Gene Polymorphisms LEP, LEPR, 5HT2A, GHRL, NPY, and FTO-Obesity Biomarkers in Metabolic Risk Assessment: A Retrospective Pilot Study in Overweight and Obese Population in Romania
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Genotyping
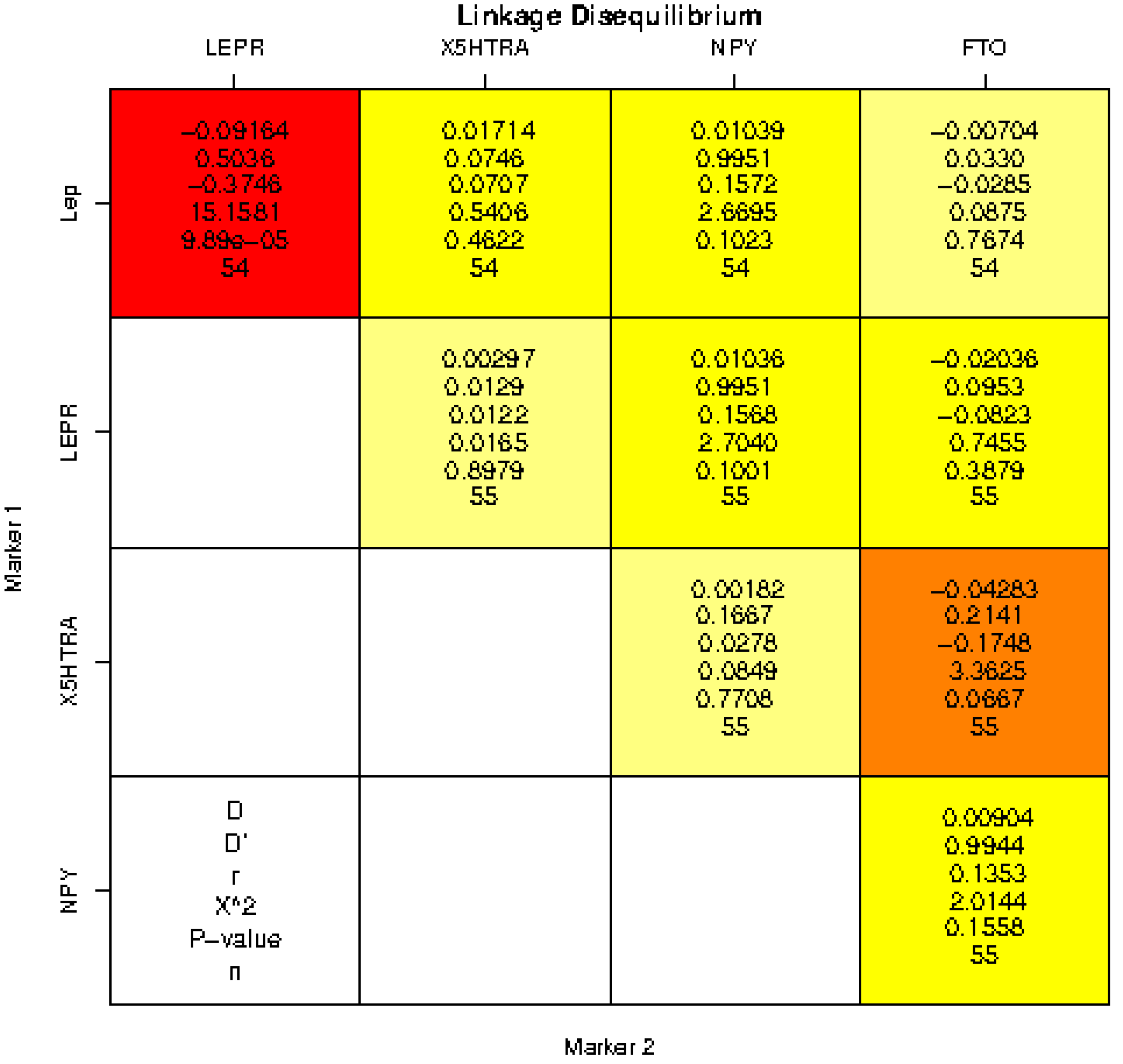
2.2. Statistical Analysis
3. Results
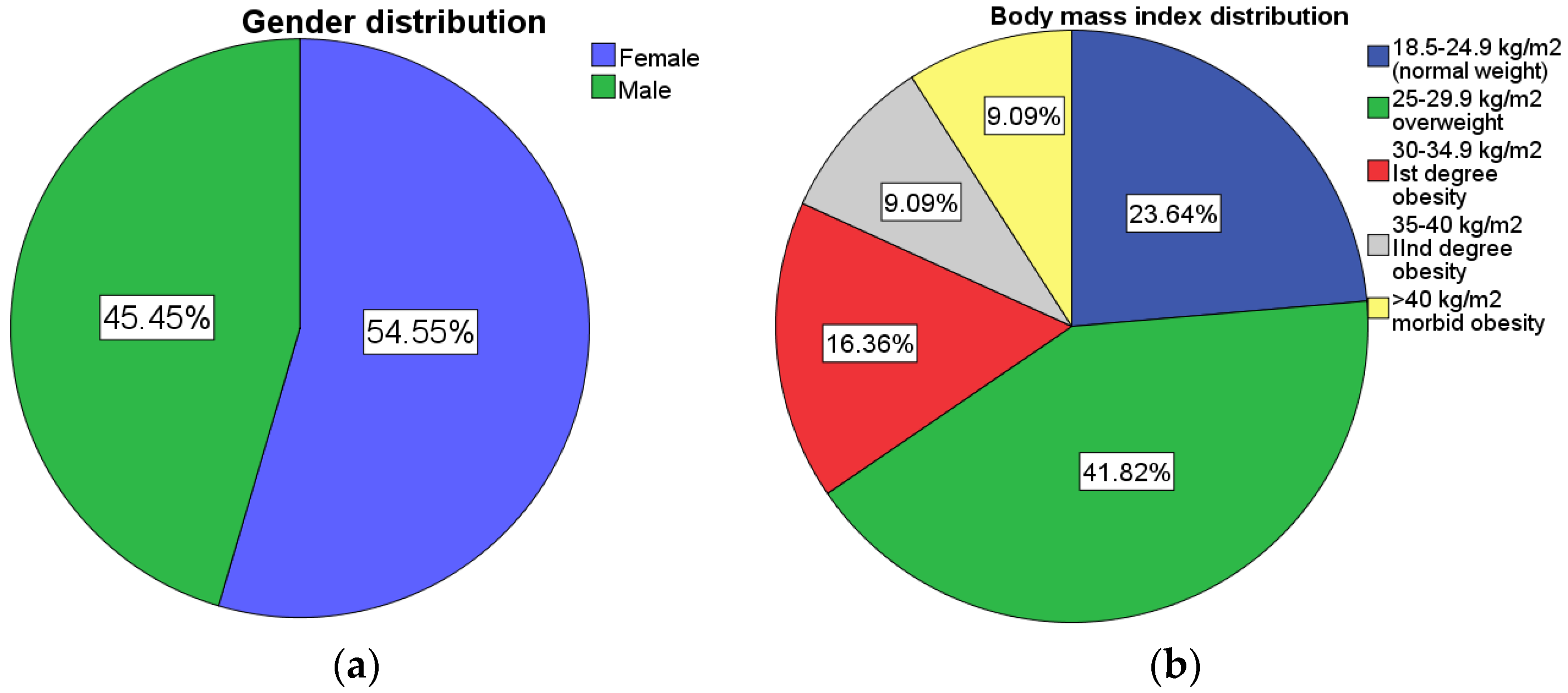
3.1. Socio-Demographic Data
3.2. LEP Gene
3.3. Leptin Receptor Gene
3.4. 5HTR2A Gene
3.5. GHRL Gene
3.6. NPY Gene
3.7. FTO Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pepe, R.B.; Lottenberg, A.M.; Fujiwara, C.T.; Beyruti, M.; Cintra, D.E.; Machado, R.M.; Rodrigues, A.; Jensen, N.S.; Caldas, A.P.; Fernandes, A.E.; et al. Position statement on nutrition therapy for overweight and obesity: Nutrition department of the Brazilian association for the study of obesity and metabolic syndrome (ABESO—2022). Diabetol. Metab. Syndr. 2023, 15, 1–53. [Google Scholar]
- Nordang, G.B.N.; Busk, Ø.L.; Tveten, K.; Hanevik, H.I.; Fell, A.K.M.; Hjelmesæth, J.; Holla, Ø.L.; Hertel, J.K. Next-generation sequencing of the monogenic obesity genes LEP, LEPR, MC4R, PCSK1 and POMC in a Norwegian cohort of patients with morbid obesity and normal weight controls. Mol. Genet. Metab. 2017, 121, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Cuda, S.; Censani, M.; Kharofa, R.; Williams, D.R.; O’Hara, V.; Karjoo, S.; Paisley, J.; Browne, N.T. Social consequences and genetics for the child with overweight and obesity: An obesity medicine association (OMA) clinical practice statement 2022. Obes. Pillars. 2022, 3, 100032. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.; Mora, S. