Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria
Abstract
1. Introduction
2. Materials and Methods
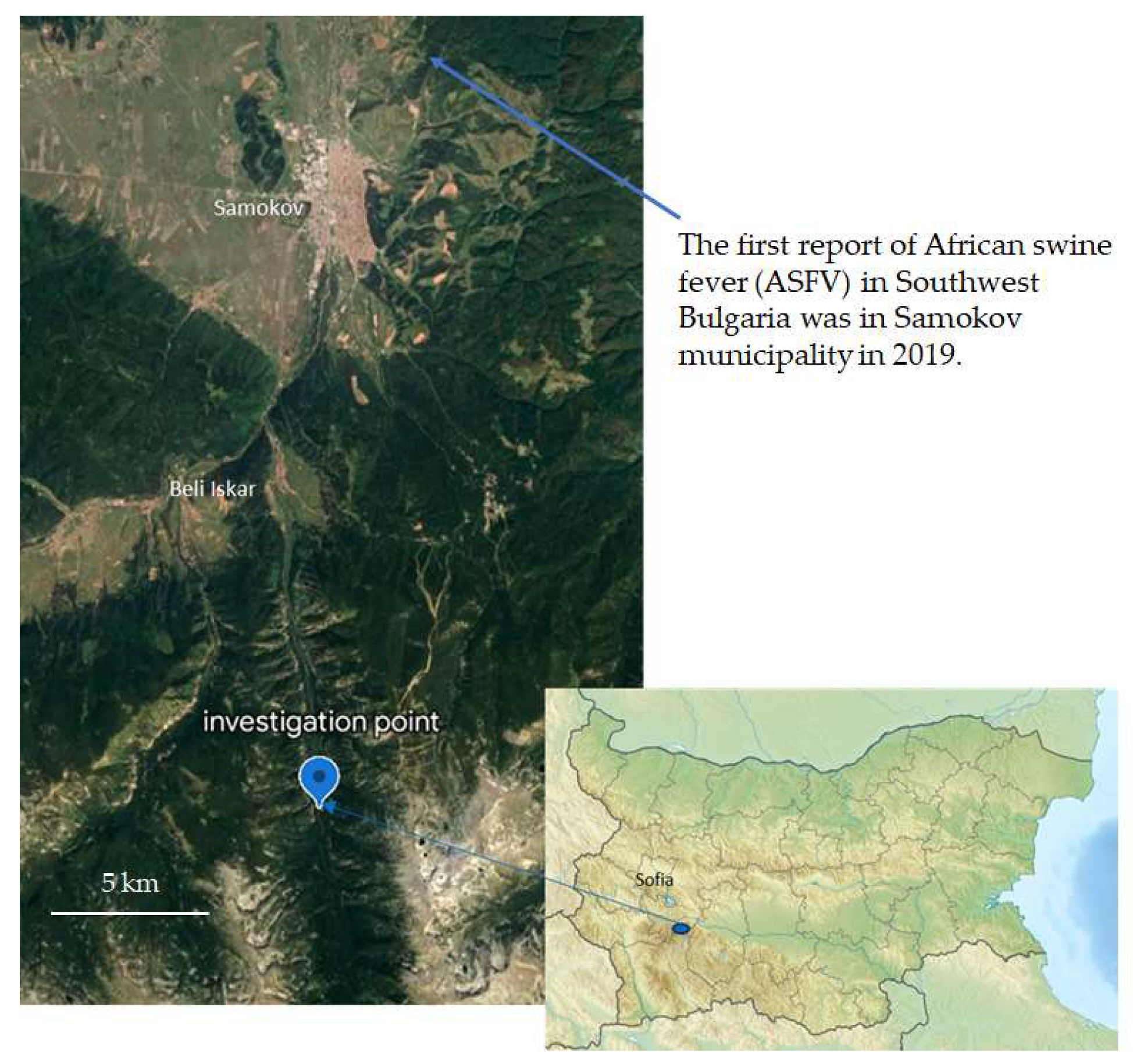
2.1. Sample Collection
2.2. Isolation of Single Bacterial Cultures
2.3. Biochemical Identification
2.3.1. Identification with BD PhoenixTM
2.3.2. Identification with API 20 E of the Gram-Negative Bacteria
2.3.3. Confirmation of Identification of Escherichia coli and Yersinia enterocolitica by Traditional Polymerase Chain Reaction (PCR)
