Abstract
Lactic acid bacteria (LAB) comprise a group of microorganisms responsible for developing the sensory and chemical characteristics of several foods and fermented products, particularly cheese. For this reason, after isolation and identification of LAB, validated protocols and procedures for their long-term preservation without compromising its integrity and technological properties, as well as methodologies aiming to assess their viability and integrity are paramount. This study aimed to isolate and identify autochthonous LAB from artisanal Adobera cheese and determine the effect of LAB cryopreservation with thioglycolate broth and glycerol on their viability, membrane integrity, and kinetics. Sixteen LAB were isolated and genetically identified from artisanal cheese samples; eleven of those strains were selected (genus Lactobacillus, Leuconostoc, Streptococcus, and Lactococcus) and included in the cryo-preservation assay. The initial average concentration of the bacterial suspensions was 6.89 log10 CFU mL−1; increasing to 8.9 log10 CFU mL−1 21 days later and slightly reduced at day 42 post-preservation (losses below one logarithm). About 77% of the cells maintained their membrane potential 180 days after their preservation and showed normal Kinetic parameters, maintaining normal adaptation times (Lag phase) and Log phases (9 h average), before reaching the stationary phase. The proposed protocol constitutes a viable alternative to the long-term preservation of different LAB genera because it keeps their viability and integrity. Using flow cytometry allowed the enumeration of viable LAB and provide evidence of the integrity of their membrane.
1. Introduction
Raw milk cheeses are known for the complex microbial communities involved in their production. Thus, several studies have been carried out to describe the bacterial communities of several artisanal cheeses based on high-throughput sequencing-based technologies, including some genuine Mexican cheeses [1,2]. In particular, the microbiota of Adobera cheese from the Los Altos region in México, a raw-milk, fresh, semihard and unripe cheese [3], is strongly represented by lactic acid bacteria-related genera, including Streptococcus, Lactococcus, and Lactobacillus [4]. On the other hand, some scientific reports have been published to identify and characterize some of the microorganisms involved in producing artisanal cheese from the technological and functional point of view [5,6,7,8]. Lactic acid bacteria (LAB) comprise a group of Gram-positive, non-sporulated, catalase-negative, cytochromes-free, microaerophilic, acid-tolerant and strictly fermentative microorganisms. This group is characterized by its production of lactic acid as a by-product during metabolic activities [9]. Furthermore, LAB are classified by the Food and Drugs Administration as GRAS (Generally Recognized as Safe) and they play a key role in the production of fermented foods, beverages, silages, and other fermented products [10,11,12,13,14,15].
Additionally, LAB could be classified based on their ability to ferment carbohydrates (homofermentative or heterofermentative), acid production characteristics, and growth temperatures [9]. The LAB group includes both bacilli (Lactobacillus and Carnobacterium) and cocci (rest of the LAB genera) [12,16,17]; however, the most important LAB genera are Carnobacterium, Lactobacillus, Aerococcus, Enterococcus, Lactococcus, Vagococcus, Leuconostoc, Oenococcus, Pediococcus, Streptococcus, Tetragenococcus, and Weissella [5,10,11,12,13,18]. LAB are closely related to the development of sensorial attributes such as the aroma, flavor, and texture of cheese and other fermented foods and, in some cases, they protect against pathogens and confer probiotic properties [19,20,21,22,23,24].
Research associated with the isolation, characterization, and preservation of LAB is necessary to take advantage of these microorganisms through their use as starter cultures in fermentation processes, among other biotechnological developments [25,26]. In this context, the study of bacteria must begin with their isolation and purification in selective and differential culture media, then their identification, followed by their selection, based on the technological, and biochemical features of each strain (acid production, texture and flavor development, ability to inhibit pathogens, proteolytic and lipolytic activities, etc.) [27,28,29].
Identification and characterization can be performed through phenotypic and/or genotypic procedures; phenotyping comprises classic microbiologic methods based on cell morphology and metabolism [30,31]; meanwhile, genotyping is based on the analysis of nucleic acids, mainly DNA [32,33,34]. Genetic identification is generally supported by the amplification and sequencing of different molecular markers, allowing a more accurate differentiation and identification of the strains at different phylogenetic levels, including species, sub-species, or strains [28,33,35,36,37]. Once the identification is complete, it is essential to develop and validate strategies for preserving bacterial strains to establish viable and available culture collections of these microorganisms for subsequent research or biotechnological applications [38].
Several protocols for the short- and long-term preservation of microorganisms are available. Short-term preservation is based on the sub-culture of bacteria in selective media, while long-term preservation can be achieved via freeze-drying preservation (lyophilization) or, more commonly by cryo-freezing, either at −195 °C by using liquid nitrogen or at −70 °C by freezing and using cryo-protectors, seeking to guarantee the microorganism’s integrity and genetic stability and its viability for extended periods of time [39,40]. Nonetheless, freezing represents a stress condition that could affect cell physiology, reducing viability and changing bacteria metabolism and technological properties, even for LAB [41,42,43].
There is not solid information about the standard preservation protocol for LAB. However, most of the studies related to the preservation of this group of microorganisms are based on the use of bacterial suspensions in Man, Rogosa, and Sharpe (MRS) broth [44] and glycerol as cryo-protector, at temperatures ranging from −20 °C to −80 °C [2,45,46]. For this reason, it is necessary to generate evidence of the long-term viability of cryopreserved LAB, which guarantees the maintenance of their technological properties [47]. Thus, supporting strategies based on effective methodologies are important, allowing us to generate quick and accurate evidence of LAB’s cell viability, integrity, and technological competence during preservation. Therefore, this study focused on the isolation and genetic identification of LAB from artisanal Adobera cheese from Los Altos de Jalisco, México. It also aimed to propose and validate a freezing-based preservation protocol for those LAB, based on the assessment of their viability and integrity using culture-dependent and -independent techniques.
2. Materials and Methods
2.1. Bacterial Strains Isolation and Purification
Two commercial samples of a high-moisture content and acidified artisanal cheese, obtained from two cheese factories located in Los Altos de Jalisco region, were serially diluted and plated on MRS agar for LAB isolation. Plates were incubated under microaerophilic conditions for 24 h at 37 °C, using the BD BBL™ GasPak™ system (according to the manufacturer’s instructions. Franklin Lakes, NJ, USA), and 16 colonies, compatible with the typical LAB morphology (round colonies from 1–2 mm of diameter, creamy-to-white color, entire edges, convex or raised surface and consistency butyric and moist) were isolated for further identification and characterization [48]. Bacterial isolates were deposited in the Microbial Collection of the Genetic Resources National Center (CM-CNRG).