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274. [Google Scholar] [PubMed]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Lindström, J. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Aruoma, O.I.; Hausman-Cohen, S.; Pizano, J.; Schmidt, M.A.; Minich, D.M.; Joffe, Y.; Brady, D.M. Personalized nutrition: Translating the science of nutrigenomics into practice: Proceedings from the 2018 American College of Nutrition Meeting. J. Am. Coll. Nutr. 2019, 38, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Sanderson, E.; Elsworth, B.; Tilling, K.; Smith, G.D. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: Mendelian randomisation study. BMJ 2020, 369, m1203. [Google Scholar] [CrossRef]
- Coc, L.M.C.; Lacatusu, I.; Badea, N.; Penes, O.; Cobelschi, C.P.; Pop, A.; Meghea, A. Curcumin co-loaded with a lipid mediator in the same nanostructured lipid delivery system. Farmacia 2022, 70, 932–943. [Google Scholar] [CrossRef]
- Bhattacharya, T.; Dutta, S.; Akter, R.; Rahman, M.H.; Karthika, C.; Nagaswarupa, H.P.; Murthy, H.C.A.; Fratila, O.; Brata, R.; Bungau, S. Role of Phytonutrients in Nutrigenetics and Nutrigenomics Perspective in Curing Breast Cancer. Biomolecules 2021, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.W.; Barclay, L.J.; Burrage, L.C. Inherited Metabolic Disorders: Aspects of Chronic Nutrition Management. Nutr. Clin. Pract. 2015, 30, 502–510. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jamka, M.; Kaczmarek, N.; Mądry, E.; Krzyżanowska-Jankowska, P.; Bajerska, J.; Kręgielska-Narożna, M.; Bogdański, P.; Walkowiak, J. Metabolic Health in Obese Subjects-Is There a Link to Lactoferrin and Lactoferrin Receptor-Related Gene Polymorphisms? Nutrients 2020, 12, 2843. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liao, C.; Glodowski, C.R.; Fan, C.; Liu, J.; Mott, K.R.; Kaushik, A.; Vu, H.; Locasale, J.W.; McBrayer, S.K.; DeBerardinis, R.J.; et al. Integrated Metabolic Profiling and Transcriptional Analysis Reveals Therapeutic Modalities for Targeting Rapidly Proliferating Breast Cancers. Cancer Res. 2022, 82, 665–680. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lewis, R. Genome-wide association studies. In Human Genetics: Concepts and Application, 11th ed.; Lewis, R., Ed.; Series “WCB Cell & Molecular Biology”; McGraw–Hill Education: New York, NY, USA, 2015; pp. 139–142. [Google Scholar]
- Peneş, N.O.; Weber, B.; Păun, S.D. Role of genetic polymorphism in nutritional supplementation therapy in personalized medicine. Rom. J. Morphol. Embryol. 2017, 58, 53–58. [Google Scholar] [PubMed]
- Barreiro, L.B.; Laval, G.; Quach, H.; Patin, E.; Quintana-Murci, L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008, 40, 340–345. [Google Scholar] [CrossRef]
- Lin, J.; Xie, Z.; Lan, B.; Guo, Z.; Tang, W.F.; Liu, C.; Zhang, S.; Chen, G.; Guo, F.; Chen, Y. Investigation of Leptin and its receptor (LEPR) for single nucleotide polymorphisms in colorectal cancer: A case-control study involving 2306 subjects. Am. J. Transl. Res. 2020, 12, 3613–3628. [Google Scholar] [PubMed] [PubMed Central]
- Ruiz-Castell, M.; Le Coroller, G.; Landrier, J.-F.; Kerkour, D.; Weber, B.; Fagherazzi, G.; Appenzeller, B.M.R.; Vaillant, M.; Bohn, T. Micronutrients and Markers of Oxidative Stress and Inflammation Related to Cardiometabolic Health: Results from the EHES-LUX Study. Nutrients 