2.4. Antibiotic Resistance/Susceptibility Elucidation
2.4.1. BD PhoenixTM
2.4.2. Disk Diffusion Method
2.5. Searching for the Ail Gene of Pathogenicity in Y. enterocolitica
2.5.1. Traditional PCR
2.5.2. Droplet Digital PCR (ddPCR)
2.6. Searching for ASFV with Conventional PCR
3. Results
3.1. Isolation of Single Bacterial Cultures
3.2. Biochemical Identification
3.3. Confirmation of Identification of E. coli and Y. enterocolitica by Traditional PCR
3.4. Antibiotic Resistance/Susceptibility Elucidation
3.4.1. BD PhoenixTM
3.4.2. Disk Diffusion Method
3.5. No Ail Gene of Pathogenicity in Y. enterocolitica Was Found
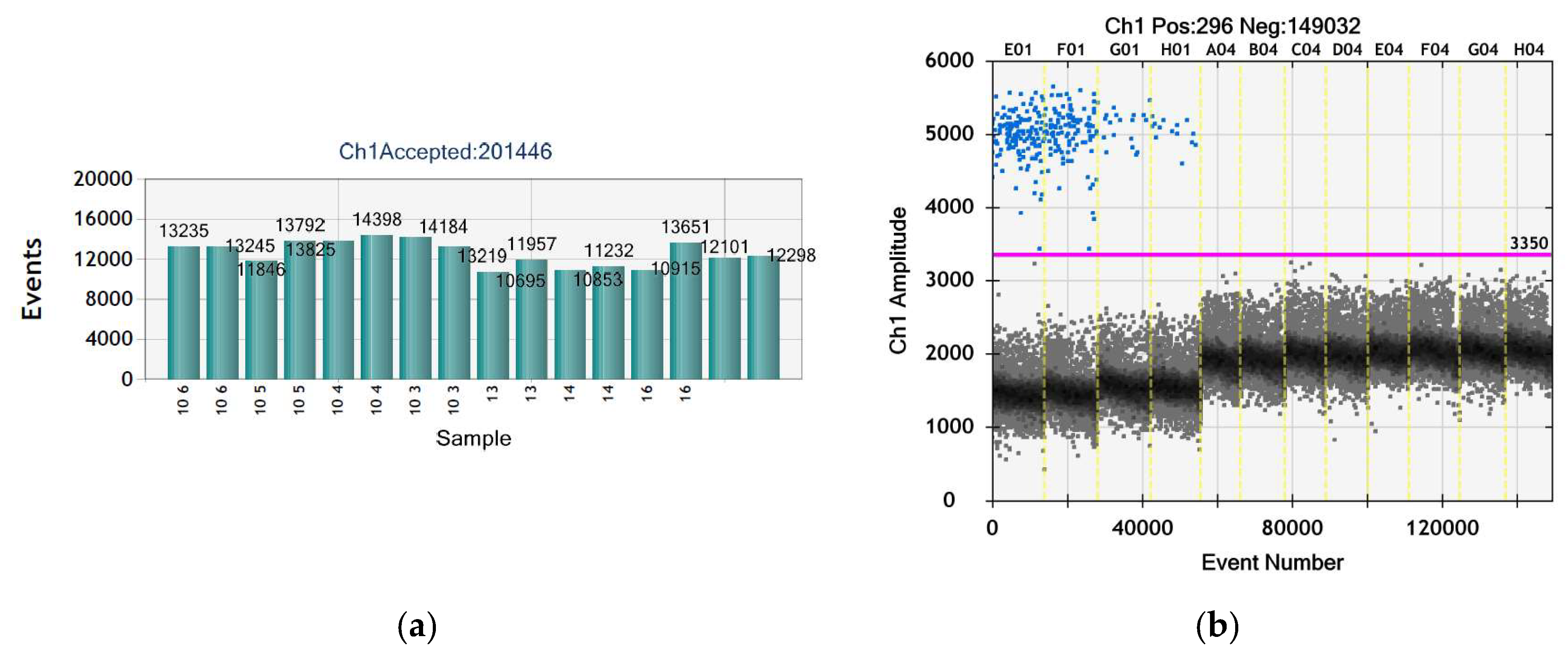
3.6. No ASFV Was Detected
4. Discussion
4.1. Previous Studies on the GM of Ap. flavicollis (Yellow-Necked Mice)
4.2. Previous Studies on GM of M. glareolus (Bank Voles)
4.3. Previous Studies on the Antimicrobial Resistance of M. glareolus and Ap. flavicollis
4.4. Biotechnological and Practical Potential of the Isolated Bacteria
4.5. Can Other Animals Spread ASFV?
4.6. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walter:, J.; Armet, A.M.; Finlay, B.B.; Shanahan, F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell 2020, 180, 221–232. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Tukalenko, E.; Mousseau, T.A.; Thompson, L.R.; Knight, R.; Mappes, T.; Watts, P.C. Two Hundred and Fifty-Four Metagenome-Assembled Bacterial Genomes from the Bank Vole Gut Microbiota. Sci. Data 2020, 7, 312. [Google Scholar] [CrossRef] [PubMed]
- Mamuad, L.L.; Kim, S.H.; Biswas, A.A.; Yu, Z.; Cho, K.-K.; Kim, S.-B.; Lee, K.; Lee, S.S. Rumen Fermentation and Microbial Community Composition Influenced by Live Enterococcus faecium Supplementation. AMB Express 2019, 9, 123. [Google Scholar] [CrossRef]
- Arentsen, T.; Qian, Y.; Gkotzis, S.; Femenia, T.; Wang, T.; Udekwu, K.; Forssberg, H.; Diaz Heijtz, R. The Bacterial Peptidoglycan-Sensing Molecule Pglyrp2 Modulates Brain Development and Behavior. Mol. Psychiatry 2017, 22, 257–266. [Google Scholar] [CrossRef] [PubMed]
- González-Arancibia, C.; Urrutia-Piñones, J.; Illanes-González, J.; Martinez-Pinto, J.; Sotomayor-Zárate, R.; Julio-Pieper, M.; Bravo, J.A. Do Your Gut Microbes Affect Your Brain Dopamine? Psychopharmacology 2019, 236, 1611–1622. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 2017, 171, 1015–1028.e13. [Google Scholar] [CrossRef] [PubMed]
- Viney, M. The Gut Microbiota of Wild Rodents: Challenges and Opportunities. Lab. Anim. 2019, 53, 252–258. [Google Scholar] [CrossRef]
- Pearce, J.; Venier, L. Small Mammals as Bioindicators of Sustainable Boreal Forest Management. For. Ecol. Manag. 2005, 208, 153–175. [Google Scholar] [CrossRef]
- Gwenzi, W.; Chaukura, N.; Muisa-Zikali, N.; Teta, C.; Musvuugwa, T.; Rzymski, P.; Abia, A.L.K. Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance. Antibiot. Basel Switz. 2021, 10, 68. [Google Scholar] [CrossRef]
- de Jonge, N.; Carlsen, B.; Christensen, M.H.; Pertoldi, C.; Nielsen, J.L. The Gut Microbiome of 54 Mammalian Species. Front. Microbiol. 2022, 13, 886252. [Google Scholar] [CrossRef]
- Deutz, A. Paratuberkulose Bei Wildtieren—(Ernährungsbedingte) Krankheit Der Zukunft? In Proceedings of the Tagung für die Jägerschaft, Federal Institute for Alpine Agriculture Gumpenstein, A-8952 Irdning, Gumpenstein, Austria, 16–17 February 2004. [Google Scholar]
- Kreisinger, J.; Bastien, G.; Hauffe, H.C.; Marchesi, J.; Perkins, S.E. Interactions between Multiple Helminths and the Gut Microbiota in Wild Rodents. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140295. [Google Scholar] [CrossRef] [PubMed]
- Lavrinienko, A.; Hämäläinen, A.; Hindström, R.; Tukalenko, E.; Boratyński, Z.; Kivisaari, K.; Mousseau, T.A.; Watts, P.C.; Mappes, T. Comparable Response of Wild Rodent Gut Microbiome to Anthropogenic Habitat Contamination. Mol. Ecol. 2021, 30, 3485–3499. [Google Scholar] [CrossRef] [PubMed]
- Hanhimäki, E.; Watts, P.C.; Koskela, E.; Koteja, P.; Mappes, T.; Hämäläinen, A.M. Evolved High Aerobic Capacity Has Context-Specific Effects on Gut Microbiota. Front. Ecol. Evol. 2022, 10, 934164. [Google Scholar] [CrossRef]
- Kohl, K.D.; Sadowska, E.T.; Rudolf, A.M.; Dearing, M.D.; Koteja, P. Experimental Evolution on a Wild Mammal Species Results in Modifications of Gut Microbial Communities. Front. Microbiol. 2016, 7, 634. [Google Scholar] [CrossRef] [PubMed]
- Brila, I.; Lavrinienko, A.; Tukalenko, E.; Kallio, E.R.; Mappes, T.; Watts, P.C. Idiosyncratic Effects of Coinfection on the Association between Systemic Pathogens and the Gut Microbiota of a Wild Rodent, the Bank Vole Myodes glareolus. J. Anim. Ecol. 2023, 92, 826–837. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Tukalenko, E.; Mappes, T.; Watts, P.C. Skin and Gut Microbiomes of a Wild Mammal Respond to Different Environmental Cues. Microbiome 2018, 6, 209. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Tukalenko, E.; Kesäniemi, J.; Kivisaari, K.; Masiuk, S.; Boratyński, Z.; Mousseau, T.A.; Milinevsky, G.; Mappes, T.; Watts, P.C. Applying the Anna Karenina Principle for Wild Animal Gut Microbiota: Temporal Stability of the Bank Vole Gut Microbiota in a Disturbed Environment. J. Anim. Ecol. 2020, 89, 2617–2630. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Mappes, T.; Tukalenko, E.; Mousseau, T.A.; Møller, A.P.; Knight, R.; Morton, J.T.; Thompson, L.R.; Watts, P.C. Environmental Radiation Alters the Gut Microbiome of the Bank Vole Myodes glareolus. ISME J. 2018, 12, 2801–2806. [Google Scholar] [CrossRef]
- Brila, I.; Lavrinienko, A.; Tukalenko, E.; Ecke, F.; Rodushkin, I.; Kallio, E.R.; Mappes, T.; Watts, P.C. Low-Level Environmental Metal Pollution Is Associated with Altered Gut Microbiota of a Wild Rodent, the Bank Vole (Myodes glareolus). Sci. Total Environ. 2021, 790, 148224. [Google Scholar] [CrossRef]
- Furness, L.E.; Campbell, A.; Zhang, L.; Gaze, W.H.; McDonald, R.A. Wild Small Mammals as Sentinels for the Environmental Transmission of Antimicrobial Resistance. Environ. Res. 2017, 154, 28–34. [Google Scholar] [CrossRef]
- Guenther, S.; Grobbel, M.; Heidemanns, K.; Schlegel, M.; Ulrich, R.G.; Ewers, C.; Wieler, L.H. First Insights into Antimicrobial Resistance among Faecal Escherichia coli Isolates from Small Wild Mammals in Rural Areas. Sci. Total Environ. 2010, 408, 3519–3522. [Google Scholar] [CrossRef] [PubMed]
- Skarżyńska, M.; Zając, M.; Kamińska, E.; Bomba, A.; Żmudzki, J.; Jabłoński, A.; Wasyl, D. Salmonella and Antimicrobial Resistance in Wild Rodents-True or False Threat? Pathog. Basel Switz. 2020, 9, 771. [Google Scholar] [CrossRef] [PubMed]
- Kmet’, V.; Čuvalová, A.; Stanko, M. Small Mammals as Sentinels of Antimicrobial-Resistant Staphylococci. Folia Microbiol. 2018, 63, 665–668. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Leekitcharoenphon, P.; Hendriksen, R.S.; Aarestrup, F.M.; Wasyl, D. A Metagenomic Glimpse into the Gut of Wild and Domestic Animals: Quantification of Antimicrobial Resistance and More. PLoS ONE 2020, 15, e0242987. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://bnr.bg/post/101521898/poroda-na-raba-na-izchezvaneto (accessed on 8 August 2023).
