Identification of CoDREB Genes for Drought and Cold Tolerance in Camellia oleifera
Abstract
1. Introduction
2. Materials and Methods
2.1. Homologs Sequence Identification of DREB Genes in C. oleifera
2.2. Phylogenetic Analysis of DREB Proteins and Nomenclature
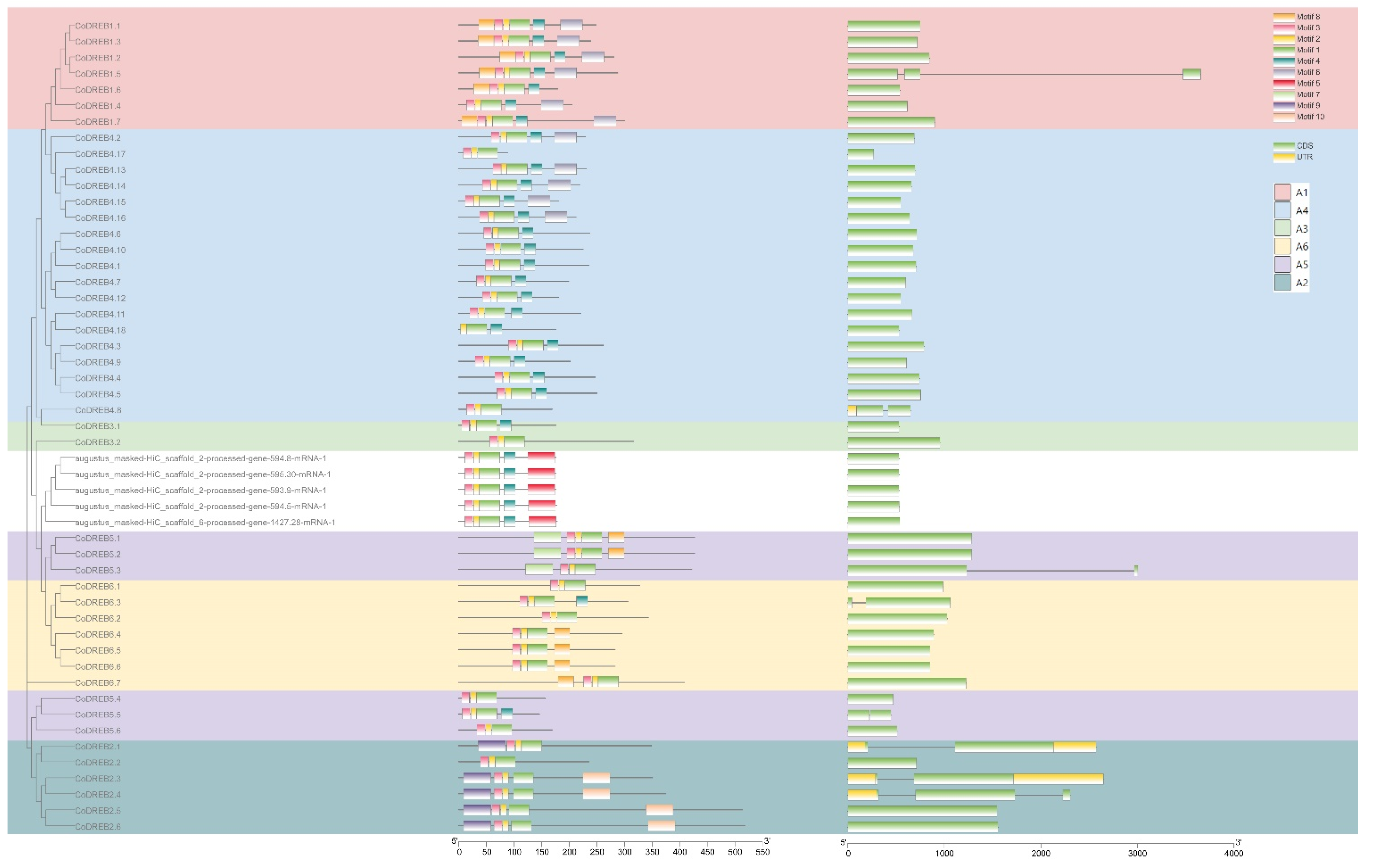
2.3. Gene Structure and MEME Conserved Motif Analysis
2.4. RNA-Seq Data
3. Results
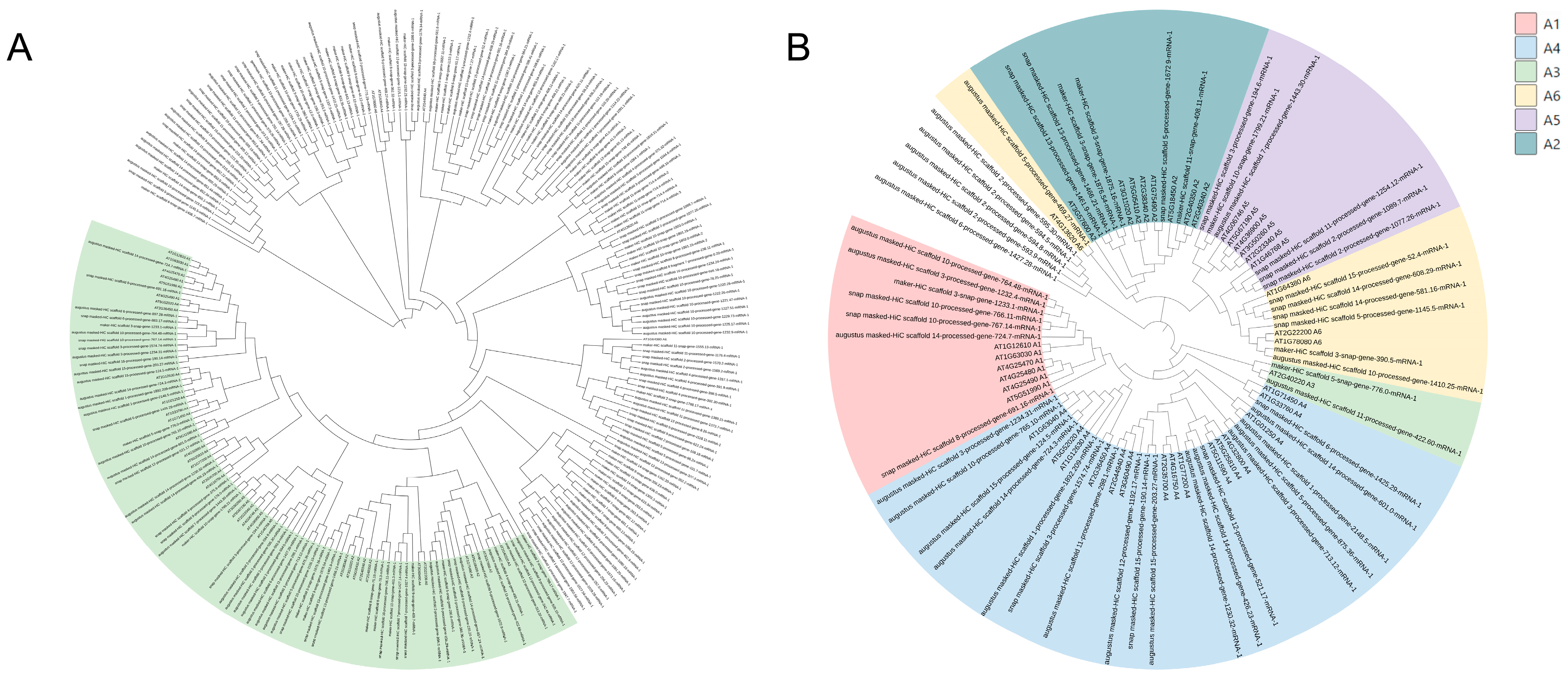
3.1. Genome-Wide Identification and Phylogenetic Analysis of DREB Genes in C. oleifera
3.2. Phylogenetic Relationship, Conserved Motifs and Genes Structure Analysis and Nomenclature of CoDREBs
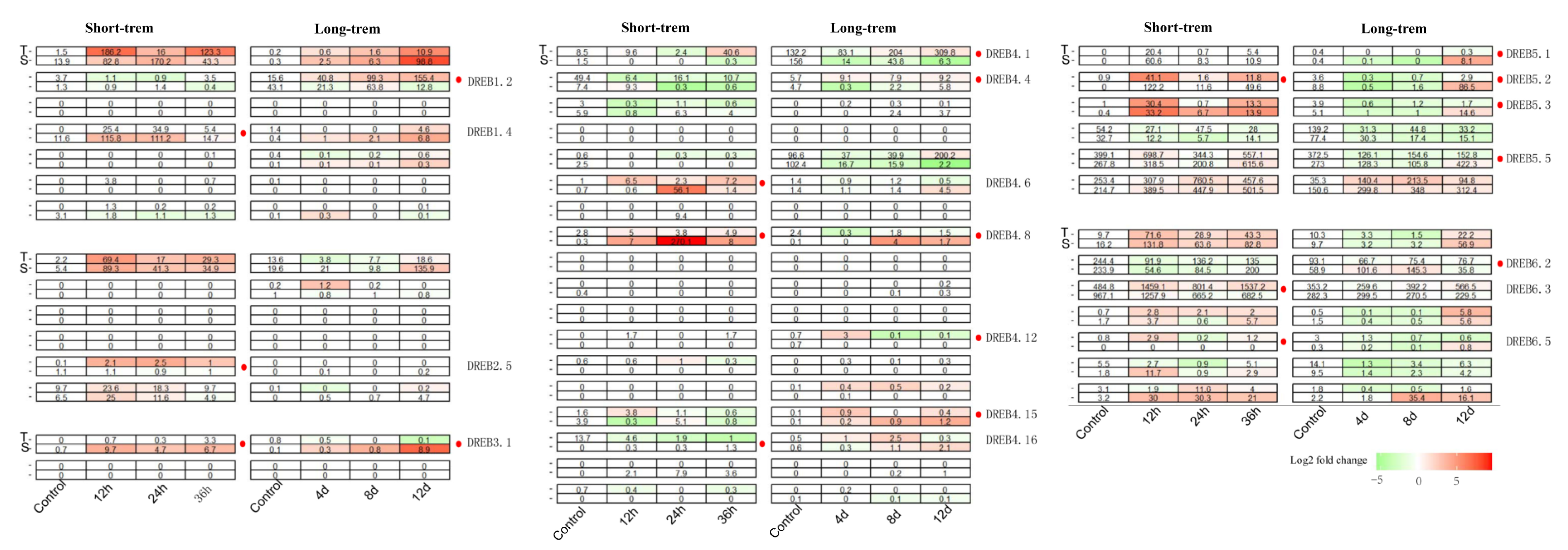
3.3. Expression Profiles of 46 CoDREB Genes in Response to Short-/Long-Term Drought Stress
3.4. CoDREBs in Response to Mild, Severe Drought and Followed by Recovery
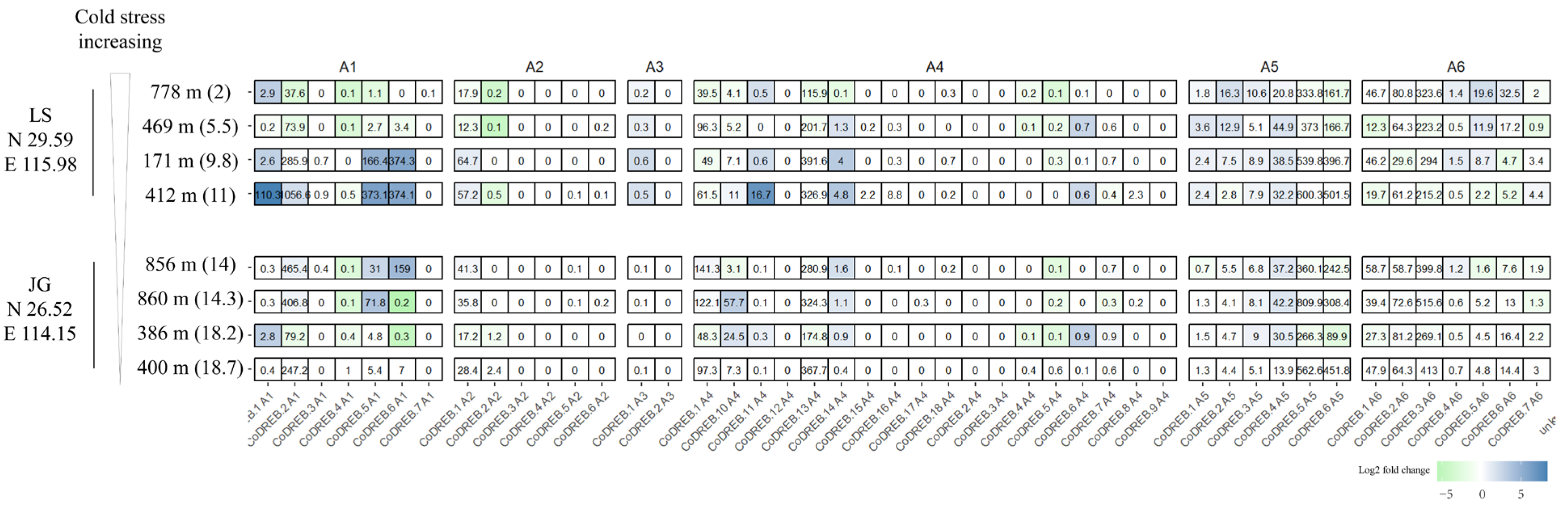
3.5. Transcriptome Analysis of Wild C. oleifera Grown from Different Latitudes and Elevations to Discover CoDREBs Involved in Cold Acclimation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lenaerts, B.; Collard, B.C.Y.; Demont, M. Review: Improving global food security through accelerated plant breeding. Plant Sci. 2019, 287, 110207. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Vanderschuren, H.; Qaim, M.; Mahfouz, M.M.; Kohli, A.; Mansoor, S.; Tester, M. New plant breeding technologies for food security. Science 2019, 363, 1390–1391. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Lu, J.; Zhang, Z.; Ma, L.; Liu, C.; Chen, Y. Global transcriptome and correlation analysis reveal cultivar-specific molecular signatures associated with fruit development and fatty acid determination in Camellia oleifera Abel. Int. J. Genom. 2020, 2022, 6162802. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yang, X.; Huang, X.; Duan, S.; Long, C.; Chen, J.; Rong, J. Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genom. 2017, 18, 211. [Google Scholar] [CrossRef]