2021, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Alfaqih, M.A.; Elsalem, L.; Nusier, M.; Mhedat, K.; Khader, Y.; Ababneh, E. Serum Leptin Receptor and the rs1137101 Variant of the LEPR Gene Are Associated with Bladder Cancer. Biomolecules 2023, 13, 1498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mazahreh, T.S.; Alfaqih, M.; Saadeh, R.; Al-Zoubi, N.A.; Hatamleh, M.; Alqudah, A.; Aleshawi, A.J.; Alzoubi, A. The Effects of Laparoscopic Sleeve Gastrectomy on the Parameters of Leptin Resistance in Obesity. Biomolecules 2019, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Battaglia, D.M.; Foster, T.P.; Nichols, C.D. Serotonin 5-HT2A receptor activity mediates adipocyte differentiation through control of adipogenic gene expression. Sci. Rep. 2021, 11, 19714. [Google Scholar] [CrossRef]
- Halder, I.; Muldoon, M.F.; Ferrell, R.E.; Manuck, S.B. Serotonin receptor 2A (HTR2A) gene polymorphisms are associated with blood pressure, central adiposity, and the metabolic syndrome. Metab. Syndr. Relat. Disord. 2007, 5, 323–330. [Google Scholar] [CrossRef]
- Genis-Mendoza, A.D.; Ruiz-Ramos, D.; López-Narvaez, M.L.; Tovilla-Zárate, C.A.; Rosa Garcia, A.; Cortes Meda, G.; Nicolini, H. Genetic association analysis of 5-HTR2A gene variants in eating disorders in a Mexican population. Brain Behav. 2019, 9, e01286. [Google Scholar] [CrossRef] [PubMed]
- Al-Nbaheen, M.S. Relationship between single nucleotide polymorphism studies in ghrelin gene with obesity subjects. J. King Saud Univ. -Sci. 2023, 35, 102393. [Google Scholar] [CrossRef]
- Human Assembly GRCh37. Available online: https://grch37.ensembl.org/Homo_sapiens/Gene/Ontologies/biological_process?db=core;g=ENSG00000122585;r=7:24323782-24331484 (accessed on 20 November 2023).