- Available online: https://www.vesti.bg/bulgaria/svinskata-chuma-stigna-do-iuzhna-bylgariia-6098131 (accessed on 21 August 2023).
- Available online: https://btvnovinite.bg/bulgaria/parvi-sluchaj-na-afrikanska-chuma-po-svinete-v-sofijska-oblast.html (accessed on 21 August 2023).
- Available online: https://bnr.bg/horizont/post/101152445/potvardiha-nalichieto-na-afrikanska-chuma-v-lovno-stopanstvo-iskar (accessed on 21 August 2023).
- Bej, A.K.; DiCesare, J.L.; Haff, L.; Atlas, R.M. Detection of Escherichia coli and Shigella spp. in Water by Using the Polymerase Chain Reaction and Gene Probes for Uid. Appl. Environ. Microbiol. 1991, 57, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Clifford, R.J.; Milillo, M.; Prestwood, J.; Quintero, R.; Zurawski, D.V.; Kwak, Y.I.; Waterman, P.E.; Lesho, E.P.; Mc Gann, P. Detection of Bacterial 16S rRNA and Identification of Four Clinically Important Bacteria by Real-Time PCR. PLoS ONE 2012, 7, e48558. [Google Scholar] [CrossRef]
- Neubauer, H.; Hensel, A.; Aleksic, S.; Meyer, H. Identification of Yersinia enterocolitica within the Genus Yersinia. Syst. Appl. Microbiol. 2000, 23, 58–62. [Google Scholar] [CrossRef]
- Lambertz, S.T.; Nilsson, C.; Hallanvuo, S.; Lindblad, M. Real-Time PCR Method for Detection of Pathogenic Yersinia enterocolitica in Food. Appl. Environ. Microbiol. 2008, 74, 6060–6067. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 3rd ed.; Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009. [Google Scholar]
- The European Committee on Antimicrobil Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0. 2022. Available online: https://www.eucast.org/ (accessed on 1 August 2023).
- Fernández-Pinero, J.; Gallardo, C.; Elizalde, M.; Robles, A.; Gómez, C.; Bishop, R.; Heath, L.; Couacy-Hymann, E.; Fasina, F.O.; Pelayo, V.; et al. Molecular Diagnosis of African Swine Fever by a New Real-Time PCR Using Universal Probe Library. Transbound. Emerg. Dis. 2013, 60, 48–58. [Google Scholar] [CrossRef]
- GenBank Overview. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 1 August 2023).
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA Sequence-Based Analysis of the Pseudomonas Species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef]
- Arnold, T.; Neubauer, H.; Nikolaou, K.; Roesler, U.; Hensel, A. Identification of Yersinia enterocolitica in Minced Meat: A Comparative Analysis of API 20E, Yersinia Identification Kit and a 16S rRNA-Based PCR Method. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Kwaga, J.; Iversen, J.O.; Misra, V. Detection of Pathogenic Yersinia enterocolitica by Polymerase Chain Reaction and Digoxigenin-Labeled Polynucleotide Probes. J. Clin. Microbiol. 1992, 30, 2668–2673. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Powderly, W.G.; Opal, S.M. Infectious Diseases, 4th ed.; Elsevier: London, UK, 2017. [Google Scholar]
- Richwagen, N.; Lyles, J.T.; Dale, B.L.F.; Quave, C.L. Antibacterial Activity of Kalanchoe mortagei and K. fedtschenkoi Against ESKAPE Pathogens. Front. Pharmacol. 2019, 10, 67. [Google Scholar] [CrossRef]
- Cooney, S.; O’Brien, S.; Iversen, C.; Fanning, S. Bacteria: Other Pathogenic Enterobacteriaceae—Enterobacter and Other Genera; Elsevier: Amsterdam, The Netherlands, 2014; pp. 433–441. [Google Scholar]
- Fàbrega, A.; Vila, J. Yersinia enterocolitica: Pathogenesis, Virulence and Antimicrobial Resistance. Enfermedades Infecc. Microbiol. Clínica 2012, 30, 24–32. [Google Scholar] [CrossRef]
- Mahlen, S.D. Serratia Infections: From Military Experiments to Current Practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Bollet, C.; Grimont, P.; Gainnier, M.; Geissler, A.; Sainty, J.M.; De Micco, P. Fatal Pneumonia Due to Serratia proteamaculans Subsp. Quinovora. J. Clin. Microbiol. 1993, 31, 444–445. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, Genomics, and Clinical Significance of the Pseudomonas fluorescens Species Complex, an Unappreciated Colonizer of Humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Mascini, E.M.; Troelstra, A.; Beitsma, M.; Blok, H.E.M.; Jalink, K.P.; Hopmans, T.E.M.; Fluit, A.C.; Hene, R.J.; Willems, R.J.L.; Verhoef, J.; et al. Genotyping and Preemptive Isolation to Control an Outbreak of Vancomycin-Resistant Enterococcus faecium. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006, 42, 739–746. [Google Scholar] [CrossRef][Green Version]
- Dutkiewicz, J.; Mackiewicz, B.; Kinga Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A Mysterious Bacterium of Evil and Good. Part III. Deleterious Effects: Infections of Humans, Animals and Plants. Ann. Agric. Environ. Med. AAEM 2016, 23, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, 3. [Google Scholar] [CrossRef]
- Keller, R.; Pedroso, M.Z.; Ritchmann, R.; Silva, R.M. Occurrence of Virulence-Associated Properties in Enterobacter cloacae. Infect. Immun. 1998, 66, 645–649. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Pagès, J.-M. Enterobacter aerogenes and Enterobacter cloacae; Versatile Bacterial Pathogens Confronting Antibiotic Treatment. Front. Microbiol. 2015, 6, 392. [Google Scholar] [CrossRef]