2.2. Genetic Identification of Lactic Acid Bacteria
Gram-positive and catalase- and oxidase-negative isolated strains were cross-streaked on agar MRS and incubated under microaerophilic conditions at 37 °C for 24 h. After the incubation period, three colonies of each strain were suspended in TE-50:20 buffer to perform genomic DNA extraction. The remaining biomass was reserved for the preservation procedure. DNA extraction was carried out through the methodology suggested by Wilson [49]. DNA integrity was confirmed via electrophoresis in agarose gel and stored at −20 °C until its amplification. Genetic identification was performed via PCR-amplification of the 16S rRNA gene, using the universal primers 27f (5-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′TACGGYTACCTTGTTACGACTT-3′) [50]. PCR products were resolved by electrophoresis (1.5% of agarose gel in TAE buffer: 2 mol/L of Tris, 1 mol/L of acetic acid, 0.05 mol/L de EDTA pH 8.0), and visualized through UV-vis. Amplicons were stored at –20 °C, followed by a Sanger-based sequencing performed by Macrogen (Seoul, South Korea). In silico analysis of the sequences included the edition and generation of consensus sequences, followed by identity assignation using GenBank and the Ribosomal data project. The matching criteria for identity assignment was ≥ 98% of similarity at genus level and ≥99% of similarity at species level.
Species-level identification of the Plantarum group members, to differentiate between Lactiplantibacillus plantarum, Lactobacillus paraplantarum and Lactobacillus pentosus was complemented using the partial amplification of the recA gene, as suggested by [51]. Identities were confirmed via phylogenetic reconstruction using the Mega software v11 [52] based on the 16S rDNA sequences of the strains and using homologous sequences using the Maximum Likelihood evolutionary method and the Jukes–Cantor model [53]; the inferred tree was generated from a bootstrap consensus of 1000 replicates [54].
2.3. Preservation of Lactic Acid Bacteria and Efficiency Evaluation
Bacterial suspensions were prepared with fresh biomass culture of each strain in sterile indicator-free thioglycolate broth without dextrose (Difco™, Fisher Sc. Hampton, NH, USA) until reaching an approximate concentration of 9-log10 CFU mL−1, according to the McFarland standard. Briefly, a 500 μL aliquot of each cell suspension was deposited into 2 mL cryo-tubes and mixed with 500 μL of 20% sterile glycerol using a vortex. The cryo-tubes were stored at −20 °C for 2 h and preserved at −80 °C until the analysis was performed. In order to assess the efficacy of the preservation protocol, we proposed a scheme that allows the evaluation of survival and integrity in both short- and long-term of LAB during cryopreservation, as follows.
2.3.1. Evaluation of Viability: Short-Term Survival
Cell viability was estimated through plate count using the drop method suggested by Miles et al., with some modifications [55,56]. Ten-fold serial dilutions were prepared from each cryo-preserved bacterial suspension previously thawed at room temperature. Briefly, 10 μL of each dilution was dropped on MRS agar. Plates were incubated in microaerophilic conditions at 37 °C for 24 h for the subsequent count of colonies developed at each drop. Viability was assessed at days 0, 21 and 42 of preservation and expressed as log CFU mL−1. The analysis was performed in duplicate.
2.3.2. Flow Cytometry: Long-Term Survival and Integrity
To generate more evidence about the long-term survival and integrity of cryo-preserved LAB, membrane potential was assessed at the beginning of the assay and 180 days after preservation using an Attune™ flow cytometer (Applied Biosystems, Waltham, MS, USA) and the commercial kit BacLight™ B34950 (Molecular Probes®, Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Each preserved suspension was thawed at room temperature and 500 μL were transferred to a 1.5 mL sterile tube. Biomass was harvested via centrifugation (13,000 rpm for 5 min at 25 °C) and washed twice with a PBS buffer (200 mL of 10 mM sodium phosphate, 145 mM sodium chloride, pH 7.4). The recovered biomass was briefly suspended in PBS buffer until it reached a concentration of 1 × 106 cells mL−1, then 1 mL was transferred to cytometer tubes. Two cell suspensions of each strain were prepared and stained with 10 μL of DiOC2(3) (3,3′-Diethyloxacarbocyanine Iodide) and incubated at room temperature for 15 min. Each stain was prepared in duplicate. A negative control consisting of an inactive cell suspension depolarized with 10 μL CCCP (3,3′ carbonyl-cyanide-m-chlorophenylhydrazone) was included, as well as a positive control that comprised a cell suspension without staining or depolarization. Briefly, each sample was placed on the cytometer, beginning with the positive control to adjust the detection threshold, followed by the depolarized suspension and the samples. The analysis was performed by exciting the samples using a 488 nm laser, and fluorescence emitted in the red and green channels was recorded. From each sample, frontal dispersion, lateral dispersion, and fluorescence were obtained with amplification of the logarithmic signal. The magnitude of the membrane potential was assessed based on the red–green fluorescence ratio using the density plots obtained via flow cytometry. Percentage (%) of membrane-active cells, was estimated as the red–red+green fluorescence ratio. Each cell suspension was analyzed in duplicate.
2.3.3. Recovery and Re-Adaptability Abilities in the Long-Term of Cryo-Preserved Lactic Acid Bacteria
After 240 days of preservation, bacterial suspensions were thawed at room temperature and used to inoculate culture tubes with MRS broth to an approximate concentration of 1 × 106 CFU mL−1. Inoculated tubes were incubated at 37 °C for 24 h under agitation at 100 rpm. Optical density was registered every 60 min by a UV-vis spectrophotometer at 660 nm. Data were plotted against time to determine the kinetic parameters of the bacteria.
2.4. Statistical Analysis
Data obtained from plate count and flow cytometry were analyzed using the SAS™ software v.9.3 (SAS Institute Inc., Cary, NC, USA), through the general linear model (GLM) procedure, analysis of variance, and means comparison via the Tukey test with a significance level of 0.05.