- Pesonen, U. NPY L7P polymorphism and metabolic diseases. Regul. Pept. 2008, 149, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.H.; Zhang, C.; Chen, J.; Bowers, K.; Hu, F.B.; Kang, G.; Qi, L. Polymorphisms in the neuropeptide Y gene and the risk of obesity: Findings from two prospective cohorts. J. Clin. Endocrinol. Metab. 2011, 96, E2055-62. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Katus, U.; Villa, I.; Ringmets, I.; Veidebaum, T.; Harro, J. Neuropeptide Y gene variants in obesity, dietary intake, blood pressure, lipid and glucose metabolism: A longitudinal birth cohort study. Peptides 2021, 139, 170524. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.; Ausmeier, K.; Dildrop, R.; Rüther, U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mamm. Genome. 2002, 13, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.-J.; Zhu, H.; He, H.; Wu, K.-H.; Li, J.; Chen, X.-D.; Zhang, J.-G.; Shen, H.; Tian, Q.; Krousel-Wood, M.; et al. Replication of 6 obesity genes in a meta-analysis of genome-wide association studies from diverse ancestries. PLoS ONE 2014, 9, e96149. [Google Scholar] [CrossRef] [PubMed]
- Dastgheib, S.A.; Bahrami, R.; Setayesh, S.; Salari, S.; Mirjalili, S.R.; Noorishadkam, M.; Sadeghizadeh-Yazdi, J.; Akbarian, E.; Neamatzadeh, H. Evidence from a meta-analysis for association of MC4R rs17782313 and FTO rs9939609 polymorphisms with susceptibility to obesity in children. Diabetes Metab. Syndr. 2021, 15, 102234. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; McCarthy, M.I. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Scuteri, A.; Sanna, S.; Chen, W.M.; Uda, M.; Albai, G.; Strait, J.; Abecasis, G.R. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Speakman, J.R.; Loos RJ, F.; O’Rahilly, S.; Hirschhorn, J.N.; Allison, D.B. GWAS for BMI: A treasure trove of fundamental insights into the genetic basis of obesity. Int. J. Obes. 2018, 42, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Ağagündüz, D.; Gezmen-Karadağ, M. Association of FTO common variant (rs9939609) with body fat in Turkish individuals. Lipids Health Dis. 2019, 18, 212. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bratti LD, O.S.; Nunes, B.F.; Gorges, D.M.; Filippin-Monteiro, F.B. In silico approach to identify non-synonymous missense variants in human obesity-related genes: Comprehensive analyses in variants reported in Brazilian databases. Hum. Gene 2023, 36, 201174. [Google Scholar] [CrossRef]
- Constantin, A.; Costache, G.; Sima, A.V.; Glavce, C.S.; Vladica, M.; Popov, D.L. Leptin G-2548A and leptin receptor Q223R gene polymorphisms are not associated with obesity in Romanian subjects. Biochem. Biophys. Res. Commun. 2010, 391, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Nasui, B.A.; Toth, A.; Popescu, C.A.; Penes, O.N.; Varlas, V.N.; Ungur, R.A.; Ciuciuc, N.; Silaghi, C.A.; Silaghi, H.; Pop, A.L. Comparative Study on Nutrition and Lifestyle of Information Technology Workers from Romania before and during COVID-19 Pandemic. Nutrients 2022, 14, 1202. [Google Scholar] [CrossRef]
- WHO European Regional Obesity Report 2022. Available online: https://iris.who.int/bitstream/handle/10665/353747/9789289057738-eng.pdf?sequence=1 (accessed on 4 November 2023).