- Kaleva, M.D.; Ilieva, Y.; Zaharieva, M.M.; Dimitrova, L.; Kim, T.C.; Tsvetkova, I.; Georgiev, Y.; Orozova, P.; Nedev, K.; Najdenski, H. Antimicrobial Resistance and Biofilm Formation of Escherichia coli Isolated from Pig Farms and Surroundings in Bulgaria. Microorganisms 2023, 11, 1909. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, L.; Kaleva, M.; Zaharieva, M.M.; Stoykova, C.; Tsvetkova, I.; Angelovska, M.; Ilieva, Y.; Kussovski, V.; Naydenska, S.; Najdenski, H. Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria. Antibiot. Basel Switz. 2021, 10, 940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, P.; Zhu, Y.; Zhang, G.; Xu, Y.; Yang, Q. Performance Evaluation of BD Phoenix NMIC-413 Antimicrobial Susceptibility Testing Panel for Imipenem, Meropenem, and Ertapenem Against Clinical Carbapenem-Resistant and Carbapenem-Susceptible Enterobacterales. Front. Med. 2021, 8, 643194. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, E.; Miholova, D.; Cibulka, J.; Mader, P.; Slameva, A. Small Mammals as Bioindicators for Terrestrial Ecosystems in Bohemia. In Biomarkers: A Pragmatic Basis for Remediation of Severe Pollution in Eastern Europe; Peakall, D.B., Walker, C.H., Migula, P., Eds.; NATO Science Series; Springer: Dordrecht, The Netherlands, 1999; pp. 308–309. ISBN 978-94-011-4550-3. [Google Scholar]
- Gaukler, S.M.; Murphy, S.M.; Berryhill, J.T.; Thompson, B.E.; Sutter, B.J.; Hathcock, C.D. Investigating Effects of Soil Chemicals on Density of Small Mammal Bioindicators Using Spatial Capture-Recapture Models. PLoS ONE 2020, 15, e0238870. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.A.; González-Barrio, D.; Tenorio, C.; Ruiz-Fons, F.; Torres, C. Antimicrobial Resistance in Faecal Escherichia coli Isolates from Farmed Red Deer and Wild Small Mammals. Detection of a Multiresistant E. coli Producing Extended-Spectrum Beta-Lactamase. Comp. Immunol. Microbiol. Infect. Dis. 2016, 45, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Allan, N.; Knotts, T.A.; Pesapane, R.; Ramsey, J.J.; Castle, S.; Clifford, D.; Foley, J. Conservation Implications of Shifting Gut Microbiomes in Captive-Reared Endangered Voles Intended for Reintroduction into the Wild. Microorganisms 2018, 6, 94. [Google Scholar] [CrossRef]
- Metcheva, R.; Beltcheva, M.; Chassovnikarova, T. The Snow Vole (Chionomys nivalis) as an Appropriate Environmental Bioindicator in Alpine Ecosystems. Sci. Total Environ. 2008, 391, 278–283. [Google Scholar] [CrossRef]
- Curtis, J.T.; Assefa, S.; Francis, A.; Köhler, G.A. Fecal Microbiota in the Female Prairie Vole (Microtus ochrogaster). PLoS ONE 2018, 13, e0190648. [Google Scholar] [CrossRef]
- Cabral, L.; Persinoti, G.F.; Paixão, D.A.A.; Martins, M.P.; Morais, M.A.B.; Chinaglia, M.; Domingues, M.N.; Sforca, M.L.; Pirolla, R.A.S.; Generoso, W.C.; et al. Gut Microbiome of the Largest Living Rodent Harbors Unprecedented Enzymatic Systems to Degrade Plant Polysaccharides. Nat. Commun. 2022, 13, 629. [Google Scholar] [CrossRef] [PubMed]
- Weldon, L.; Abolins, S.; Lenzi, L.; Bourne, C.; Riley, E.M.; Viney, M. The Gut Microbiota of Wild Mice. PLoS ONE 2015, 10, e0134643. [Google Scholar] [CrossRef]
- Johnson, R.B.; Peterson, D.A.; Tolbert, B.M. Cellulose Metabolism in the Rat. J. Nutr. 1960, 72, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Cano-Ramírez, C.; Santiago-Hernández, A.; Rivera-Orduña, F.N.; García-Huante, Y.; Zúñiga, G.; Hidalgo-Lara, M.E. Expression, Purification and Characterization of an Endoglucanase from Serratia proteamaculans CDBB-1961, Isolated from the Gut of Dendroctonus Adjunctus (Coleoptera: Scolytinae). AMB Express 2016, 6, 63. [Google Scholar] [CrossRef]
- Chantarasiri, A.; Boontanom, P.; Nuiplot, N. Isolation and Characterization of Lysinibacillus sphaericus BR2308 from Coastal Wetland in Thailand for the Biodegradation of Lignin. AACL Bioflux 2017, 10, 200–209. [Google Scholar]
- Anand, A.A.P.; Vennison, S.J.; Sankar, S.G.; Prabhu, D.I.G.; Vasan, P.T.; Raghuraman, T.; Geoffrey, C.J.; Vendan, S.E. Isolation and Characterization of Bacteria from the Gut of Bombyx Mori That Degrade Cellulose, Xylan, Pectin and Starch and Their Impact on Digestion. J. Insect Sci. 2010, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.; Ngo, T.-D.; Kumar, A.; Ayranci, C.; Tang, T. Cleaning Carbohydrate Impurities from Lignin Using Pseudomonas fluorescens. Green Chem. 2019, 21, 1648–1659. [Google Scholar] [CrossRef]
- Shil, R.K.; Mojumder, S.; Sadida, F.F.; Uddin, M.; Sikdar, D. Isolation and Identification of Cellulolytic Bacteria from the Gut of Three Phytophagus Insect Species. Braz. Arch. Biol. Technol. 2014, 57, 927–932. [Google Scholar] [CrossRef]
- Lin, L.; Kan, X.; Yan, H.; Wang, D. Characterization of Extracellular Cellulose-Degrading Enzymes from Bacillus thuringiensis Strains. Electron. J. Biotechnol. 2012, 15, 2. [Google Scholar] [CrossRef]
- Pang, J.; Liu, Z.-Y.; Hao, M.; Zhang, Y.-F.; Qi, Q.-S. An Isolated Cellulolytic Escherichia coli from Bovine Rumen Produces Ethanol and Hydrogen from Corn Straw. Biotechnol. Biofuels 2017, 10, 165. [Google Scholar] [CrossRef]
- Goswami, R.; Mukherjee, S.; Chakraborty, A.K.; Balachandran, S.; Sinha Babu, S.P.; Chaudhury, S. Optimization of Growth Determinants of a Potent Cellulolytic Bacterium Isolated from Lignocellulosic Biomass for Enhancing Biogas Production. Clean Technol. Environ. Policy 2016, 18, 1565–1583. [Google Scholar] [CrossRef]
- Opere, B.; Abiala, M.A.; Dosumu, A. Cellulolytic Soil Bacteria Exhibited Tolerance to Heavy Metals. Int. J. Microbiol. Res. 2018, 9, 7–15. [Google Scholar]
- Iwobi, A. Uncovering Novel Pathogenicity-Associated Loci among Yersinia enterocolitica Species by Subtractive Hybridization. Available online: https://edoc.ub.uni-muenchen.de/998/ (accessed on 2 August 2023).