3. Results and Discussions
Microorganisms, including the LAB, are very important to the production of fermented foods required for animals and humans, and through their ability to regulate several processes, they maintain the balance in different environments, reduce or degrade polluting compounds, fix nutrients, limit the survival and proliferation of pathogens and produce several important compounds such as vitamins, antibiotics, enzymes, alcohol, organic acids, among others [5,57,58,59]. These bacteria require correct ex situ preservation. This premise has favored the establishment of many culture collections worldwide, at different scales and for different purposes; however, they all keep the microorganisms viable and pure, maintaining their metabolic abilities [60].
3.1. Isolation and Identification of Lactic Acid Bacteria
From two samples of high-moisture content and acidified artisanal cheese, 16 colonies corresponding to Gram-positive bacteria, with negative catalase and oxidase activities, were isolated and purified. Amplicons of 1500 bp were obtained via PCR-amplification from the genomic DNA of each isolate. Identities were assigned through Sanger-based sequencing and bioinformatic analyses, resulting in 56% of the strains being assigned to the genus Lactobacillus, 25% to Leuconostoc, 12.5% to Lactococcus, and 6.5% to Streptococcus (Table 1). Taxonomic assignations were corroborated by a phylogenetic reconstruction (Figure 1).

Table 1.
Genetic identification of lactic acid bacteria isolated from artisanal Adobera cheese from Los Altos de Jalisco.
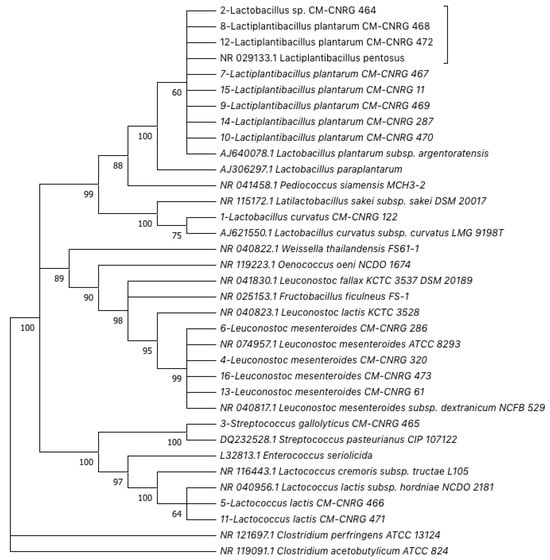
Figure 1.
Phylogenetic reconstruction using the 16S rDNA partial sequences of lactic acid bacteria isolated from artisanal Adobera cheese from Los Altos de Jalisco and homologous sequences obtained from Genebank. The Maximum Likelihood Method and Jukes–Cantor model were used (Bootstrap consensus from 1000 replicates).
Our results are similar to those of a previous publication that reported the isolation of these LAB genera from different fresh raw-milk cheeses [7,61]. Specifically, regarding Mexican artisanal fresh cheeses, it has been reported that predominant LAB genera obtained through culture-dependent isolation are Lactobacillus, Lactococcus, Leuconostoc, and Enterococcus [2]; moreover, the main genera reported for raw-milk Adobera cheese from the Los Altos region in México included Streptococcus, Lactococcus, and Lactobacillus [4]. In this context, several studies have highlighted the probiotic potential of numerous LAB strains, mainly Lactobacillus strains isolated from artisanal fermented foods, including artisanal cheeses [5,32]; therefore, it is essential to devise strategies that allow their isolation, study and use, and their correct conservation to keep them available for use.
3.2. Preservation and Evaluation of Lactic Acid Bacteria (LAB) Viability
LAB Preservation is usually carried out using bacterial suspensions prepared in skimmed milk [62,63] or MRS broth [42] and maintained at different temperatures (20, 35, 45 °C), defined by the objective of preservation or the available resources. MRS broth was initially designed as a medium for the selective isolation and growth of different Lactobacillus in substitution of tomato juice agar; nonetheless, it may be used for the cultivation of most LAB [64]. Therefore, MRS broth has become the most frequently used medium for growing this type of LAB, either with or without modifications in its composition. However, the specific growth conditions required by individual members of the LAB group represent a challenge to evaluate non-common media for the isolation, growth, and preservation of this group of microorganisms [65], which also offer easy access, storage, preparation, and handling. In this context, thioglycolate broth is a general-purpose liquid enrichment medium based on the incorporation of sodium thioglycolate as a reducing agent to remove traces of oxygen in the medium (slightly viscous) to allow the recovery and growth of several types of microorganisms, including aerobic, facultative anaerobes, and aerotolerant and microaerophilic bacteria, like LAB [66,67,68]. Furthermore, as it is not a selective medium, it is a cheaper alternative that does not require special storage conditions and its preparation is simple and comparable to that of any other non-selective culture medium. For these reasons, we proposed a preservation method for LAB based on the conservation of bacterial suspensions maintained in thioglycolate broth under cryo-preservation.
From the 16 LAB strains isolated and genetically identified, 11 strains were selected to be included in the cryo-preservation assay using thioglycolate broth and glycerol 20%, followed by an effectivity evaluation expressed as viability and cell integrity. Cell viability of LAB during cryo-preservation, monitored through the plate count, showed that the initial average concentration was 6.979 ± 0.72 log10 CFU mL−1, which increased at day 21 after preservation (8.936 ± 0.49 log10 CFU mL−1). After 42 days of preservation, cell density remained close to the initial count (7.899 ± 0.37 log10 CFU mL−1), but 11% lower than cell viability at day 21; nonetheless, it is important to foreground that the methodology used allows to estimate the viable count of the bacteria through drop-based inoculation so, although it has been reported that is a profitable technique [55], there will always be a certain degree of error associated with the technique. However, the survival rate observed is within the recovery levels suggested for freeze-based conservation, which should be between 50 and 75 % of preserved cells when ultra-low temperature preservation methods, such as cryo-preservation and freeze-drying, are used [69,70]. Soto [71] reported that under cooling conditions using MRS broth, LAB maintained their viability for 84 days, while freeze-based preservation using the same MRS broth allowed the recovery of viable cells up to at least 360 days later; nevertheless, the percentage of viability was affected in the three strains included in that study (Lactobacillus casei, Lactobacillus salivarius, and Pediococcus acidilactici) and the major sensibility was exhibited by the Lactobacillus strains that they included in the study.
