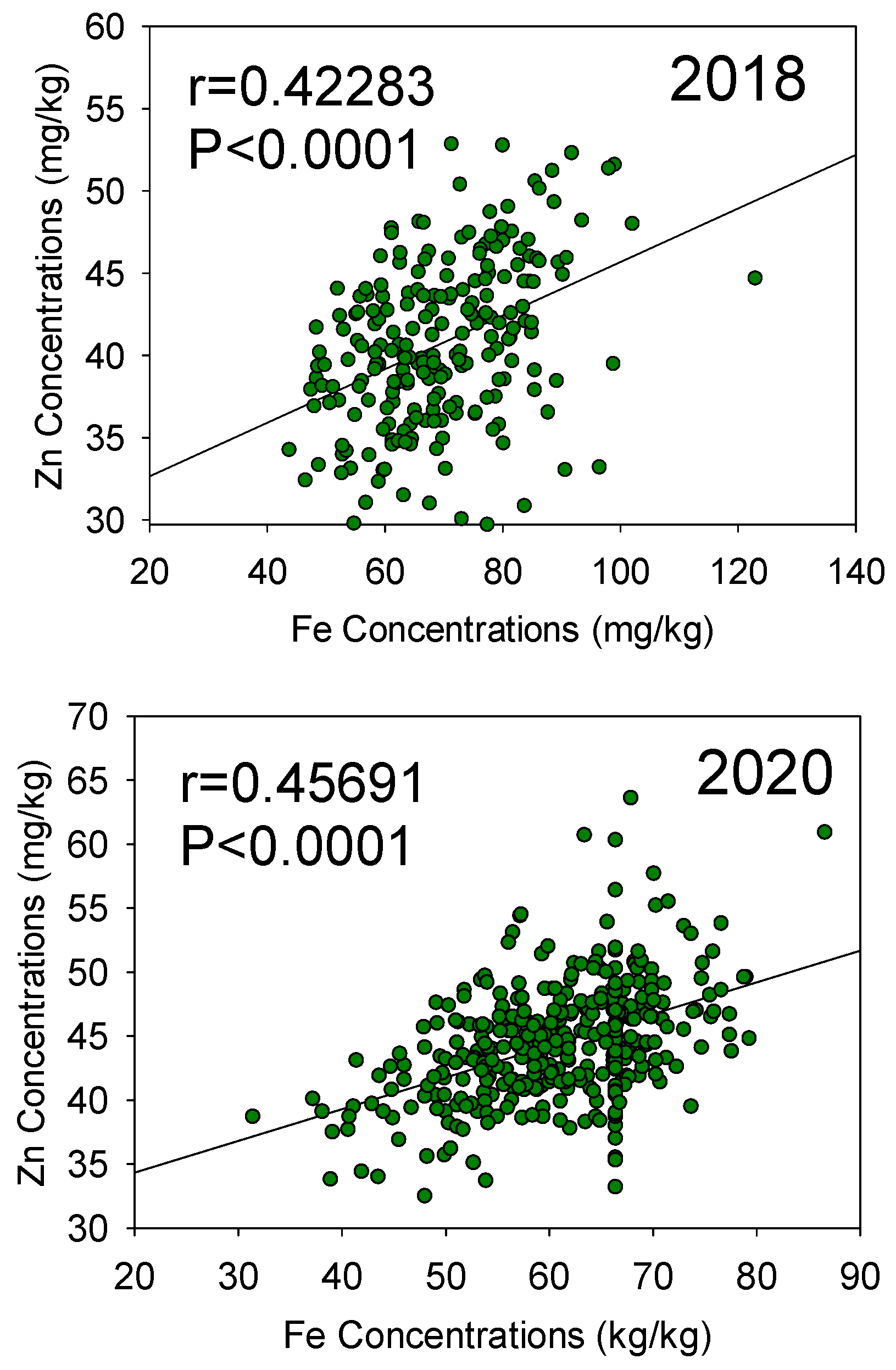
| Year | Variable | Forrest Parent1 | Williams 82 Parent2 | Mean | Maximum | Minimum | Median | SE | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | Fe | 69.56 | 67.73 | 69.49 | 122.99 | 43.76 | 68.16 | 0.82 | 0.26 | 2.54 |
| Zn | 40.90 | 39.07 | 40.67 | 52.85 | 29.71 | 40.26 | 0.32 | 0.16 | 2.58 | |
| Year | Variable | Forrest Parent1 | Williams 82 Parent2 | Mean | Maximum | Minimum | Median | SE | Skewness | Kurtosis |
| 2020 | Fe | 72.2 | 58.5 | 60.74 | 86.60 | 31.40 | 61.10 | 0.42 | −0.56 | 3.47 |
| Zn | 48.7 | 50.1 | 44.44 | 63.60 | 32.50 | 44.25 | 0.23 | 0.66 | 4.48 |
| Nutrient Concentration in Mature Seeds | Source | Sum Square | Mean Square | H2 |
|---|---|---|---|---|
| Line | 28,724.4 | 95.75 | −0.125 | |
| Iron | Year | 2765.7 | 2765.66 | |
| Line:Year | 19,545.7 | 107.39 | ||
| Nutrient Concentration in Mature Seeds | Source | Sum Square | Mean Square | H2 |
| Line | 7433.2 | 24.78 | 0.304 | |
| Zinc | Year | 1702.3 | 1702.33 | |
| Line:Year | 3137.1 | 17.24 |
| Spring Lake, NC (2018) | |||||||
|---|---|---|---|---|---|---|---|
| Trait | QTL | Chr. | Marker | Pos. (cM) | LOD | R2 | Add. Eff. |
| Fe | qFe-01-[NC-2018] | 1 | Gm01_4968769-Gm01_4932276 | 85.1–88.9 | 2.76 | 4.69 | 2.5358 |
| qFe-02-[NC-2018] | 2 | Gm02_9925870 | 140.1–142.2 | 2.53 | 6.63 | −4.9400 | |
| qFe-03-[NC-2018] | 6 | Gm06_1584748-Gm06_3361566 | 177.1–198.9 | 7.05 | 12.75 | 4.1433 | |
| Zn | qZn-01-[NC-2018] | 2 | Gm02_1037321-Gm02_1020061 | 138.4–139.8 | 3.50 | 6.87 | −3.3498 |
| qZn-02-[NC-2018] | 3 | Gm03_4198497-Gm03_4479032 | 153.1–164.1 | 3.52 | 7.18 | −1.3534 | |
| qZn-03-[NC-2018] | 7 | Gm07_2121760-Gm07_1092699 | 104.4–114.9 | 4.45 | 8.79 | 1.7485 | |
| qZn-04-[NC-2018] | 19 | Gm19_5032228 | 186.5–188.9 | 2.50 | 4.73 | −1.0899 | |
| Carbondale, IL (2020) | |||||||
| Trait | QTL | Chr. | Marker | Pos. (cM) | LOD | R2 | Add. Eff. |
| Fe | qFe-01-[IL-2020] | 1 | Gm01_5324236-Gm01_5264250 | 61.2–61.6 | 2.98 | 3.31 | −1.50 |
| qFe-02-[IL-2020] | 2 | Gm02_5102501-Gm02_1481798 | 133.4–133.5 | 5.27 | 5.97 | 3.84 | |