- Li, J.; Yuan, X.; Desta, S.T.; Dong, Z.; Mugabe, W.; Shao, T. Characterization of Enterococcus faecalis JF85 and Enterococcus faecium Y83 Isolated from Tibetan Yak (Bos Grunniens) for Ensiling Pennisetum Sinese. Bioresour. Technol. 2018, 257, 76–83. [Google Scholar] [CrossRef]
- Nurliana, N.; Siregar, B.H.; Sari, W.E.; Helmi, T.Z.; Sugito, S. Identification of Cellulolytic Lactic Acid Bacteria from the Intestines of Laying Hens given AKBISprob Based on 16S Ribosomal Ribonucleic Acid Gene Analysis. Vet. World 2022, 15, 1650–1656. [Google Scholar] [CrossRef] [PubMed]
- Watts, P.C.; Mappes, T.; Tukalenko, E.; Mousseau, T.A.; Boratyński, Z.; Møller, A.P.; Lavrinienko, A. Interpretation of Gut Microbiota Data in the ‘Eye of the Beholder’: A Commentary and Re-Evaluation of Data from ‘Impacts of Radiation Exposure on the Bacterial and Fungal Microbiome of Small Mammals in the Chernobyl Exclusion Zone’. J. Anim. Ecol. 2022, 91, 1535–1545. [Google Scholar] [CrossRef]
- DNA Barcoding Identifies the Plants a Person Has Eaten: Reliable Technique Should Improve Clinical Trials, Nutrition Studies and Historical Research—ScienceDaily. Available online: https://www.sciencedaily.com/releases/2023/06/230627141950.htm (accessed on 23 August 2023).
- Snider, A.M.; Bonisoli-Alquati, A.; Pérez-Umphrey, A.A.; Stouffer, P.C.; Taylor, S.S. Metabarcoding of Stomach Contents and Fecal Samples Provide Similar Insights about Seaside Sparrow Diet. Ornithol. Appl. 2022, 124, duab060. [Google Scholar] [CrossRef]
- Gladnishka, T.; Christova, I.; Kantardjiev, T. First PCR Detection of Francisella tularensis in Reservoir Animals from Endemic Area in Bulgaria. Probl. Infect. Parasit. Dis. 2004, 32, 33–34. [Google Scholar]
- Nikolova, S.; Tzvetkov, Y.; Najdenski, H.; Vesselinova, A. Isolation of Pathogenic Yersiniae from Wild Animals in Bulgaria. J. Vet. Med. B Infect. Dis. Vet. Public Health 2001, 48, 203–209. [Google Scholar] [CrossRef]
- Maurice, C.F.; CLKnowles, S.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked Seasonal Variation in the Wild Mouse Gut Microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef]
- Knowles, S.C.L.; Eccles, R.M.; Baltrūnaitė, L. Species Identity Dominates over Environment in Shaping the Microbiota of Small Mammals. Ecol. Lett. 2019, 22, 826–837. [Google Scholar] [CrossRef]
- Kunicki-Goldfinger, W.; Kunicka-Goldfinger, W. Intestinal microflora of Sorex araneus araneus L. and Clethrionomys glareolus glareolus Schreb. in natural conditions. I. Quantitative and qualitative characteristics of the intestinal microflora. Acta Microbiol. Pol. 1962, 11, 43–75. [Google Scholar]
- Kunicki-Goldfinger, W.; Kunicka-Goldfinger, W. Intestinal microflora of Sorex araneus araneus L. and Clethrionomys glareolus glareolus Schreb. in natural conditions. II. General characteristics of separate strains. Acta Microbiol. Pol. 1962, 11, 77–91. [Google Scholar]
- Kunicki-Goldfinger, W.; Kunicka-Goldfinger, W. Intestinal microflora of Sorex araneus araneus L. and Clethrionomys glareolus glareolus Schreb. in nature conditions. III. Seasonal variations. Acta Microbiol. Pol. 1962, 11, 93–110. [Google Scholar]
- Zaneveld, J.R.; McMinds, R.; Vega Thurber, R. Stress and Stability: Applying the Anna Karenina Principle to Animal Microbiomes. Nat. Microbiol. 2017, 2, 17121. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in Tropical Rain Forests and Coral Reefs. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.E.; Janecko, N.; Pearl, D.L.; Boerlin, P.; Reid-Smith, R.J.; Jardine, C.M. Comparison of Escherichia coli Recovery and Antimicrobial Resistance in Cecal, Colon, and Fecal Samples Collected from Wild House Mice (Mus Musculus). J. Wildl. Dis. 2013, 49, 432–436. [Google Scholar] [CrossRef][Green Version]
- Mounier, J.; Monnet, C.; Vallaeys, T.; Arditi, R.; Sarthou, A.-S.; Hélias, A.; Irlinger, F. Microbial Interactions within a Cheese Microbial Community. Appl. Environ. Microbiol. 2008, 74, 172–181. [Google Scholar] [CrossRef]
- Hardyhall Bt Corn: Is It Worth the Risk? SCQ. Available online: https://www.scq.ubc.ca/bt-corn-is-it-worth-the-risk/ (accessed on 23 August 2023).
- Ohba, M.; Mizuki, E.; Uemori, A. Parasporin, a New Anticancer Protein Group from Bacillus thuringiensis. Anticancer Res. 2009, 29, 427–433. [Google Scholar]
- Berry, C. The Bacterium, Lysinibacillus sphaericus, as an Insect Pathogen. J. Invertebr. Pathol. 2012, 109, 1–10. [Google Scholar] [CrossRef]
- Santana-Martinez, J.C.; Silva, J.J.; Dussan, J. Efficacy of Lysinibacillus sphaericus against Mixed-Cultures of Field-Collected and Laboratory Larvae of Aedes aegypti and Culex quinquefasciatus. Bull. Entomol. Res. 2019, 109, 111–118. [Google Scholar] [CrossRef]
- Lozano, L.C.; Dussán, J. Metal Tolerance and Larvicidal Activity of Lysinibacillus sphaericus. World J. Microbiol. Biotechnol. 2013, 29, 1383–1389. [Google Scholar] [CrossRef]
- Selenska-Pobell, S.; Panak, P.; Miteva, V.; Boudakov, I.; Bernhard, G.; Nitsche, H. Selective Accumulation of Heavy Metals by Three Indigenous Bacillus Strains, B. cereus, B. megaterium and B. sphaericus, from Drain Waters of a Uranium Waste Pile. FEMS Microbiol. Ecol. 1999, 29, 59–67. [Google Scholar] [CrossRef]
- Horizontal arsC Gene Transfer among Microorganisms Isolated from Arsenic Polluted Soil. Available online: https://repository.urosario.edu.co/items/c52e482f-470c-4ff8-b8f3-468a60aeaac2 (accessed on 23 August 2023).