Similar results were reported by Zamora [43] in a study aimed to evaluate the effect of two preservation methods on the strains viability with pathogen-antagonist capacity. The author reported that using 12 LAB strains suspended in MRS broth and preserved under freezing conditions at −80 °C, strains maintained 90% of their viability (75 to 100%), a higher value in comparison to the viability obtained through preservation using freeze-drying, which resulted in a loss of viability of up to 97%. The author pointed out that the loss of viability using freeze-drying may occur during the first steps of freezing and dehydration. On the other hand, the loss of viable cells through freezing could be associated with different stages of the process, including an inadequate cooling rate (related to the formation of intra- and extracellular ice or gradual increase of solutes in the suspension) as well as the storage temperature and thawing conditions, but also the nature and the concentration of the cryoprotective agent [72].
3.3. Effect of Cryo-Preservation on Membrane Potential of Lactic Acid Bacteria
The basic function of bacterial conservation is to maintain the viability, purity, stability, and characteristics of the original bacteria [40,72]. Nonetheless, due to the scarce available information related to the effects of conservation methods on the bacteria’s integrity, there is a paramount need to generate evidence of metabolic cell competence in the long term during the preservation. Since membrane potential is involved in several processes of active cells (i.e., autolysis, nutrient transport, and survival under low pH) [73], it could be considered an indicator of cytoplasmic membrane integrity, and therefore of the metabolic competence of the bacteria. In this context, the red–green fluorescence ratio of bacteria stained with diethyloxacarbocyanine (DiOC2(3)), measured through flow cytometry, may be used as an indicator of the magnitude of the membrane potential (Figure 2) [74].
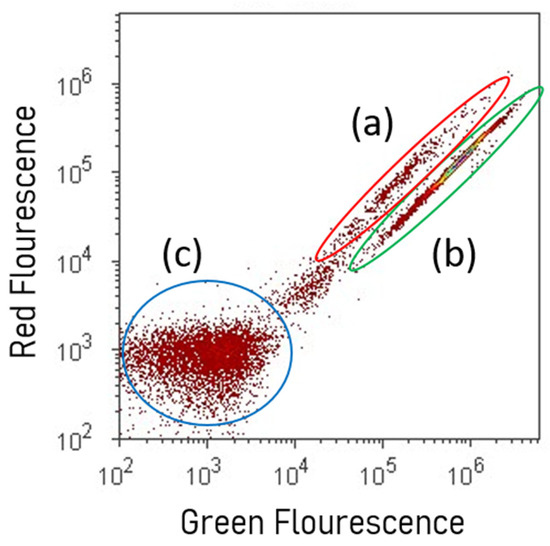
Figure 2.
Density graph of sub-populations of cells assessed via flow cytometry. (a) Viable cells with non-depolarized cytoplasmic membranes stained with DiOC2(3); (b) cells with cytoplasmic membrane depolarized with CCCP; (c) unstained cells auto-fluorescence.
In this study, LAB suspensions were stained with DiOC2(3) immediately after bacterial suspensions’ preparation (thioglycolate broth mixed with glycerol as cryoprotectant) and 180 days after cryo-preservation under −80 °C (Figure 3) to assess changes in the cytoplasmic membrane potential through fluorescence measurement via flow cytometry. Reductions in the magnitude of membrane potential of LAB (based on red–green fluorescence ratio) were observed to be minimal after 180 days under cryo-preservation, equivalent to a 12% average in comparison with fresh cell suspensions. Data confirmed that cell suspensions not only remained viable but also preserved their integrity at the cytoplasmic membrane level. Except for Streptococcus gallolyticus subp. Pasteurianus, all the strains included in the assay maintained their viability at least six months later, expressed as a percentage of cells with active cytoplasmic membranes (64 to 94%). Notwithstanding that not all strains responded in the same way to the same conservation method [42], we hypothesized that low fluorescence values observed with the Streptococcus strain could be related to the ability of the cell to allow the entry of dyes, which could interfere in the evaluation by fluorescence-based techniques, as flow cytometry.
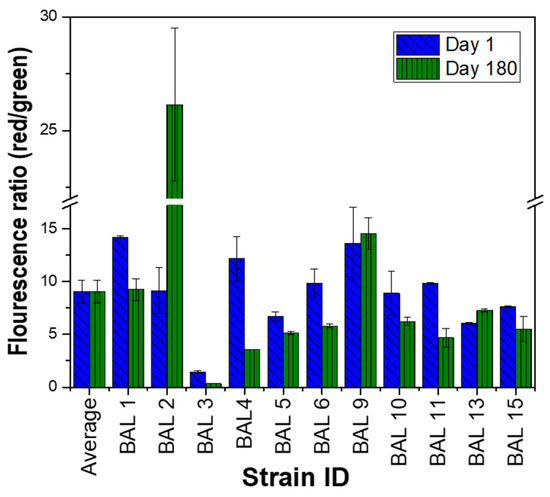
Figure 3.
Membrane potential of lactic acid bacteria before and 180 days after cryo-preservation. Red–green ratios were calculated using population mean fluorescence intensities assessed via flow cytometry.
However, when the integrity of the cytoplasmic membrane is compromised, development is inhibited, even cell death can occur [42]. In that context, some of the assessed strains exhibited a slight increase in the red–green fluorescence ratio that could be related to the ability of bacteria to adapt to the conditions under which they were preserved, and the recovery of possible injured cells induced by sodium thioglycolate as a reducing agent. Regarding that, oxygen stress could be induced to LAB during the cell’s suspension preparation. Nonetheless, one of the advantages of reducing agents such as sodium thioglycolate is the conferred ability to recover injured cells, like Bifidobacterium strains [75]. Also, sodium thioglycolate allows the growth of anaerobic bacteria, even cultivated under aerobic conditions, e.g., Clostridium perfringens strains [76].
3.4. Kinetic Parameters of Lactic Acid Bacteria Recovered from Cryopresevation
Finally, to confirm the metabolic capacity of preserved LAB, the growth curve of each strain was obtained 240 days after preservation, through their reactivation on MRS agar and monitoring by UV spectroscopy (Figure 4). The results suggest that the viability and metabolic capacity of the bacteria were not compromised during conservation. In general terms, we observed that the adaptation phase (phase delay) occurred in the first 3–4 h (Figure 4A–F). From there, LAB showed an exponential increase in optical density (exponential phase) that lasted over 9 h, which demonstrates the reactivation and duplication capacity of the bacteria. Our results are consistent with some previous kinetic parameters reported by several authors for lactic acid bacteria (Figure 4G) [77]. Specifically, for the Streptococcus strain, a typical growth curve was observed (Figure 4C), which confirms its viability and reinforces the hypothesis that the low levels of fluorescence observed by evaluating the membrane potential may be associated with the permeability of the strain to the carbocyanine used for flow cytometric evaluation.