- He, Z.; Liu, C.; Zhang, Z.; Wang, R.; Chen, Y. Integration of MRNA and MiRNA analysis reveals the differentially regulatory network in two different Camellia oleifera cultivars under drought stress. Front. Plant Sci. 2022, 13, 1001357. [Google Scholar] [CrossRef]
- Zhou, G.; Wei, X.; Wu, Y.; Liu, S.; Huang, Y.; Yan, J.; Zhang, D.; Zhang, Q.; Liu, J.; Meng, Z. Quantifying the hydrological responses to climate change in an intact forested small watershed in southern China. Glob. Change Biol. 2011, 17, 3736–3746. [Google Scholar] [CrossRef]
- Dong, B.; Wu, B.; Hong, W.; Li, X.; Li, Z.; Xue, L.; Huang, Y. Transcriptome analysis of the tea oil camellia (Camellia oleifera) reveals candidate drought stress genes. PLoS ONE 2017, 12, e0181835. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Jaleel, C.A.; Manivannan, P.; Wahid, A.; Farooq, M.; Al-Juburi, J.; Somasundaram, R.; Panneerselvam, R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int. J. Agric. Biol. 2009, 11, 100–105. [Google Scholar]
- Muhammad, I.; Shalmani, A.; Ali, M.; Yang, Q.H.; Ahmad, H.; Li, F.B. Mechanisms regulating the dynamics of photosynthesis under abiotic stresses. Front. Plant Sci. 2021, 11, 615942. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, E.T.; Campbell, M.M. Genome-wide responses to drought in forest trees. Forestry 2011, 84, 273–283. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, X.; Zhang, Y.; Du, Z.; Chen, X.; Kong, X.; Sun, W.; Chen, C. Drought stress modify cuticle of tender tea leaf and mature leaf for transpiration barrier enhancement through common and distinct modes. Sci. Rep. 2020, 10, 6696. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, H.Y.; Yang, X.N.; Zhan, J.J.; Zhou, H.; Wang, C.; Yu, Y.; Lu, X.Y.; Chen, Y.Z.; Tian, Y. Transcriptomic analysis of Camellia oleifera in response to drought stress using high throughput RNA-seq. Russ. J. Plant Physiol. 2017, 64, 728–737. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high-salt and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Lin, P.; Wang, K.; Wang, Y.; Hu, Z.; Yan, C.; Huang, H.; Ma, X.; Cao, Y.; Long, W.; Liu, W.; et al. The genome of Oil-Camellia and population genomics analysis provide insights into seed Oil Domestication. Genome Biol. 2022, 23, 14. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; Eddy, S.R.; Birney, E.; Bateman, A.; Durbin, R. Pfam: Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998, 26, 320–322. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (ITOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Akhtar, R.; Birney, E.; Bower, L.; Cerdeno-Tarraga, A.; Cheng, Y.; Cleland, I.; Faruque, N.; Goodgame, N.; Gibson, R. The european nucleotide archive. Nucleic Acids Res. 2011, 39, D28–D31. [Google Scholar] [CrossRef] [PubMed]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J.; The Bioconda Team. Bioconda: Sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; pp. 12–21. [Google Scholar]
- Silva, E.C.D.; Albuquerque, M.; Neto, A.A.; Junior, S. Drought and its consequences to plants—From individual to ecosystem. In Responses of Organisms to Water Stress; IntechOpen: London, UK, 2013; pp. 17–47. [Google Scholar]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription Factor family in crop improvement: Functions of the AP2/EREBP family. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, T. Expansion and stress responses of the AP2/EREBP super family in cotton. BMC Genom. 2017, 18, 118. [Google Scholar]
- Sun, J.; Peng, X.; Fan, W.; Tang, M.; Liu, J.; Shen, S. Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene 2014, 535, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.; Liu, Y.; Gao, H.; Wang, Z.; Sun, G. CkDREB gene in caragana korshinskii is involved in the regulation of stress response to multiple abiotic stresses as an AP2/EREBP transcription factor. Mol. Biol. Rep 2011, 38, 2801–2811. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Sun, J.; Liu, Y.; Chen, F.; Shen, S. Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Mol. Biol. 2007, 63, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Jia, J.; Kong, D.; Zhang, Z.; Song, S.; Li, Y.; Pang, X. Genome-wide identification and analysis of the DREB genes and their expression profiles under abiotic stresses in Chinese Jujube (Ziziphus jujuba Mill.). J. For. Res. 2019, 30, 1277–1287. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Li, Z.; Wang, G.; Liu, X.; Wang, Z.; Zhang, M.; Zhang, J. Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genom. 2021, 22, 456. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Guo, X.; Wu, C.; Xiang, Y.; Duan, S.; Yang, W.; Li, W.; Cao, F.; Liu, L. Genome-wide identification and expression analysis of DREB genes in Alfalfa (Medicago sativa) in response to cold stress. Plant Signal. Behav. 2022, 17, 2081420. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lv, H.; Li, L.; Liu, J.; Mu, S.; Li, X.; Gao, J. Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2015, 10, e0126657. [Google Scholar] [CrossRef]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Li, H.; Wang, Y.X.; Zhuang, J. Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Funct. Integr. Genom. 2015, 15, 741–752. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G. Draft genome sequence of Camellia sinensis Var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Perumal, V. Signaling and Communication in Plants, 1st ed.; Springer: Dordrecht, The Netherlands, 2020; pp. 249–267. [Google Scholar]
- Li, G.; Jin, L.; Sheng, S. Genome-wide identification of BHLH transcription factor in medicago sativa in response to cold stress. Genes 2022, 13, 2371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lei, L.; Lai, J.; Zhao, H.; Song, W. Effects of drought stress and water recovery on physiological responses and gene expression in maize seedlings. BMC Plant Biol. 2018, 18, 68. [Google Scholar] [CrossRef]