- AlHoufie, S.T.S.; Foster, H.A. Effects of Sub-Lethal Concentrations of Mupirocin on Global Transcription in Staphylococcus Aureus 8325-4 and a Model for the Escape from Inhibition. J. Med. Microbiol. 2016, 65, 858–866. [Google Scholar] [CrossRef]
- Al-Qaysi, S.A.S.; Al-Haideri, H.; Al-Shimmary, S.M.; Abdulhameed, J.M.; Alajrawy, O.I.; Al-Halbosiy, M.M.; Moussa, T.A.A.; Farahat, M.G. Bioactive Levan-Type Exopolysaccharide Produced by Pantoea agglomerans ZMR7: Characterization and Optimization for Enhanced Production. J. Microbiol. Biotechnol. 2021, 31, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Smits, T.H.; Montesinos, E.; Frey, J.E.; Duffy, B. Genotypic Comparison of Pantoea agglomerans plant and Clinical Strains. BMC Microbiol. 2009, 9, 204. [Google Scholar] [CrossRef]
- Guinat, C.; Gogin, A.; Blome, S.; Keil, G.; Pollin, R.; Pfeiffer, D.U.; Dixon, L. Transmission Routes of African Swine Fever Virus to Domestic Pigs: Current Knowledge and Future Research Directions. Vet. Rec. 2016, 178, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Baldacchino, F.; Muenworn, V.; Desquesnes, M.; Desoli, F.; Charoenviriyaphap, T.; Duvallet, G. Transmission of Pathogens by Stomoxys Flies (Diptera, Muscidae): A Review. Parasite Paris Fr. 2013, 20, 26. [Google Scholar] [CrossRef]
- Frant, M.P.; Gal-Cisoń, A.; Bocian, Ł.; Ziętek-Barszcz, A.; Niemczuk, K.; Szczotka-Bochniarz, A. African Swine Fever (ASF) Trend Analysis in Wild Boar in Poland (2014–2020). Animals 2022, 12, 1170. [Google Scholar] [CrossRef]
- Bonnet, S.I.; Bouhsira, E.; De Regge, N.; Fite, J.; Etoré, F.; Garigliany, M.-M.; Jori, F.; Lempereur, L.; Le Potier, M.-F.; Quillery, E.; et al. Putative Role of Arthropod Vectors in African Swine Fever Virus Transmission in Relation to Their Bio-Ecological Properties. Viruses 2020, 12, 778. [Google Scholar] [CrossRef]
- Lv, T.; Xie, X.; Song, N.; Zhang, S.; Ding, Y.; Liu, K.; Diao, L.; Chen, X.; Jiang, S.; Li, T.; et al. Expounding the Role of Tick in Africa Swine Fever Virus Transmission and Seeking Effective Prevention Measures: A Review. Front. Immunol. 2022, 13, 1093599. [Google Scholar] [CrossRef]
- Sanchez Botija, C.; Badiola, C. Presencie of the African swine pest virus in Haematopinus suis. Bull.-Off. Int. Epizoot. 1966, 66, 699–705. [Google Scholar]
- Mellor, P.S.; Kitching, R.P.; Wilkinson, P.J. Mechanical Transmission of Capripox Virus and African Swine Fever Virus by Stomoxys calcitrans. Res. Vet. Sci. 1987, 43, 109–112. [Google Scholar] [CrossRef]
- Vergne, T.; Andraud, M.; Bonnet, S.; De Regge, N.; Desquesnes, M.; Fite, J.; Etore, F.; Garigliany, M.-M.; Jori, F.; Lempereur, L.; et al. Mechanical Transmission of African Swine Fever Virus by Stomoxys calcitrans: Insights from a Mechanistic Model. Transbound. Emerg. Dis. 2021, 68, 1541–1549. [Google Scholar] [CrossRef]
- European Commission, Mission of the Community Veterinary Emergency Team to Lithuania. Available online: http://ec.europa.eu/food/animals/docs/reg-com_ahw_20140821_pres_asf_lithuania_cvet.pdf (accessed on 8 March 2016).
- Hakobyan, S.A.; Ross, P.A.; Bayramyan, N.V.; Poghosyan, A.A.; Avetisyan, A.S.; Avagyan, H.R.; Hakobyan, L.H.; Abroyan, L.O.; Harutyunova, L.J.; Karalyan, Z.A. Experimental Models of Ecological Niches for African Swine Fever Virus. Vet. Microbiol. 2022, 266, 109365. [Google Scholar] [CrossRef]
- Hess, W.R.; Endris, R.G.; Haslett, T.M.; Monahan, M.J.; McCoy, J.P. Potential Arthropod Vectors of African Swine Fever Virus in North America and the Caribbean Basin. Vet. Parasitol. 1987, 26, 145–155. [Google Scholar] [CrossRef]
- Yoon, H.; Hong, S.-K.; Lee, I.; Choi, D.-S.; Lee, J.-H.; Lee, E.; Wee, S.-H. Arthropods as Potential Vectors of African Swine Fever Virus Outbreaks in Pig Farms in the Republic of Korea. Vet. Med. Sci. 2021, 7, 1841–1844. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H.; Amendt, J.; Blome, S.; Depner, K.; Kampen, H. Evaluation of Blowfly Larvae (Diptera: Calliphoridae) as Possible Reservoirs and Mechanical Vectors of African Swine Fever Virus. Transbound. Emerg. Dis. 2018, 65, e210–e213. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Depner, K.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Schmidt, C.; et al. Research Priorities to Fill Knowledge Gaps in the Control of African Swine Fever: Possible Transmission of African Swine Fever Virus by Vectors. EFSA J. Eur. Food Saf. Auth. 2021, 19, e06676. [Google Scholar] [CrossRef]
- Hess, W.R. African Swine Fever Virus. Virol. Monogr. Virusforsch. Einzeldarst. 1971, 9, 1–33. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Qi, W.; Yang, Y.; Liu, Z.; An, T.; Wu, X.; Chen, J. Prevention and Control Strategies of African Swine Fever and Progress on Pig Farm Repopulation in China. Viruses 2021, 13, 2552. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.F.; Schade-Weskott, M.L.; Harris, H.J.; Heath, L.; Kriel, G.J.P.; de Klerk-Lorist, L.-M.; van Schalkwyk, L.; Buss, P.; Trujillo, J.D.; Crafford, J.E.; et al. Extension of Sylvatic Circulation of African Swine Fever Virus in Extralimital Warthogs in South Africa. Front. Vet. Sci. 2021, 8, 746129. [Google Scholar] [CrossRef] [PubMed]
- Leite Velho, E. Observations Sur La Peste Porcine En Angola. Bull.-Off. Int. Épizoot. 1956, 46, 335–340. [Google Scholar]
- Mendes, A.M. The Lapinization of the Virus of African Swine Fever. Bull.-Off. Int. Épizoot. 1962, 58, 699–705. [Google Scholar]
- Sanchez Botija, C. Estudios Sobre La Peste Porcina Africana En Espana. Bull.-Off. Int. Épizoot. 1962, 58, 707–727. [Google Scholar]
- Eustace Montgomery, R. On A Form of Swine Fever Occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- SHIC Grant-Funded Study in Vietnam Indicates Rodents May Be Low Risk ASF Vector. Swine Health Information Center. Available online: https://www.swinehealth.org/shic-grant-funded-study-in-vietnam-indicates-rodents-may-be-low-risk-asf-vector/ (accessed on 8 August 2023).