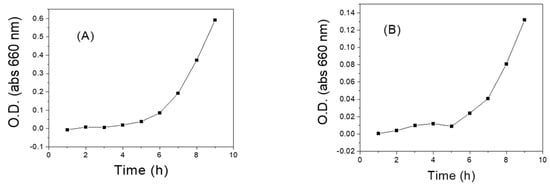
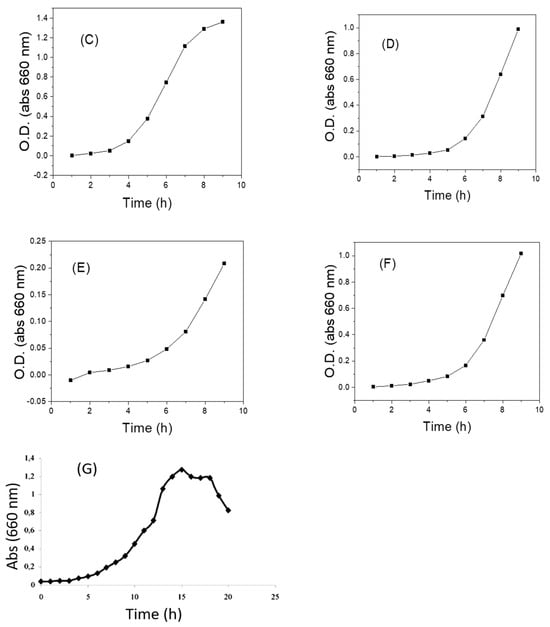
Figure 4.
Growth curves of some LAB recovered from cryopreservation in thioglycolate broth and 20% glycerol at –80 °C. (A) LAB 1 (Q2C3-2) Lactobacillus curvatus; (B) LAB 2 (Q2C3-1B) Lactobacillus sp.; (C) LAB 3 (Q2C8-2) Streptococcus gallolyticus subsp. pasteurianus; (D) LAB 4 (Q2C11) Leuconostoc mesenteroides (E); LAB 5 (Q2C14-2) Lactococcus lactis; (F) LAB 10 (Q4C11-3) Lactobacillus plantarum subs. plantarum. (G) Growth curve of LAB recovered from preservation in MRS broth with mineral oil and reactivated in MRS broth [77].
4. Conclusions
Lactic acid bacteria (LAB) strains from artisanal Adobera cheese from the Los Altos region in México corresponded to common bacteria genera reported for raw-milk cheeses. Preservation of LAB using cryo-freezing in thioglycolate broth and glycerol is a viable alternative to preserve this group of bacteria, guaranteeing the viability and integrity of the microorganism. Flow cytometry is a sensitive and accurate tool that easily allows the discrimination of viable LAB with active cytoplasmic membranes, separating them from dead or inactive cells at the cytoplasmic membrane level. Therefore, the use of flow cytometry as a tool for monitoring the integrity of the cytoplasmic membranes could be an excellent and technologically viable strategy to be implemented to monitor the long-term preservation efficiency of LAB.
Author Contributions
Conceptualization, R.I.A.-G., R.J.D.-M., L.M.A.-E. and J.M.R.-G.; methodology, C.I.C.-C., Z.V. and S.G.-C.; software, L.J.G.-G. and J.M.R.-G.; validation, R.I.A.-G. and J.M.R.-G.; formal analysis, L.M.A.-E. and J.M.R.-G.; investigation, C.I.C.-C., S.G.-C. and J.M.R.-G.; resources, R.I.A.-G. and R.J.D.-M.; data curation, L.M.A.-E., Z.V. and J.M.R.-G.; writing—original draft preparation, L.M.A.-E., Z.V., L.X.Z.-M. and J.M.R.-G.; writing—review and editing, L.M.A.-E., Z.V. and J.M.R.-G.; visualization, R.I.A.-G. and J.M.R.-G.; supervision, R.I.A.-G. and J.M.R.-G.; project administration, R.I.A.-G. and J.M.R.-G.; funding acquisition, R.I.A.-G. and J.M.R.-G. All authors have read and agreed to the published version of the manuscript.
Funding
National Institute of Forestry, Agricultural and Livestock Research, México (INIFAP, 1034034810, 12112434143).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset used and/or analyzed in the current study is available from the corresponding author upon reasonable request.