- Selote, D.S.; Bharti, S.; Khanna-Chopra, R. Drought acclimation reduces O2− accumulation and lipid peroxidation in wheat seedlings. Biochem. Biophys. Res. Commun. 2004, 314, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The Water-Water Cycle in Chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Hayano-Kanashiro, C.; Calderón-Vázquez, C.; Ibarra-Laclette, E.; Herrera-Estrella, L.; Simpson, J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 2009, 4, e7531. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of stomatal closure in plants exposed to drought and cold stress. In Survival Strategies in Extreme Cold and Desiccation: Adaptation Mechanisms and Their Applications; Springer: Singapore, 2018. [Google Scholar]
- Zheng, C.; Wang, Y.; Ding, Z.; Zhao, L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front. Plant Sci. 2016, 7, 1858. [Google Scholar] [CrossRef] [PubMed]

| Symbol | Seq Id | 14th | 19th | Source | Strand | Start | End |
|---|---|---|---|---|---|---|---|
| CoDREB1.1 | augustus_masked-HiC_scaffold_10-processed-gene-764.48-mRNA-1 | V | E | HiC_scaffold_10 | − | 76409702 | 76410451 |
| CoDREB1.2 | augustus_masked-HiC_scaffold_3-processed-gene-1232.4-mRNA-1 | V | E | HiC_scaffold_3 | − | 123283087 | 123283932 |
| CoDREB1.3 | snap_masked-HiC_scaffold_10-processed-gene-766.11-mRNA-1 | V | E | HiC_scaffold_10 | − | 76623619 | 76624338 |
| CoDREB1.4 | augustus_masked-HiC_scaffold_14-processed-gene-724.7-mRNA-1 | V | E | HiC_scaffold_14 | + | 72478180 | 72478797 |
| CoDREB1.5 | maker-HiC_scaffold_3-snap-gene-1233.1-mRNA-1 | V | E | HiC_scaffold_3 | − | 123362118 | 123365776 |
| CoDREB1.6 | snap_masked-HiC_scaffold_10-processed-gene-767.14-mRNA-1 | V | E | HiC_scaffold_10 | − | 76705366 | 76705905 |
| CoDREB1.7 | snap_masked-HiC_scaffold_8-processed-gene-691.16-mRNA-1 | V | E | HiC_scaffold_8 | − | 69154923 | 69155825 |