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Manju, S.K.; Anilkumar, T.R.; Vysakh, G.; Leena, B.K.; Lekshminarayan, V.; Kumar, P.G.; Shenoy, T.K. A Case-Control Study of the Association of Leptin Gene Polymorphisms with Plasma Leptin Levels and Obesity in the Kerala Population. J. Obes. 2022, 2022, 1040650. [Google Scholar] [CrossRef] [PubMed]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Korner, A.; Jacobson, P.; Carlsson, L.M.; Kiess, W.; Vatin, V.; Lecoeur, C.; et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef]
- Portolés, O.; Sorlí, J.V.; Francés, F.; Coltell, O.; González, J.I.; Sáiz, C.; Corella, D. Effect of genetic variation in the leptin gene promoter and the leptin receptor gene on obesity risk in a population-basedcase-control study in Spain. Eur. J. Epidemiol. 2006, 21, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Mammès, O.; Betoulle, D.; Aubert, R.; Herbeth, B.; Siest, G.; Fumeron, F. Association of the G-2548A polymorphism in the 5′ region of the LEP gene with overweight. Ann. Hum. Genet. 2000, 64, 391–394. [Google Scholar] [CrossRef]
- Sabi, E.M.; Bin Dahman, L.S.; Mohammed, A.K.; Sumaily, K.M.; Al-Daghri, N.M. −2548G>A LEP Polymorphism Is Positively Associated with Increased Leptin and Glucose Levels in Obese Saudi Patients Irrespective of Blood Pressure Status. Medicina 2022, 58, 346. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Bains, V.; Sharma, T.; Badaruddoza. Relationship between leptin gene variants (–2548G> A and 19A> G) and obesity among north Indian Punjabi population. J. Genet. 2022, 102, 6. [Google Scholar] [CrossRef]
- Bilge, S.; Yılmaz, R.; Karaslan, E.; Özer, S.; Ateş, Ö.; Ensari, E.; Demir, O. The Relationship of Leptin (+19) AG, Leptin (2548) GA, and Leptin Receptor Gln223Arg Gene Polymorphisms with Obesity and Metabolic Syndrome in Obese Children and Adolescents. Pediatr. Gastroenterol. Hepatol. Nutr. 2021, 24, 306–315. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, L.; Yuan, L.H.; Xiao, Y.; Lu, M.Y.; Zhang, L.J.; Wang, Y. Association of leptin gene -2548 G/A polymorphism with obesity: A meta-analysis. Ann. Nutr. Metab. 2014, 64, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Ricca, V.; Nacmias, B.; Cellini, E.; Di Bernardo, M.; Rotella, C.M.; Sorbi, S. 5-HT2A receptor gene polymorphism and eating disorders. Neurosci. Lett. 2002, 323, 105–108. [Google Scholar] [CrossRef]
- Kring, S.I.I.; Werge, T.; Holst, C.; Toubro, S.; Astrup, A.; Hansen, T.; Pedersen, O.; Sørensen, T.I.A. Polymorphisms of serotonin receptor 2A and 2C genes and COMT in relation to obesity and type 2 diabetes. PLoS ONE 2009, 4, e6696. [Google Scholar] [CrossRef] [PubMed]
- O’Rahilly, S.; Farooqi, I.S. Genetics of obesity. Philos. Trans. R Soc. Lond B Biol. Sci. 2006, 361, 1095–1105. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Gene | Chromosome | SNP | rsID | Amino Acid | Biochemical Structure |
|---|---|---|---|---|---|
| LEP | 7 | A-2548G | rs7799039 | - | Leptin |
| LEPR | 4 | A-223G | rs1137101 | Q223R | Leptin receptor |
| 5HTR2A | 13 | G-1439A | rs6311 | - | Serotonin 2A receptor |
| GHRL | 3 | G-72T | rs696217 | L72M | Ghrelin |
| NPY | 4 | T-29063C | rs16139 | L7P | Neuropeptide Y |
| FTO | 16 | A-T | rs9939609 | - | Fat mass and obesity protein |
| Total (N) | 55 | ||
|---|---|---|---|
| Age (X ± SD, y/o.) | 37.4 ± 12 | ||
| Gender (n) | M = 25, F = 30 | ||
| Weight (X ± SD, kg) | 87 ± 27 kg | ||
| Height (X ± SD, cm) | 172 ± 11.4 cm | ||
| BMI (X ± SD, kg/m2) | 29.4 ± 8 kg/m2 | ||
| BMI (kg/m2) | 18.6–24.9 | 25–29.9 | >30 |