Acknowledgments
Thanks to the Subcommittee of Microbial and Invertebrate Genetic Resources for Food and Agriculture, especially to the Macro-network of the Food industry.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in poro cheese, an artisanal mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Saxer, S.; Schwenninger, S.M.; Lacroix, C. Characterization of the microflora of industrial Mexican cheeses produced without added chemical preservatives. LWT-Food Sci. Technol. 2013, 53, 314–320. [Google Scholar] [CrossRef]
- Ruvalcaba-Gómez, J.M.; Ruiz-Espinosa, H.; Arteaga-Garibay, R.I.; Rojas-López, M.; Amador-Espejo, G.G.; Anaya-Esparza, L.M.; Delgado-Macuil, R.J. Texture, Physicochemical and sensory properties of artisanal adobera cheese from Los Altos de Jalisco, a genuine Mexican cheese. Int. J. Dairy Technol. 2020, 73, 411–420. [Google Scholar] [CrossRef]
- Ruvalcaba-Gómez, J.M.; Delgado-Macuil, R.J.; Zelaya-Molina, L.X.; Maya-Lucas, O.; Ruesga-Gutiérrez, E.; Anaya-Esparza, L.M.; Villagrán-de la Mora, Z.; López-de la Mora, D.A.; Arteaga-Garibay, R.I. Bacterial succession through the artisanal process and seasonal effects eefining bacterial communities of raw-milk Adobera cheese revealed by high throughput DNA sequencing. Microorganisms 2020, 9, 24. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Scatassa, M.L.; Gaglio, R.; Cardamone, C.; Macaluso, G.; Arcuri, L.; Todaro, M.; Mancuso, I. Anti-listeria activity of lactic acid bacteria in two traditional Sicilian cheeses. Ital. J. Food Saf. 2017, 6, 6191. [Google Scholar] [CrossRef][Green Version]
- Garabal, J.I.; Rodríguez-Alonso, P.; Centeno, J.A. Characterization of lactic acid bacteria isolated from raw cows’ milk Cheeses Currently Produced in Galicia (NW Spain). LWT-Food Sci. Technol. 2008, 41, 1452–1458. [Google Scholar] [CrossRef]
- de Aguiar e Câmara, S.P.; Maduro Dias, C.; Rocha, L.; Dapkevicius, A.; Duarte Rosa, H.J.; de Borba, A.E.S.; Silveira, M.d.G.; Malcata, F.X.; de Lurdes Enes Dapkevicius, M. Assessment of autochthonous lactic acid bacteria as starter cultures for improved manufacture of pico cheese using a cheese model. Int. Dairy J. 2022, 128, 105294. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; Silva, R.C.; Ibrahim, S.A. Lactic acid bacteria: Food safety and human health applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Ingram, M. The lactic acid bacteria—A broad view. In Lactic Acid Bacteria in Beverages and Food; Carr, J.G., Cutting, C.V., Whiting, G.C., Eds.; Academic Press: London, UK, 1975; Volume 1, pp. 1–13. [Google Scholar]
- Almanza, F.; Barrera, E. Tecnología de la Leche y Derivados; Unisur: Bogotá, Colombia, 1991. [Google Scholar]
- Axelsson, L. Lactic Acid Bacteria. In Lactic Acid Bacteria; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Parra Huertas, R.A. Bacterias Ácido Lácticas: Papel Funcional En Los Alimentos. Biotecnol. El Sect. Agropecu. Agroindustrial (BSAA) 2010, 8, 93–105. [Google Scholar]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic acid bacteria in wine: Technological advances and evaluation of their functional role. Front. Microbiol. 2021, 11, 612118. [Google Scholar] [CrossRef] [PubMed]
- Anaya-Esparza, L.M.; Villagrán, Z.; Ruvalcaba-Gómez, J.M. Bacterias Ácido-Lácticas: Propiedades biotecnológicas y aplicaciones. In Bacterias Ácido-Lácticas; Ruvalcaba-Gómez, J.M., Arteaga-Garibay, R.I., López-de la Mora, D.A., Eds.; 2022; Volume 1, pp. 95–120. [Google Scholar]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Samelis, J.; Metaxopoulos, J.; Wallbanks, S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: Description of a new genus Weissella for the Leuconostoc Paramesenteroides Group of Species. J. Appl. Bacteriol. 1993, 75, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ruvalcaba-Gómez, J.M.; Arteaga-Garibay, R.I. Bacterias acido-lácticas: Definición, clasificación y taxonomía. In Bacterias Ácido-Lácticas; Ruvalcaba-Gómez, J.M., Arteaga-Garibay, R.I., López-de la Mora, D.A., Eds.; Universidad de Guadalajara: Guadalajara, Jalisco, México, 2022; Volume 1, pp. 13–40. [Google Scholar]
- Pavunc, A.L.; Beganovic, J.; Kos, B.; Uroic, K.; Blazic, M.; Suskovic, J. Characterization and application of autochthonous starter cultures for fresh cheese production. Food Technol. Biotechnol. 2012, 50, 141–151. [Google Scholar]
- Speranza, B.; Bevilacqua, A.; Corbo, M.R.; Altieri, C.; Sinigaglia, M. Selection of autochthonous strains as promising starter cultures for Fior Di Latte, a traditional cheese of southern Italy. J. Sci. Food Agric. 2015, 95, 88–97. [Google Scholar] [CrossRef]
- Ramírez-López, C.; Vélez-Ruiz, J.F. Aislamiento, caracterización y selección de bacterias lácticas autóctonas de leche y queso fresco artesanal de cabra. Inf. Technol 2016, 27, 115–128. [Google Scholar] [CrossRef][Green Version]
- Kharnaior, P.; Tamang, J.P. Probiotic Properties of lactic acid bacteria isolated from the spontaneously fermented soybean foods of the eastern himalayas. Fermentation 2023, 9, 461. [Google Scholar] [CrossRef]
- Zareie, Z.; Moayedi, A.; Garavand, F.; Tabar-Heydar, K.; Khomeiri, M.; Maghsoudlou, Y. Probiotic properties, safety assessment, and aroma-generating attributes of some lactic acid bacteria isolated from Iranian traditional cheese. Fermentation 2023, 9, 338. [Google Scholar] [CrossRef]
- Ruvalcaba-Gómez, J.M.; Ruiz-Espinosa, H.; Méndez-Robles, M.D.; Arteaga-Garibay, R.I.; Anaya-Esparza, L.M.; Villagrán, Z.; Delgado-Macuil, R.J. Use of Autochthonous lactic acid bacteria as starter culture of pasteurized milk adobera cheese. Fermentation 2022, 8, 234. [Google Scholar] [CrossRef]
- Abedin, M.M.; Chourasia, R.; Phukon, L.C.; Sarkar, P.; Ray, R.C.; Singh, S.P.; Rai, A.K. Lactic acid bacteria in the functional food industry: Biotechnological properties and potential applications. Crit. Rev. Food Sci. Nutr. 2023, 1–19. [Google Scholar] [CrossRef]
- Fan, Q.; Zeng, X.; Wu, Z.; Guo, Y.; Du, Q.; Tu, M.; Pan, D. Nanocoating of lactic acid bacteria: Properties, protection mechanisms, and future trends. Crit. Rev. Food Sci. Nutr. 2023, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.A. Caracterización de Bacterias Lácticas Aisladas a Partir de Cultivos Iniciadores Comerciales y su Conservación. Ph.D. Thesis, Instituto Politécnico Nacional, Ciudad de México, Mexico, 1989. [Google Scholar]
- Daniel, A.A.; Egbebi, A.O.; Onasanya, A.A. Isolation and identification of lactic acid bacteria from spontaneously fermented Kunun-Zaki using RAPD-PCR analysis. ABUAD Int. J. Nat. Appl. Sci. 2023, 3, 34–42. [Google Scholar] [CrossRef]
- Okoye, C.O.; Gao, L.; Wu, Y.; Li, X.; Wang, Y.; Jiang, J. Identification, characterization and optimization of culture medium conditions for organic acid-producing lactic acid bacteria strains from Chinese fermented vegetables. Prep. Biochem. Biotechnol. 2023, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.M. Identificación Bioquímica de Bacterias Acido-Lácticas Aisladas a Partir de Productos Lácteos en el Estado de Hidalgo. Bachelor’s Thesis, Universidad Autónoma del Estado de Hidalgo, Pachuca de Soto, México, 2006. Available online: http://dgsa.uaeh.edu.mx:8080/bibliotecadigital/handle/231104/381 (accessed on 1 October 2023).