| CoDREB2.1 | maker-HiC_scaffold_11-snap-gene-408.11-mRNA-1 | V | E | HiC_scaffold_11 | + | 40854256 | 40856829 |
| CoDREB2.2 | snap_masked-HiC_scaffold_5-processed-gene-1672.9-mRNA-1 | V | E | HiC_scaffold_5 | + | 167280494 | 167281204 |
| CoDREB2.3 | maker-HiC_scaffold_3-snap-gene-1875.16-mRNA-1 | V | E | HiC_scaffold_3 | − | 187547455 | 187550105 |
| CoDREB2.4 | maker-HiC_scaffold_3-snap-gene-1876.54-mRNA-1 | V | E | HiC_scaffold_3 | − | 187645199 | 187647503 |
| CoDREB2.5 | snap_masked-HiC_scaffold_13-processed-gene-1466.21-mRNA-1 | V | E | HiC_scaffold_13 | − | 146604874 | 146606418 |
| CoDREB2.6 | snap_masked-HiC_scaffold_13-processed-gene-1461.9-mRNA-1 | V | E | HiC_scaffold_13 | + | 146118529 | 146120085 |
| CoDREB3.1 | maker-HiC_scaffold_5-snap-gene-776.0-mRNA-1 | V | E | HiC_scaffold_5 | + | 77602969 | 77603618 |
| CoDREB3.2 | augustus_masked-HiC_scaffold_11-processed-gene-422.60-mRNA-1 | V | E | HiC_scaffold_11 | + | 42215560 | 42216513 |
| CoDREB4.1 | augustus_masked-HiC_scaffold_14-processed-gene-1230.32-mRNA-1 | V | E | HiC_scaffold_14 | − | 123064670 | 123065377 |
| CoDREB4.10 | snap_masked-HiC_scaffold_12-processed-gene-521.17-mRNA-1 | V | E | HiC_scaffold_12 | - | 52181645 | 52182322 |
| CoDREB4.11 | augustus_masked-HiC_scaffold_3-processed-gene-713.12-mRNA-1 | V | A | HiC_scaffold_3 | + | 71361910 | 71362575 |
| CoDREB4.12 | augustus_masked-HiC_scaffold_14-processed-gene-601.0-mRNA-1 | V | E | HiC_scaffold_14 | + | 60115087 | 60115632 |
| CoDREB4.13 | augustus_masked-HiC_scaffold_3-processed-gene-1234.31-mRNA-1 | V | E | HiC_scaffold_3 | − | 123398472 | 123399167 |
| CoDREB4.14 | augustus_masked-HiC_scaffold_10-processed-gene-765.10-mRNA-1 | V | E | HiC_scaffold_10 | − | 76529118 | 76529777 |
| CoDREB4.15 | augustus_masked-HiC_scaffold_15-processed-gene-124.5-mRNA-1 | V | E | HiC_scaffold_15 | + | 12454548 | 12455093 |
| CoDREB4.16 | augustus_masked-HiC_scaffold_14-processed-gene-724.3-mRNA-1 | V | E | HiC_scaffold_14 | + | 72440566 | 72441204 |