| qFe-03-[IL-2020] | 12 | Gm12_553862-Gm12_1632399 | 177.3–187.3 | 6.44 | 7.38 | −2.83 | |
| Zn | qZn-01-[IL-2020] | 5 | Gm05_3674925-Gm05_3361872 | 29.4–29.8 | 2.51 | 3.31 | 1.38 |
| qZn-02-[IL-2020] | 8 | Gm08_1247584-Gm08_1572868 | 98.41–100.4 | 2.54 | 3.91 | 0.92 | |
| Trait | Environment | QTL | Genomic Interval | Candidate Genes | Reference Genome |
|---|---|---|---|---|---|
| Spring Lake, NC 2018 | qFe-01-[NC-2018] | Gm01_4968769-Gm01_4932276 | GlymaLee.01G024000.1/GlymaLee.01G049700.1 | Glyma4.0 | |
| Spring Lake, NC 2018 | qFe-02-[NC-2018] | Gm02_9925870 | GlymaLee.02G078100.1 | Glyma4.0 | |
| Fe | Spring Lake, NC 2018 | qFe-03-[NC-2018] | Gm06_1584748-Gm06_3361566 | Glyma.06G021000 | Glyma4.0 |
| Carbondale, IL 2020 | qFe-01-[IL-2020] | Gm01_5324236-Gm01_5264250 | GlymaLee.01G024000.1/GlymaLee.01G049700.1 | Glyma4.0 | |
| Carbondale, IL 2020 | qFe-02-[IL-2020] | Gm02_5102501-Gm02_1481798 | Glyma.02g124700 | Glyma4.0 | |
| Carbondale, IL 2020 | qFe-03-[IL-2020] | Gm12_553862-Gm12_1632399 | GlymaLee.12G121900.1 | Glyma4.0 | |
| Spring Lake, NC 2018 | qZn-01-[NC-2018] | Gm02_1037321-Gm02_1020061 | Glyma.02G013000 | Glyma4.0 | |
| Spring Lake, NC 2018 | qZn-02-[NC-2018] | Gm03_4198497-Gm03_4479032 | Glyma.03G033750 | Glyma4.0 | |
| Zn | Spring Lake, NC 2018 | qZn-03-[NC-2018] | Gm07_2121760-Gm07_1092699 | GlymaLee.10G030400.1 | Glyma4.0 |
| Spring Lake, NC 2018 | qZn-04-[NC-2018] | Gm19_5032228 | Glyma.19G037800 | Glyma4.0 | |
| Carbondale, IL 2020 | qZn-01-[IL-2020] | Gm05_3674925-Gm05_3361872 | GlymaLee.05G051100.1 | Glyma4.0 | |
| Carbondale, IL 2020 | qZn-02-[IL-2020] | Gm08_1247584-Gm08_1572868 | Glyma.08G016500 | Glyma4.0 |
| Candidate Genes | Functional Annotation |
|---|---|
| GlymaLee.01G024000.1/GlymaLee.01G049700.1 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein/probable 2-oxoglutarate/Fe(II)-dependent dioxygenase |
| GlymaLee.02G078100.1 | Fe superoxide dismutase 2 |
| Glyma.06G021000 | Iron ion binding/oxidoreductase |
| GlymaLee.01G024000.1/GlymaLee.01G049700.1 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein/probable 2-oxoglutarate/Fe(II)-dependent dioxygenase |