- Zamanpour, S.; Rezvani, R.; Jafarzadeh Isfahani, A.; Afshari, A. Isolation and some basic characteristics of lactic acid bacteria from beetroot (Beta vulgaris L.)—A preliminary study. Canrea J. Food Technol. Nutr. Culin. J. 2023, 42–56. [Google Scholar] [CrossRef]
- Hadef, S.; Idoui, T.; Sifour, M.; Genay, M.; Dary-Mourot, A. Screening of wild lactic acid bacteria from Algerian traditional cheeses and goat butter to develop a new probiotic starter culture. Probiotics Antimicrob. Proteins 2023, 15, 387–399. [Google Scholar] [CrossRef]
- Karahan, A.G.; Başyiğit Kılıç, G.; Kart, A.; Şanlıdere Aloğlu, H.; Öner, Z.; Aydemir, S.; Erkuş, O.; Harsa, Ş. Genotypic identification of some lactic acid bacteria by amplified fragment length polymorphism analysis and investigation of their potential usage as starter culture combinations in beyaz cheese manufacture. J. Dairy Sci. 2010, 93, 1–11. [Google Scholar] [CrossRef]
- Aquilanti, L.; Silvestri, G.; Zannini, E.; Osimani, A.; Santarelli, S.; Clementi, F. Phenotypic, genotypic and technological characterization of predominant lactic acid bacteria in pecorino cheese from central Italy. J. Appl. Microbiol. 2007, 103, 948–960. [Google Scholar] [CrossRef]
- de las Rivas, B.; Marcobal, Á.; Muñoz, R. Development of a multilocus sequence typing method for analysis of lactobacillus plantarum strains. Microbiology 2006, 152, 85–93. [Google Scholar] [CrossRef]
- González-García, L.N.; Vanegas-López, M.; Riaño-Pachón, D.M. Comparing the potential for identification of lactobacillus Spp. of 16S RDNA variable regions. Acta Biol. Colomb. 2013, 18, 349–364. [Google Scholar]
- Valdéz-Alarcón, J.J.; Ruvalcaba-Gómez, J.M.; Chávez-Bárcenas, A.T.; Arteaga-Garibay, R.I. Identificación molecular y genotipificación de bacterias acido-lácticas. In Bacterias Ácido-Lácticas; Ruvalcaba-Gómez, J.M., Arteaga-Garibay, R.I., López-de la Mora, D.A., Eds.; Universidad de Guadalajara: Guadalajara, México, 2022; Volume 1, pp. 67–94. [Google Scholar]
- Smith, D. Culture Collections. Adv. Appl. Microbiol. 2012, 79, 73–118. [Google Scholar]
- Fonseca, F.; Girardeau, A.; Passot, S. Freeze-drying of lactic acid bacteria: A stepwise approach for developing a freeze-drying protocol based on physical properties. Methods Mol. Biol. 2021, 2180, 703–719. [Google Scholar] [PubMed]
- Arencibia, A.D.F.; Rosario, F.L.A.; Gámez, M.R. Métodos generales de conservación de microorganismos. In Proceedings of the I Taller Científico de Los Laboratorios Liorad, VI Taller de Colecciones de Cultivos Microbianos y Otros Materiales Biológicos; Ediciones Finlay: La Habana, Cuba, 2008; Available online: https://www.researchgate.net/institution/Centro-Nacional-de-Investigaciones-Cientificas (accessed on 1 October 2023).
- Wang, G.-Q.; Pu, J.; Yu, X.-Q.; Xia, Y.-J.; Ai, L.-Z. Influence of freezing temperature before freeze-drying on the viability of various lactobacillus plantarum strains. J. Dairy Sci. 2020, 103, 3066–3075. [Google Scholar] [CrossRef] [PubMed]
- Rault, A.; Béal, C.; Ghorbal, S.; Ogier, J.-C.; Bouix, M. Multiparametric flow cytometry allows rapid assessment and comparison of lactic acid bacteria viability after freezing and during frozen storage. Cryobiology 2007, 55, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Zamora, R.L.M. Aislamiento, Identificación y Conservación de Cultivos de Bacterias Lácticas Antagonistas de Microbiota Contaminante de Sangre de Matadero; Universitat de Girona: Girona, España, 2003. [Google Scholar]
- Corry, J.E.L.; Curtis, G.D.W.; Baird, R.M. De Man, Rogosa and Sharpe (MRS) Agar. Prog. Ind. Microbiol. 2003, 37, 511–513. [Google Scholar]
- Di Cagno, R.; Cardinali, G.; Minervini, G.; Antonielli, L.; Rizzello, C.G.; Ricciuti, P.; Gobbetti, M. Taxonomic structure of the yeasts and lactic acid bacteria microbiota of pineapple (Ananas comosus L. Merr.) and use of autochthonous starters for minimally processing. Food Microbiol. 2010, 27, 381–389. [Google Scholar] [CrossRef]
- Domingos-Lopes, M.F.P.; Stanton, C.; Ross, P.R.; Dapkevicius, M.L.E.; Silva, C.C.G. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal pico cheese. Food Microbiol. 2017, 63, 178–190. [Google Scholar] [CrossRef]
- Sadat, N.N.; Rezaeizadeh, G.; Sarami, F.; Khayam-Nekooii, M.S.; Khosravi-Darani, K. A comparison between different methods for the preservation of lactic acid bacteria for usage as a starter culture. Biol. J. Microorg. 2022, 11, 77–94. [Google Scholar]
- Jackson, M.S.; Bird, A.R.; McOrist, A.L. Comparison of two selective media for the detection and enumeration of lactobacilli in human faeces. J. Microbiol. Methods 2002, 51, 313–321. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic <scp>DNA</Scp> from bacteria. Curr. Protoc. Mol. Biol. 2001, 56. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus Plantarum, L. Pentosus, and L. Paraplantarum by RecA gene sequence analysis and multiplex PCR assay with RecA gene-derived primers. Appl. Env. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Hedges, A.J. Estimating the Precision of Serial Dilutions and Viable Bacterial Counts. Int. J. Food Microbiol. 2002, 76, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The Estimation of the Bactericidal Power of the Blood. Epidemiol. Infect. 1938, 38, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, F. Lactic Acid Bacteria. In Encyclopedia of Food and Health; Elsevier: Amsterdam, The Netherlands, 2016; pp. 501–508. [Google Scholar]