- Scientific Opinion on African Swine Fever. EFSA. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/3628 (accessed on 9 August 2023).
- Available online: https://www.dw.com/bg/%D0%BA%D0%BE%D0%B9-%D0%B5-%D0%B2%D0%B8%D0%BD%D0%BE%D0%B2%D0%B5%D0%BD-%D0%B7%D0%B0-%D1%81%D0%B2%D0%B8%D0%BD%D1%81%D0%BA%D0%B0%D1%82%D0%B0-%D0%BA%D0%B0%D1%81%D0%B0%D0%BF%D0%BD%D0%B8%D1%86%D0%B0-%D0%B2-%D0%B1%D1%8A%D0%BB%D0%B3%D0%B0%D1%80%D0%B8%D1%8F/a-49860730 (accessed on 23 August 2023).
- Barash, I.; Manulis-Sasson, S. Recent Evolution of Bacterial Pathogens: The Gall-Forming Pantoea agglomerans Case. Annu. Rev. Phytopathol. 2009, 47, 133–152. [Google Scholar] [CrossRef]
- Wang, J.; Chen, T.; Xue, L.; Wei, X.; White, J.F.; Qin, Z.; Li, C. A New Bacterial Leaf Blight Disease of Oat (Avena sativa) Caused by Pantoea agglomerans in China. Plant Pathol. 2022, 71, 470–478. [Google Scholar] [CrossRef]

| Primers | Sequences | Tm | Amplicon | References |
|---|---|---|---|---|
| Escherichia coli uidA F | 5′-AAA ACG GCA AGA AAA AGC AG-3′ | 55 °C | 147 bp 1 | [30] |
| Escherichia coli uidA R | 5′-ACG CGT GGT TAC AGT CTT GCG-3′ | |||
| E. coli yccT F | 5’-GCA TCG TGA CCA CCT TGA-3’ | 56 °C | 59 bp | [31] |
| E. coli yccT R | 5’-CAG CGT GGT GGC AAA A-3’ | |||
| Yersinia enterocolitica 16S rRNA F (YeI-16SrRNA) | 5′-ATA CCG CAT AAC GTC TTC G-3′ | 47 °C | 330 bp | [32] |
| Yersinia enterocolitica 16S rRNA R (YeII-16SrRNA) | 5′-TTC TTC TGC GAG TAA CGT C-3′ | |||
| Y. enterocolitica ail F (real10A) | 5′–ATG ATA ACT GGG GAG TAA TAG GTT CG-3′ | 55 °C | 163 bp | [33] |
| Y. enterocolitica ail R (real9A) | 5′-CCC AGT AAT CCA TAA AGG CTA ACA TAT-3′ | |||
| ASFV VP72 F | 5′-ACCACAAGATCAGCCGTAGTG-3′ | 60 °C | 420 bp | Designed for this study |
| ASFV VP72 R | 5′-AGATTGGCACAAGTTCGGACA-3′ |
| Animal Sample | Isolate | Species |
|---|---|---|
| I ♀ Ap. flavicollis 1 | 1, 2, 3, 4 | E. coli3 |
| II ♂ Ap. flavicollis | 8, 11 | E. coli3 |
| 5, 6, 7, 9, 10, 12 | Hafnia alvei | |
| III ♀ Ap. flavicollis | 13, 14, 15, 16, 17 | Y. enterocolitica3 |
| IV ♂ Ap. flavicollis | 18, 20, 21, 22 (90%), 23, 24, 25 | E. coli3 |
| 19, 53 | Yersinia kristensenii | |
| V ♂ M. glareolus 2 | 26 | Lysinibacillus sphaericus |
| 27, 28, 31 | Serratia liquefaciens | |
| 29 (95%), 32 (95%) | Pseudomonas fluorescens group | |
| 30 | Serratia proteamaculans | |
| VI ♂ Ap. flavicollis | 33 (94%) | Enterococcus hirae |
| 34, 35 (90%), 36 | Enterococcus faecalis | |
| VII ♂ M. glareolus | 38 | Enterococcus faecalis |
| 39 | Lysinibacillus sphaericus | |
| 37 (96%), 40 (92%) | Bacillus thuringiensis | |
| VIII ♂ Ap. flavicollis | 41 | Enterococcus faecium |
| 42, 43, 44, 45 | Pantoea agglomerans | |
| IX ♂ Ap. flavicollis | 47, 48 (96%), 50 (95%) | Serratia marcescens |
| 46 (97%) | Pseudescherichia vulneris | |
| 49, 52 | Klebsiella pneumoniae ssp. ozaenae | |
| 51 | Enterobacter cloacea |
| Bacteria | E. coli | H. alvei | Y. enterocolitica | Y. kristensenii | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic Class | Antimicrobial (Antibiotic)/Isolate | 1 | 2 | 3 | 4 | 8 | 11 | 18 | 20 | 21 | 22 | 23 | 24 | 25 | 5 | 6 | 7 | 9 | 10 | 12 | 13 | 14 | 15 | 16 | 17 | 19 | 53 |
| Penicillins | Amoxicillin-Clavulanate | S | S | S | S | S | S | S | S | S | S | S | S | S | R | R | R | R | R | R | R | R | R | R | R | R | R |
| Ampicillin | S | S | S | S | S | S | S | S | S | S | S | S | S | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| Piperacillin-Tazobactam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S | |
| Cephalosporins | Cefazolin | I | I | I | I | I | I | I | I | I | I | I | I | I | R | R | R | R | R | R | R | R | R | R | R | R | >4 |
| Cefotaxime | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Ceftazidime | S | S | S | S | S | S | S | S | S | S | S | S | S | I | I | I | I | I | I | S | S | S | S | I | R | I | |
| Cefuroxime | I | I | I | I | I | I | I | I | I | I | I | I | I | 8 | 8 | >8 | 8 | >8 | >8 | 8 | 8 | 8 | 8 | 8 | R | 8 | |
| Cephalexin | S | S | S | X | S | X | S | X | X | X | S | X | X | X | X | X | X | X | X | R | R | R | R | R | R | X | |
| Carbapenems | Ertapenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | n.t. | n.t. | n.t. | n.t. | n.t. | R | R |
| Imipenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | |
| Meropenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Monobactams | Aztreonam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Fluoroquinolones | Ciprofloxacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Aminoglycosides | Amikacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Gentamicin | S | S | S | S | S | S | S | R | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Tobramycin | S | S | S | S | S | S | S | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Polymyxins | Colistin | X | X | X | X | X | X | X | X | X | X | X | X | X | R | R | R | R | R | R | X | X | X | X | X | X | X |
| Other | Nitrofurantoin | S | S | S | X | S | X | S | X | X | X | S | X | X | X | X | X | X | X | X | 32 | X | 32 | X | 64 | 32 | X |
| Trimethoprim | S | S | S | X | S | X | S | X | X | X | S | X | X | X | X | X | X | X | X | S | X | S | X | S | S | X | |
| Trimethoprim-Sulfamethoxazole | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Fosfomycin w/G6P | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Bacteria | S. liquefaciens | S. proteamaculans | S. marcescens | P. agglomerans | P. vulneris | K. pneumoniae ssp. ozaenae | E. cloacae | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic Class | Antimicrobial/Isolate | 27 | 28 | 31 | 30 | 47 | 48 | 50 | 42 | 43 | 44 | 45 | 46 | 49 | 52 | 51 |