| CoDREB4.17 | augustus_masked-HiC_scaffold_1-processed-gene-1892.209-mRNA-1 | V | E | HiC_scaffold_1 | + | 189269071 | 189269337 |
| CoDREB4.18 | augustus_masked-HiC_scaffold_5-processed-gene-875.36-mRNA-1 | V | A | HiC_scaffold_5 | − | 87523685 | 87524215 |
| CoDREB4.2 | snap_masked-HiC_scaffold_3-processed-gene-1574.74-mRNA-1 | V | E | HiC_scaffold_3 | − | 157422246 | 157422935 |
| CoDREB4.3 | augustus_masked-HiC_scaffold_12-processed-gene-1192.17-mRNA-1 | V | E | HiC_scaffold_12 | − | 119269647 | 119270435 |
| CoDREB4.4 | snap_masked-HiC_scaffold_15-processed-gene-190.14-mRNA-1 | V | E | HiC_scaffold_15 | + | 19021440 | 19022183 |
| CoDREB4.5 | augustus_masked-HiC_scaffold_15-processed-gene-203.27-mRNA-1 | V | E | HiC_scaffold_15 | − | 20340240 | 20340995 |
| CoDREB4.6 | augustus_masked-HiC_scaffold_14-processed-gene-426.23-mRNA-1 | V | E | HiC_scaffold_14 | + | 42621443 | 42622156 |
| CoDREB4.7 | augustus_masked-HiC_scaffold_1-processed-gene-2148.5-mRNA-1 | V | E | HiC_scaffold_1 | + | 214865752 | 214866351 |
| CoDREB4.8 | snap_masked-HiC_scaffold_6-processed-gene-1425.29-mRNA-1 | V | E | HiC_scaffold_6 | − | 142541920 | 142542450 |
| CoDREB4.9 | augustus_masked-HiC_scaffold_11-processed-gene-298.1-mRNA-1 | V | E | HiC_scaffold_11 | − | 29817495 | 29818103 |
| CoDREB5.1 | snap_masked-HiC_scaffold_2-processed-gene-1077.26-mRNA-1 | V | L | HiC_scaffold_2 | − | 107715224 | 107716507 |
| CoDREB5.2 | snap_masked-HiC_scaffold_2-processed-gene-1089.7-mRNA-1 | V | L | HiC_scaffold_2 | + | 108932798 | 108934081 |
| CoDREB5.3 | snap_masked-HiC_scaffold_11-processed-gene-1254.12-mRNA-1 | V | L | HiC_scaffold_11 | + | 125398486 | 125401490 |
| CoDREB5.4 | augustus_masked-HiC_scaffold_7-processed-gene-1443.30-mRNA-1 | V | E | HiC_scaffold_7 | − | 144370313 | 144370783 |
| CoDREB5.5 | maker-HiC_scaffold_10-snap-gene-1799.21-mRNA-1 | V | E | HiC_scaffold_10 | + | 179923482 | 179923929 |