| Glyma.02g124700 | 2-oxoglutarate/Fe(II)-dependent dioxygenase |
| GlymaLee.12G121900.1 | Fe-S cluster assembly protein |
| Glyma.02G013000 | RING/FYVE/PHD zinc finger superfamily protein |
| Glyma.03G033750 | C2H2-like zinc finger protein |
| GlymaLee.10G030400.1 | WRKY family transcription factor |
| Glyma.19G037800 | RING finger protein 38-like |
| GlymaLee.05G051100.1 | Cu/Zn-superoxide dismutase copper chaperone |
| Glyma.08G016500 | Zinc finger protein CONSTANS-LIK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellaloui, N.; Knizia, D.; Yuan, J.; Song, Q.; Betts, F.; Register, T.; Williams, E.; Lakhssassi, N.; Mazouz, H.; Nguyen, H.T.; et al. Genomic Regions and Candidate Genes for Seed Iron and Seed Zinc Accumulation Identified in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population. Int. J. Plant Biol. 2024, 15, 452-467. https://doi.org/10.3390/ijpb15020035
Bellaloui N, Knizia D, Yuan J, Song Q, Betts F, Register T, Williams E, Lakhssassi N, Mazouz H, Nguyen HT, et al. Genomic Regions and Candidate Genes for Seed Iron and Seed Zinc Accumulation Identified in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population. International Journal of Plant Biology. 2024; 15(2):452-467. https://doi.org/10.3390/ijpb15020035
Chicago/Turabian StyleBellaloui, Nacer, Dounya Knizia, Jiazheng Yuan, Qijian Song, Frances Betts, Teresa Register, Earl Williams, Naoufal Lakhssassi, Hamid Mazouz, Henry T. Nguyen, and et al. 2024. "Genomic Regions and Candidate Genes for Seed Iron and Seed Zinc Accumulation Identified in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population" International Journal of Plant Biology 15, no. 2: 452-467. https://doi.org/10.3390/ijpb15020035
APA StyleBellaloui, N., Knizia, D., Yuan, J., Song, Q., Betts, F., Register, T., Williams, E., Lakhssassi, N., Mazouz, H., Nguyen, H. T., Meksem, K., Mengistu, A., & Kassem, M. A. (2024). Genomic Regions and Candidate Genes for Seed Iron and Seed Zinc Accumulation Identified in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population. International Journal of Plant Biology, 15(2), 452-467. https://doi.org/10.3390/ijpb15020035