| Penicillins | Amoxicillin-Clavulanate | S | S | S | R | R | R | S | S | S | S | S | S | S | S | R |
| Ampicillin | S | S | S | R | R | R | S | S | S | R | S | R | R | R | R | |
| Piperacillin-Tazobactam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | ||
| Cephalosporins | Cefazolin | >4 | >4 | >4 | R | R | R | >4 | >4 | >4 | >4 | >4 | 2 | I | I | R |
| Cefotaxime | S | S | S | S | S | S | S | S | I | S | I | S | S | S | S | |
| Ceftazidime | S | S | S | S | S | S | S | S | I | S | S | S | S | S | S | |
| Cefuroxime | >8 | >8 | >8 | R | R | R | >8 | >8 | >8 | >8 | >8 | 2 | I | I | 8 | |
| Cephalexin | X | X | X | X | R | R | X | X | X | X | X | S | X | X | X | |
| Carbapenems | Ertapenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Imipenem | S | S | S | S | S | S | I | S | S | S | S | S | S | S | S | |
| Meropenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Monobactams | Aztreonam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Fluoroquinolones | Ciprofloxacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Aminoglycosides | Amikacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Gentamicin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Tobramycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Polymyxins | Colistin | X | X | X | R | R | R | R | X | X | X | X | X | X | X | X |
| Other | Nitrofurantoin | X | X | X | R | R | R | X | X | X | X | X | 64 | X | X | X |
| Trimethoprim | X | X | X | X | S | S | X | X | X | X | X | S | X | X | X | |
| Trimethoprim-Sulfamethoxazole | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Fosfomycin w/G6P | R | R | R | S | S | S | S | S | S | S | S | R | R | R | R | |
| Bacteria | E. hirae | E. faecalis | E. faecium | ||||
|---|---|---|---|---|---|---|---|
| Antibiotic Class | Antimicrobial (Antibiotic)/Isolate | 33 | 34 | 35 | 36 | 38 | 41 |
| Penicillins | Ampicillin | S | S | S | S | S | S |
| Oxacillin | >2 | >2 | >2 | >2 | >2 | >2 | |
| Penicillin G | n.t. | >0.25 | >0.25 | ≤0.25 | ≤0.25 | ≤0.25 | |
| Cephalosporins | Cefoxitin | >8 | R | R | R | R | R |
| Ceftaroline | 1 | R | R | R | R | R | |
| Carbapenems | Imipenem | I | I | I | I | I | I |
| Fluoroquinolones | Ciprofloxacin | n.t. | S | S | X | X | R |
| Fluoroquinolones | Moxifloxacin | 0.5 | 1 | ≤0.25 | 0.5 | 1 | 1 |
| Aminoglycosides | Gentamicin | >4 | R | R | R | R | R |
| Gentamicin-Syn | S | S | S | S | S | S | |
| Tetracyclines | Tetracycline | ≤0.5 | 1 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Tigecycline | n.t | S | S | S | S | n.t | |
| Macrolides | Clindamycin | >1 | R | R | R | R | ≤0.25 |
| Erythromycin | ≤0.25 | R | R | R | R | R | |
| Glycopeptides | Teicoplanin | S | S | S | S | S | S |
| Vancomycin | S | S | S | S | S | S | |
| Oxazolidinones | Linezolid | S | S | S | S | S | S |
| Other | Daptomycin | 4 | 2 | 4 | 2 | 4 | 2 |
| Fosfomycin w/G6P | 32 | >64 | 32 | 64 | 64 | >64 | |
| Fusidic acid | 8 | R | R | R | R | R | |
| Mupirocin High level | ≤256 | ≤256 | ≤256 | ≤256 | ≤256 | ≤256 | |
| Nitrofurantoin | 32 | S | S | X | X | 32 | |
| Trimethoprim-Sulfamethoxazole | R | R | R | R | R | R | |
| Bacteria | P. fluorescens Group | ||
|---|---|---|---|
| Antibiotic Class | Antibiotic/Isolate | 29 | 32 |
| Penicillins | Ticarcillin | R | R |
| Cephalosporins | Ceftazidime | I | I |
| Carbapenems | Meropenem | I | R |
| Monobactams | Aztreonam | R | R |
| Fluoroquinolones | Ciprofloxacin | I | S |
| Aminoglycosides | Tobramycin | S | S |
| Sample | Target Type | DNA Copies/µL | DNA Copies/20 µL (Reaction) | Average of DNA Copies/20 µL (Reaction) |
|---|---|---|---|---|
| 102 | Positive control 1 | 12.4 | 248 | 2.21 × 102 |
| 102 | 9.7 | 194 | ||
| 101 | Positive control 2 | 1.6 | 32 | 2.8 × 101 |
| 101 | 1.2 | 24 | ||
| 1 | Unknown 1 | 0 | 0 | 0 |
| 1 | 0 | 0 | ||
| 2 | Unknown 2 | 0 | 0 | 0 |
| 2 | 0 | 0 | ||
| 3 | Unknown 3 | 0 | 0 | 0 |
| 3 | 0 | 0 | ||
| NC | Negative control (nuclease-free water) | 0 | 0 | 0 |
| NC | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilieva, Y.; Zaharieva, M.M.; Dimitrova, L.; Kaleva, M.D.; Jordanova, J.; Dimitrova, M.; Beltcheva, M.; Aleksieva, I.; Georgiev, Y.; Manasiev, Y.; et al. Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria. Microbiol. Res. 2023, 14, 1788-1819. https://doi.org/10.3390/microbiolres14040123
Ilieva Y, Zaharieva MM, Dimitrova L, Kaleva MD, Jordanova J, Dimitrova M, Beltcheva M, Aleksieva I, Georgiev Y, Manasiev Y, et al. Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria. Microbiology Research. 2023; 14(4):1788-1819. https://doi.org/10.3390/microbiolres14040123
Chicago/Turabian StyleIlieva, Yana, Maya Margaritova Zaharieva, Lyudmila Dimitrova, Mila D. Kaleva, Joanna Jordanova, Maya Dimitrova, Michaela Beltcheva, Iliana Aleksieva, Yordan Georgiev, Yordan Manasiev, and et al. 2023. "Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria" Microbiology Research 14, no. 4: 1788-1819. https://doi.org/10.3390/microbiolres14040123
APA StyleIlieva, Y., Zaharieva, M. M., Dimitrova, L., Kaleva, M. D., Jordanova, J., Dimitrova, M., Beltcheva, M., Aleksieva, I., Georgiev, Y., Manasiev, Y., & Najdenski, H. (2023). Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria. Microbiology Research, 14(4), 1788-1819. https://doi.org/10.3390/microbiolres14040123