| CoDREB5.6 | snap_masked-HiC_scaffold_3-processed-gene-194.6-mRNA-1 | V | E | HiC_scaffold_3 | − | 19425828 | 19426337 |
| CoDREB6.1 | snap_masked-HiC_scaffold_5-processed-gene-1145.5-mRNA-1 | V | L | HiC_scaffold_5 | + | 114517410 | 114518396 |
| CoDREB6.2 | augustus_masked-HiC_scaffold_10-processed-gene-1410.25-mRNA-1 | V | L | HiC_scaffold_10 | − | 141067137 | 141068168 |
| CoDREB6.3 | maker-HiC_scaffold_3-snap-gene-390.5-mRNA-1 | V | L | HiC_scaffold_3 | − | 39027029 | 39028091 |
| CoDREB6.4 | snap_masked-HiC_scaffold_15-processed-gene-52.4-mRNA-1 | V | L | HiC_scaffold_15 | + | 5199287 | 5200177 |
| CoDREB6.5 | snap_masked-HiC_scaffold_14-processed-gene-581.16-mRNA-1 | V | L | HiC_scaffold_14 | − | 58133946 | 58134797 |
| CoDREB6.6 | snap_masked-HiC_scaffold_14-processed-gene-608.29-mRNA-1 | V | L | HiC_scaffold_14 | + | 60806950 | 60807801 |
| CoDREB6.7 | augustus_masked-HiC_scaffold_5-processed-gene-469.27-mRNA-1 | V | L | HiC_scaffold_5 | − | 46960303 | 46961529 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Guo, P.; Sheng, S.; Li, J.; Yan, J. Identification of CoDREB Genes for Drought and Cold Tolerance in Camellia oleifera. Int. J. Plant Biol. 2023, 14, 228-241. https://doi.org/10.3390/ijpb14010019
Wang Y, Guo P, Sheng S, Li J, Yan J. Identification of CoDREB Genes for Drought and Cold Tolerance in Camellia oleifera. International Journal of Plant Biology. 2023; 14(1):228-241. https://doi.org/10.3390/ijpb14010019
Chicago/Turabian StyleWang, Ying, Purui Guo, Song Sheng, Jian’an Li, and Jindong Yan. 2023. "Identification of CoDREB Genes for Drought and Cold Tolerance in Camellia oleifera" International Journal of Plant Biology 14, no. 1: 228-241. https://doi.org/10.3390/ijpb14010019
APA StyleWang, Y., Guo, P., Sheng, S., Li, J., & Yan, J. (2023). Identification of CoDREB Genes for Drought and Cold Tolerance in Camellia oleifera. International Journal of Plant Biology, 14(1), 228-241. https://doi.org/10.3390/ijpb14010019