| BMI subgroups (N/%) | 13 (23.6%) | 23 (41.8%) | 19 (34.6%) |
| Gene | SNP | Genotype Frequencies | * Susceptibility | ||||
|---|---|---|---|---|---|---|---|
| wtwt | wtvt | vtvt | increased | intermediate | decreased | ||
| LEP (n/%) | rs7799039 | 8 (14.5%) | 29 (52.7%) | 17 (30.9%) | 32 (58.2%) | 9 (16.4%) | 13 (23.6%) |
| LEPR (n/%) | rs1137101 | 17 (30.9) | 29 (52.7%) | 9 (16.4%) | 38 (69.1%) | 17 (30.9%) | 0 |
| 5HTR2A (n/%) | rs6311 | 23 (41.8%) | 20 (36.4%) | 12 (21.8%) | 32 (58.2%) | 23 (41.8%) | 0 |
| GHRL (n/%) | rs696217 | 45 (81.8%) | 9 (16.4%) | 1 (1.8%) | 0 | 45 (81.8%) | 10 (18.2%) |
| NPY (n/%) | rs16139 | 53 (96.4%) | 2 (3.6%) | 0 | 2 (3.6%) | 53 (96.4%) | 0 |
| FTO (n/%) | rs9939609 | 15 (27.3%) | 25 (45.5%) | 15 (27.3%) | 40 (72.7%) | 15 (27.3%) | 0 |
| Gene | Allele Frequencies | |
|---|---|---|
| LEP | A = 46 (43%) | G = 62 (57%) |
| LEPR | Q = 63 (57%) | R = 47 (43%) |
| 5HTR2A | G = 66 (60%) | A = 44 (40%) |
| GHRL | L = 100 (91%) | M = 10 (9%) |
| NPY | L = 108 (98%) | P = 2 (2%) |
| FTO | T = 55 (50%) | A = 55 (50%) |
| 0 | SNP | Genotype 1 | Genotype 2 | Genotype 3 | p-Value |
|---|---|---|---|---|---|
| LEP | rs7799039 | AA = 9 (16.4%) | GA = 28 (52%) | GG = 17 (30.9%) | p < 0.001 |
| LEPR | rs1137101 | QQ = 17 (30.9%) | QR = 29 (52.7%) | RR = 9 (16.4%) | p < 0.001 |
| 5HTR2A | rs6311 | AA = 12 (21.8%) | GA = 20 (36.4%) | GG = 23 (41.8%) | p < 0.001 |
| GHRL | rs696217 | LL = 45 (81.8%) | LM = 9 (16.4%) | MM = 1 (1.8%) | p < 0.001 |
| NPY | rs16139 | LL = 53 (96.4%) | LP = 2 (3.6%) | PP = 0 (0%) | p < 0.001 |
| FTO | rs 9939609 | AA = 15 (27.3%) | TA = 25 (45.5%) | TT = 15 (27.3%) | p < 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penes, O.N.; Weber, B.; Pop, A.L.; Bodnarescu-Cobanoglu, M.; Varlas, V.N.; Kucukberksun, A.S.; Cretoiu, D.; Varlas, R.G.; Zetu, C. Gene Polymorphisms LEP, LEPR, 5HT2A, GHRL, NPY, and FTO-Obesity Biomarkers in Metabolic Risk Assessment: A Retrospective Pilot Study in Overweight and Obese Population in Romania. Cardiogenetics 2024, 14, 93-105. https://doi.org/10.3390/cardiogenetics14020008
Penes ON, Weber B, Pop AL, Bodnarescu-Cobanoglu M, Varlas VN, Kucukberksun AS, Cretoiu D, Varlas RG, Zetu C. Gene Polymorphisms LEP, LEPR, 5HT2A, GHRL, NPY, and FTO-Obesity Biomarkers in Metabolic Risk Assessment: A Retrospective Pilot Study in Overweight and Obese Population in Romania. Cardiogenetics. 2024; 14(2):93-105. https://doi.org/10.3390/cardiogenetics14020008
Chicago/Turabian StylePenes, Ovidiu Nicolae, Bernard Weber, Anca Lucia Pop, Mihaela Bodnarescu-Cobanoglu, Valentin Nicolae Varlas, Aleksandru Serkan Kucukberksun, Dragos Cretoiu, Roxana Georgiana Varlas, and Cornelia Zetu. 2024. "Gene Polymorphisms LEP, LEPR, 5HT2A, GHRL, NPY, and FTO-Obesity Biomarkers in Metabolic Risk Assessment: A Retrospective Pilot Study in Overweight and Obese Population in Romania" Cardiogenetics 14, no. 2: 93-105. https://doi.org/10.3390/cardiogenetics14020008
APA StylePenes, O. N., Weber, B., Pop, A. L., Bodnarescu-Cobanoglu, M., Varlas, V. N., Kucukberksun, A. S., Cretoiu, D., Varlas, R. G., & Zetu, C. (2024). Gene Polymorphisms LEP, LEPR, 5HT2A, GHRL, NPY, and FTO-Obesity Biomarkers in Metabolic Risk Assessment: A Retrospective Pilot Study in Overweight and Obese Population in Romania. Cardiogenetics, 14(2), 93-105. https://doi.org/10.3390/cardiogenetics14020008