- Kieliszek, M.; Pobiega, K.; Piwowarek, K.; Kot, A.M. Characteristics of the proteolytic enzymes produced by lactic acid bacteria. Molecules 2021, 26, 1858. [Google Scholar] [CrossRef] [PubMed]
- Vasyliuk, O.M.; Skrotskyi, S.O.; Khomenko, L.A.; Babich, T.V. Probiotics based on lactic acid bacteria for aquaculture. Mikrobiol. Zh 2023, 85, 75–92. [Google Scholar] [CrossRef]
- Pritchard, D.J.; Fa, J.E.; Oldfield, S.; Harrop, S.R. Bring the captive closer to the wild: Redefining the role of ex-Situ conservation. Oryx 2012, 46, 18–23. [Google Scholar] [CrossRef]
- Poznanski, E.; Cavazza, A.; Cappa, F.; Cocconcelli, P.S. Indigenous raw milk microbiota influences the bacterial development in traditional cheese from an alpine natural park. Int. J. Food Microbiol. 2004, 92, 141–151. [Google Scholar] [CrossRef]
- Conway, P.L.; Gorbach, S.L.; Goldin, B.R. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 1987, 70, 1–12. [Google Scholar] [CrossRef]
- Zhang, L.; García-Cano, I.; Jiménez-Flores, R. Effect of milk phospholipids on the growth and cryotolerance of lactic acid bacteria cultured and stored in acid whey-based media. JDS Commun. 2020, 1, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, U.; Holzapfel, W.H. Chapter 8 Culture media for lactic acid bacteria. In Progress in Industrial Microbiology; Janet, E.L., Corry, G.D.W., Curtis, R.M.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2003; pp. 127–140. [Google Scholar]
- Renschler, M.A.; Wyatt, A.; Anene, N.; Robinson-Hill, R.; Pickerill, E.S.; Fox, N.E.; Griffith, J.A.; McKillip, J.L. Using nitrous acid-modified de Man, Rogosa, and Sharpe medium to selectively isolate and culture lactic acid bacteria from dairy foods. J. Dairy Sci. 2020, 103, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.B.; Kloos, W.E. Use of Shake Cultures in a Semisolid Thioglycolate Medium for Differentiating Staphylococci from Micrococci. Appl. Microbiol. 1972, 23, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Hollister, A.G.; Corrier, D.E.; Nisbet, D.J.; Beier, R.C.; Deloach, J.R. Effect of Lyophilization in sucrose plus dextran and rehydration in thioglycollate broth on performance of competitive exclusion cultures in broiler chicks. Poult. Sci. 1995, 74, 586–590. [Google Scholar] [CrossRef]
- Griffe, M.B.; Patterson, S.S.; Miller, C.H.; Kafrawy, A.H.; Newton, C.W. Comparison of bacterial growth in an improperly but commonly used medium versus reduced Thioglycolate with the use of an anaerobic sampling technique. Oral. Surg. Oral. Med. Oral. Pathol. 1981, 52, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Bagatolli, C.D. Validación de un Método Alternativo Para la Preservación de Bacterias. Bachelor’s Thesis, Universidad Nacional de Cuyo, Mendoza, Argentina, 2017. [Google Scholar]
- Smith, D.; Thomas, V.E. Cryogenic light microscopy and the development of cooling protocols for the cryopreservation of filamentous fungi. World J. Microbiol. Biotechnol. 1997, 14, 49–57. [Google Scholar] [CrossRef]
- Soto, L.P. Elección de un Método de Conservación Que Asegure la Viabilidad de Bacterias Indígenas Probióticas y Mejore la Efectividad de Administración a Terneros Lactantes Criados a Campo. Ph.D. Thesis, Universidad Nacional de Litoral, Santa Fe, Argentina, 2010. [Google Scholar]
- Girardeau, A.; Passot, S.; Meneghel, J.; Cenard, S.; Lieben, P.; Trelea, I.-C.; Fonseca, F. Insights into lactic acid bacteria cryoresistance using FTIR microspectroscopy. Anal. Bioanal. Chem. 2022, 414, 1425–1443. [Google Scholar] [CrossRef]
- Novo, D.; Perlmutter, N.G.; Hunt, R.H.; Shapiro, H.M. Accurate flow cytometric membrane potential measurement in bacteria using Diethyloxacarbocyanine and a Ratiometric Technique. Cytometry 1999, 35, 55–63. [Google Scholar] [CrossRef]
- Li, L.; Yang, M.; Zhu, W.; Liu, X.; Peng, X.; Li, H. Functionally ampicillin-stressed proteomics reveals that AdhE regulates alcohol metabolism for antibiotic resistance in Escherichia coli. Process Biochem. 2021, 104, 132–141. [Google Scholar] [CrossRef]
- Nebra, Y.; Jofre, J.; Blanch, A.R. The Effect of reducing agents on the recovery of injured bifidobacterium cells. J. Microbiol. Methods 2002, 49, 247–254. [Google Scholar] [CrossRef]
- Jia, Z.; Liu, Y.; Hwang, C.-A.; Huang, L. Effect of combination of oxyrase and sodium thioglycolate on growth of clostridium perfringens from spores under aerobic incubation. Food Microbiol. 2020, 89, 103413. [Google Scholar] [CrossRef] [PubMed]
- León, P.A.M.; Montoya, C.O.I.; Motato, K.E.; Granda, D.M.; Acaro, C.; Restrepo, J.M.; Echeverri, S.; Valencia, J. Colombian wild lactic acid bacteria (LAB) show good properties in Sourdough manufacture. Rev. Fac. Química Farm. 2006, 13, 26–35. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).